Abstract
PU.1, IKAROS, E2A, EBF, and PAX5 comprise a transcriptional network that orchestrates B-cell lineage specification, commitment, and differentiation. Here we identify interferon regulatory factor 8 (IRF8) as another component of this complex, and show that it also modulates lineage choice by hematopoietic stem cells (HSCs). IRF8 binds directly to an IRF8/Ets consensus sequence located in promoter regions of Sfpi1 and Ebf1, which encode PU.1 and EBF, respectively, and is associated with transcriptional repression of Sfpi1 and transcriptional activation of Ebf1. Bone marrows of IRF8 knockout mice (IRF8−/−) had significantly reduced numbers of pre-pro-B cells and increased numbers of myeloid cells. Although HSCs of IRF8−/− mice failed to differentiate to B220+ B-lineage cells in vitro, the defect could be rescued by transfecting HSCs with wild-type but not with a signaling-deficient IRF8 mutant. In contrast, overexpression of IRF8 in HSC-differentiated progenitor cells resulted in growth inhibition and apoptosis. We also found that IRF8 was expressed at higher levels in pre-pro-B cells than more mature B cells in wild-type mice. Together, these results indicate that IRF8 modulates lineage choice by HSCs and is part of the transcriptional network governing B-cell lineage specification, commitment, and differentiation.
Introduction
Hematopoietic stem cells (HSCs) undergo lineage-specific differentiation to generate myeloid and lymphoid progeny. Lineage specification is controlled by complex regulatory networks that involve stage- and cell type–specific transcription factors. One of these transcription factors is the Ets family member, PU.1.1 PU.1 regulates the development of common lymphoid progenitors (CLPs) that give rise to T cells, natural killer (NK) cells, dendritic cells (DCs), and B cells.2 There are no identifiable CLPs in embryos of Sfpi1−/− mice (Sfpi1 encodes PU.1)3 and adult bone marrow of Sfpi1 conditional knockout mice.4,5 PU.1 may also regulate the development of myeloid progenitors (MPs).3-5
Commitment to the B-cell lineage from CLPs requires another set of transcription factors including EBF, E2A, and PAX5.6 EBF acts upstream of PAX5 and can restore early B-cell development in Sfpi1−/− fetal liver HSCs.7 PAX5 activates the expression of B-lineage marker genes, such as Cd19 and Blnk, and suppresses lineage-inappropriate genes such as Csf1r and Notch1,6 thus acting as a B-lineage commitment factor. Deficiency of EBF or PAX5 results in a block of B-cell development at the pro-B-cell stage.8,9 Recently, it was shown that siRNA-mediated knockdown of Sfpi1 in embryonic stem (ES) cells induced expression of Ebf and Pax5, which correlated with enhanced B-cell developmental potential.10 These data suggested that PU.1 may play a role in the regulation of Ebf and Pax5 during B-cell commitment.
Gene activation events dependent on PU.1 frequently require coordinate regulation with members of the interferon regulatory factor (IRF) family of transcription factors including IRF4 and IRF8. IRF8, otherwise known as interferon consensus sequence–binding protein (ICSBP),11 is expressed almost exclusively in hematopoietic cells of both the myeloid and lymphoid lineages. It is best known for its multiple effects on the development of granulocytes, macrophages, Langerhans cells, and subsets of DCs,12-15 with much of this information gained from studies of mice with a null mutation of the gene (IRF8−/−).16 Recent studies of mouse and human B-lineage cells have shown that expression of IRF8 is significantly increased as they transit the germinal center (GC) and then strikingly down-regulated in terminally differentiated plasma cells.17,18 Previous studies also suggested that IRF8 could play a role in the earliest stages of B-cell development. Transformed cell lines with properties of pro-B and pre-B cells were shown to express IRF8 at high levels.19 Furthermore, studies of mice doubly deficient in IRF8 and another IRF family member, IRF4, showed that these transcription factors act together to regulate the pre-B- to B-cell transition.20
IRF8 functions as a transcriptional activator or repressor depending on the formation of different heterodimeric DNA-binding complexes that include Ets family members (PU.1, TEL),21,22 other IRF family members (IRF1, IRF2, and IRF4),23-25 as well as E47,26 NFATc1,27 and MIZ1.28 Several IRF8 target sequences also contain Ets-binding sites.29 Since PU.1 plays a critical role in the development of both lymphoid and myeloid cells,3 it seemed likely that IRF8 might also participate in the early development of both lineages. The present studies were thus undertaken to develop a fuller understanding of the roles played by IRF8 in hematopoietic differentiation with a primary focus on commitment to the B-cell lineage. Comparisons of IRF8−/− and normal mice demonstrated that IRF8 influences lineage commitment decisions by HSCs and the numbers of bone marrow (BM) pre-pro-B cells. Most importantly, IRF8 directly regulates the expression of EBF and PU.1, thereby defining novel aspects of the transcriptional regulatory network for specification and commitment to the B-cell lineage.
Methods
Mice and cell lines
IRF8 knockout mice were described previously16 and were kindly provided by Dr Keiko Ozato (National Institute of Child Health and Human Development [NICHD], National Institutes of Health [NIH], Bethesda, MD). These mice have been bred with C57BL/6 mice for at least 10 generations. Age-matched littermate mice (6-10 weeks) were used in this study. The use of mice in this study followed a protocol (LIP-4) approved by National Institute of Allergy and Infectious Diseases Animal Care and Use Committee.
The B-cell lymphoma cell line NFS-202 and subclones with siRNA-induced knockdown of IRF8 were described previously.17 The murine follicular B-cell lymphoma cell line NFS-203 expresses little or no IRF8. Plasmids containing a full-length Irf8 cDNA (pCDNA3.1-IRF8; provided by Dr Keiko Ozato, NICHD, NIH) were transfected into NFS-203 cells. Cells transfected with an empty vector (pCDNA3.1) served as a control. Neomycin-resistant cells were used for Western blotting analysis.
Chromatin immunoprecipitation assay
Western blotting
Total protein extracts from equivalent numbers of cells were resolved by standard SDS–polyacrylamide gel electrophoresis (PAGE) and blotted with rabbit antibodies against IRF8, PU.1, and actin (Santa Cruz Biotechnology, Santa Cruz, CA), followed by an HRP-labeled secondary antibody. The bands were visualized using enhanced chemiluminescence (ECL) development reagents (Amersham Pharmacia Biotech, Piscataway, NJ).
Luciferase reporter assays
The 5′ flanking region of Ebf1 (−909/+103) was PCR amplified from genomic DNA extracted from NFS-202 cells and cloned into the pGL4.1[luc2] firefly luciferase reporter vector (Promega, Madison, WI). Mutant luciferase reporter constructs were generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. The murine IRF8 and PU.1 expression vectors (pCDNA3.1-IRF8 and pCDNA3.1-PU.1) were provided by Dr Keiko Ozato. Six hundred nanograms of a reporter construct, 1 ng pGL4.75 [hRluc/CMV] (Promega), a Renilla luciferase vector used for control of transfection efficiency, and 100 ng IRF8 and/or PU.1 expression plasmids or the corresponding empty vector were cotransfected into HeLa cells using PolyFect transfection reagents (QIAGEN, Valencia, CA). Luciferase activities were measured 24 hours after transfection using the Dual-luciferase reporter assay kit (Promega) according to the manufacturer's instructions.
Flow cytometry
Cells were prepared and stained as previously reported.30 Monoclonal antibodies specific for mouse cell surface markers were obtained from BD PharMingen (San Diego, CA). Fluorescence-activated cell sorting (FACS) analyses were performed on a FACSCalibur (Becton Dickinson, Mountain View, CA) or a CyAn ADP Analyzer (Dako, Carpinteria, CA).
To sort BM HSCs, CLPs, and MPs, pooled BM cells from 5 to 6 mice were prepurified with magnetic-activated Dynal beads (Invitrogen, Carlsbad, CA) using biotinylated antibodies against mouse lineage panel (BD Biosciences, San Jose, CA) according to the manufacturer's instructions. The cells were then stained with streptavidin-Percp and antibodies to Sca-1, c-Kit, and IL-7R. Sorting was performed using a FACS Aria-Green sorter (BD Biosciences).
To sort BM B-cell subpopulations, pooled BM cells from 3 to 4 mice were pre-enriched for B cells using antibodies against CD3, CD11b, Gr-1, and Ter119 and magnetic-activated Dynal beads. The cells were then stained with antibodies to B220, CD43, CD24, and BP-1. Hardy Fraction A, B, C, and D-F cells were sorted as described in Figure 5. In all cases, the sorted populations were 90% to 95% pure as determined by reanalysis.
qPCR
Total RNA was isolated using a RNeasy Mini kit (QIAGEN) according to the manufacturer's instructions. A DNA digestion step was included before elution of RNA. Approximately 200 ng total RNA in 20 μL was reverse-transcribed with SuperScript II reverse transcriptase and random hexamer primers (Invitrogen). cDNA (10 ng) was amplified in triplicate using an ABI Prism 7900HT Sequence Detection System with the SYBR Green PCR Master Mix reagents (Applied Biosystems, Foster City, CA). The housekeeping genes, Hprt or Gapdh, were amplified as internal controls. Primer sequences for qPCR are shown in Table S1.
HSC reconstitution
About 7000 HSCs (Lin−IL-7R−c-Kit+Sca-1+) sorted from BM of IRF8−/− and WT mice were injected intravenously into lethally irradiated B6.SJL-PtprcaPepcb/BoyJ mice (The Jackson Laboratory, Bar Harbor, ME). BM cells of donor origin were distinguished by expression of CD45.2 and were analyzed by FACS at 8 weeks after reconstitution.
BM cell cultures
BM cell suspensions were cultured at 1 × 106/mL in IMDM medium supplemented with 10% fetal bovine serum (FBS) and 10 ng/mL rIL-7 (R&D Systems, Minneapolis, MN) for 5 days. The cells were then stained and analyzed by FACS.
Retroviral transduction and OP9 cocultures
OP9 stromal cells (ATCC, Manassas, VA) were plated at 1 × 104 cells/well in 24-well plates 2 to 3 days prior to the addition of sorted BM HSCs. HSCs (1 × 104/mL) were incubated with 1 mL X-vivo 15 medium supplemented with 5 ng/mL IL-7, 20 μg/mL SCF, and 10 ng/mL Flt3L (Peprotech, Rocky Hill, NJ). Cocultures were harvested at day 7 and cells were analyzed by FACS.
For enforced expression of IRF8 in HSCs, HSCs were sorted from IRF8−/− and IRF8+/+ mice and were cultured as described.31 Briefly, 1 to 3 × 104 HSCs/well were cultured in a 96-well plate in X-vivo medium containing 0.1% bovine serum albumin (BSA), IL-3 (10 ng/mL), IL-6 (10 ng/mL), SCF (40 ng/mL), and Flt3L (50 ng/mL) for 18 hours. Cells were then infected with retroviruses encoding GFP alone, IRF8-GFP, or K79E-GFP (provided by Dr Keiko Ozato) in X-vivo medium containing IL-6 (10 ng/mL), IL-3 (10 ng/mL), SCF (20 ng/mL), IL-7 (5 ng/mL), Flt3L (50 ng/mL), GM-CSF (10 ng/mL), 0.1% BSA, and 4 μg/mL polybrene. Cells were centrifuged at 1780g for 1.5 hours at 33°C and cultured for 24 hours, followed by sorting for GFP+ cells. Sorted cells were then cultured for 7 days on an OP9 monolayer in Opti-MEM containing SCF (20 ng/mL), Flt3L (50 ng/mL), and IL-7 (10 ng/mL). The myeloid and lymphoid lineage capacity of GFP+ cells was assessed by FACS.
Statistical analysis
The Student t test was used to assess the significance of the differences. P value less than .05 was considered significant.
Results
IRF8 regulates PU.1 expression
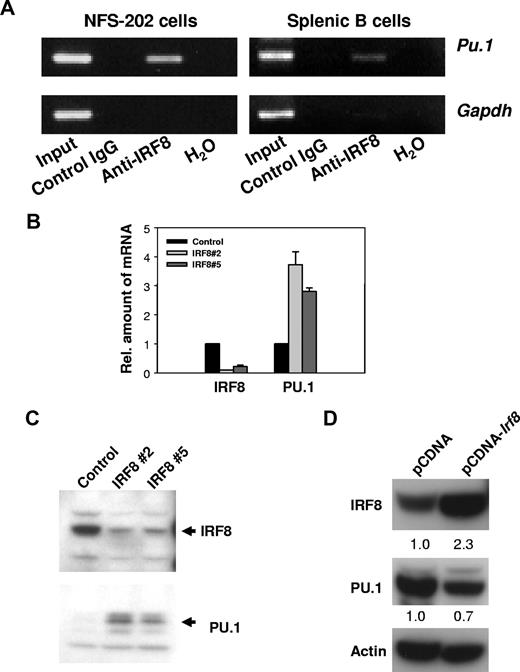
IRF8 is known to bind to any of 4 distinct target sequences in genes subject to transcriptional activation or repression by IRF8.29 Studies of the regulatory sequences of Sfpi1, which encodes PU.1, identified 2 possible IRF8 target sites. One, found about 1.6 kb upstream of the transcriptional start site, matches a novel IRF8/Ets consensus sequence termed an IECS (GAAANN(N)GGAA) recently identified in the promoters of a series of myeloid genes.29 Another sequence, found 2.2 kb upstream of the start site, is similar to an Ets/IRF response element termed an EIRE with a consensus sequence, GGAAANNGAAA.29 To determine whether IRF8 protein was bound in vivo to these 2 sequences in the regulatory region of Sfpi1, we performed ChIP analyses. Cross-linked DNA from the mouse B-cell line, NFS-202, or purified normal splenic B cells was immunoprecipitated with anti-IRF8 or control antibodies. After cross-linking was reversed, the DNA was amplified by PCR using primers designed to amplify DNA from regions flanking the 2 IRF8 consensus-binding sites as well as a nonbinding sequence 5′ of the Gapdh gene. DNA products of the expected sizes were amplified from the total input DNA and anti-IRF8–precipitated DNA containing the site at 1.6 kb, but not from DNA precipitated with the control antibody (Figure 1A). No products were amplified with primers directed to the 2.2-kb site (data not shown). Primers amplifying an unrelated Gapdh sequence did not generate a PCR product in anti-IRF8–precipitated DNA, indicating that the amplification was Sfpi1 specific. We conclude that IRF8 occupied a target site of the Sfpi1 promoter in vivo.
IRF8 suppresses PU.1 expression. (A) IRF8 is present at the promoter region of PU.1 in vivo. ChIP analyses were performed with NFS-202 cells or purified spleen B cells. Protein-DNA complexes were immunoprecipitated by addition of antibody to IRF8 and analyzed by PCR for the presence of PU.1 promoter sequences. (B) qPCR analysis of IRF8 and PU.1 expression in cells expressing a repressive IRF8 siRNA. IRF8 Nos. 2 and 5 are 2 clones from IRF8 siRNA–treated cells. (C) Western blotting analysis of IRF8 and PU.1 expression in IRF8 siRNA–expressing cells. (D) Western blotting analysis of IRF8 and PU.1 expression in NFS-203 cells expressing pCDNA-Irf8 or an empty pCDNA vector as indicated. The numbers are arbitrary units of protein intensities normalized by β-actin. All data are representative of 2 to 4 independent experiments.
IRF8 suppresses PU.1 expression. (A) IRF8 is present at the promoter region of PU.1 in vivo. ChIP analyses were performed with NFS-202 cells or purified spleen B cells. Protein-DNA complexes were immunoprecipitated by addition of antibody to IRF8 and analyzed by PCR for the presence of PU.1 promoter sequences. (B) qPCR analysis of IRF8 and PU.1 expression in cells expressing a repressive IRF8 siRNA. IRF8 Nos. 2 and 5 are 2 clones from IRF8 siRNA–treated cells. (C) Western blotting analysis of IRF8 and PU.1 expression in IRF8 siRNA–expressing cells. (D) Western blotting analysis of IRF8 and PU.1 expression in NFS-203 cells expressing pCDNA-Irf8 or an empty pCDNA vector as indicated. The numbers are arbitrary units of protein intensities normalized by β-actin. All data are representative of 2 to 4 independent experiments.
We next studied the function of IRF8 in regulating PU.1 expression. The expression of endogenous IRF8 in NFS-202 cells was inhibited by introducing either of 2 different siRNAs to IRF8.17 Using qPCR and Western blot analyses, we found that levels of PU.1 transcripts (Figure 1B) and protein (Figure 1C) were substantially increased in cells expressing interfering siRNAs. Not surprisingly, overexpression of IRF8 in the IRF8lo B-cell line, NFS-203, resulted in about a 30% reduction in PU.1 expression at the protein level (Figure 1D). From these experiments, we conclude that IRF8 acts as a transcriptional repressor for PU.1 expression in B cells.
IRF8 regulates EBF expression
In previous studies of factors governing the expression of Ebf1, Medina et al using gel shifts and ChIP assays7 identified a sequence, TTCCTCT, conserved between mice and humans and located 677-bp upstream of the start site (Figure 2A), as a target for PU.1. On re-examination, we saw that an extended sequence at this site, GAAAGAGGAA, matches the IECS consensus sequence of IRF8 described in “IRF8 regulates PU.1 expression.”29 In addition to this potential target, we identified 2 additional candidates for IRF8 binding within a 1-kb region of the Ebf1 promoter, an EICE at −585 to −594 and an ESRE at −180 to −189 (Figure 2A). Using ChIP analyses of DNA from the NFS-202 cell line, we found that cross-linked DNA precipitated with anti-IRF8 antibodies contained sequences spanning these sites but not a sequence in the Gapdh promoter (Figure 2B). Thus, IRF8 bound in vivo to these sites of the Ebf1 gene.
IRF8 regulates EBF expression. (A) Schematic arrangement of IRF8-binding sites (IECS, EICE, and ISRE) in the promoter of the Ebf1 gene. +1 corresponds to the first nucleotide of exon 1 of the gene. The nucleotides that were mutated at each IRF8-binding site are shown on the top. WT indicates wild type. (B) ChIP analysis of IRF8 binding with Ebf1. In vivo cross-linked IRF8-chromatin complexes from NFS 202 cells were analyzed using PCR primers that span the IRF8 sites in the Ebf1 gene. (C) IRF8 and PU.1 regulate EBF expression in a luciferase reporter assay. HeLa cells were cotransfected with a promoter reporter pGL4-Ebf1-WT and vectors expressing IRF8, PU.1, or both. An empty vector was used as a control. (D) Mutation of IRF8-binding sites in the Ebf1 gene impaired the expression of the Ebf1 reporter. The Ebf1 promoter reporter constructs containing mutated IRF8-binding sites were generated as illustrated in panel A and were cotransfected with plasmids expressing IRF8 and PU.1. Luciferase activities were measured after 22 hours. All data represent 3 to 4 independent experiments.
IRF8 regulates EBF expression. (A) Schematic arrangement of IRF8-binding sites (IECS, EICE, and ISRE) in the promoter of the Ebf1 gene. +1 corresponds to the first nucleotide of exon 1 of the gene. The nucleotides that were mutated at each IRF8-binding site are shown on the top. WT indicates wild type. (B) ChIP analysis of IRF8 binding with Ebf1. In vivo cross-linked IRF8-chromatin complexes from NFS 202 cells were analyzed using PCR primers that span the IRF8 sites in the Ebf1 gene. (C) IRF8 and PU.1 regulate EBF expression in a luciferase reporter assay. HeLa cells were cotransfected with a promoter reporter pGL4-Ebf1-WT and vectors expressing IRF8, PU.1, or both. An empty vector was used as a control. (D) Mutation of IRF8-binding sites in the Ebf1 gene impaired the expression of the Ebf1 reporter. The Ebf1 promoter reporter constructs containing mutated IRF8-binding sites were generated as illustrated in panel A and were cotransfected with plasmids expressing IRF8 and PU.1. Luciferase activities were measured after 22 hours. All data represent 3 to 4 independent experiments.
To evaluate the importance of these sequences to transcriptional activation of Ebf1, we introduced a 1-kb fragment from the 5′ regulatory sequences into a luciferase reporter construct to generate pGL4.1 Ebf1 WT. Cotransfection of HeLa cells with the reporter and an IRF8 expression plasmid resulted in a 2-fold increase in reporter activity (Figure 2C). Cells cotransfected with the reporter and a PU.1 expression plasmid exhibited a 5-fold luciferase activity, consistent with previous studies showing direct binding of PU.1 to these Ebf1 5′ sequences.7 Finally, cotransfections of the reporter with expression vectors for both PU.1 and the IRF8 resulted in a synergistic approximately 27-fold increase in luciferase activity (Figure 2C).
To more precisely define the 5′ sequences critical to the transcriptional regulation of Ebf1, we generated a series of truncation mutants containing wild-type sequences or sequences mutated at each of the putative IRF8 target sites, alone or in combination (Figure 2A). The effects of the truncations and mutations were tested in HeLa cells cotransfected with vectors expressing PU.1 and IRF8. The results of these studies (Figure 2D) showed that a truncation mutant containing 418-bp upstream from the start site, designated ΔKpnI, showed only 30% of the transcription activity of the wild-type (WT) full-length sequence. Mutation of the ISRE in the ΔKpnI construct (ΔKpnI M1) had no additional effect on reporter activity, indicating that the ISRE-binding site contributed little if at all to IRF8 regulation. Mutations in either the upstream targets, EICE (M2) or IECS (M3), reduced the reporter activity by 30% and 42%, respectively. A construct (M2M3) carrying both the M2 and M3 mutations exhibited a 52% reduction in reporter activity. Finally, given that M1 mutant had no significant effect on activity of the full-length sequence, it predictably had little if any effect when added in cis to the 2 other mutants in a reporter containing the full-length sequence (Figure 2D). Taken together, these results indicate that IRF8 and PU.1 act synergistically to mediate the direct transcriptional activation of Ebf1.
Heightened expression of IRF8 is associated with B-lineage commitment
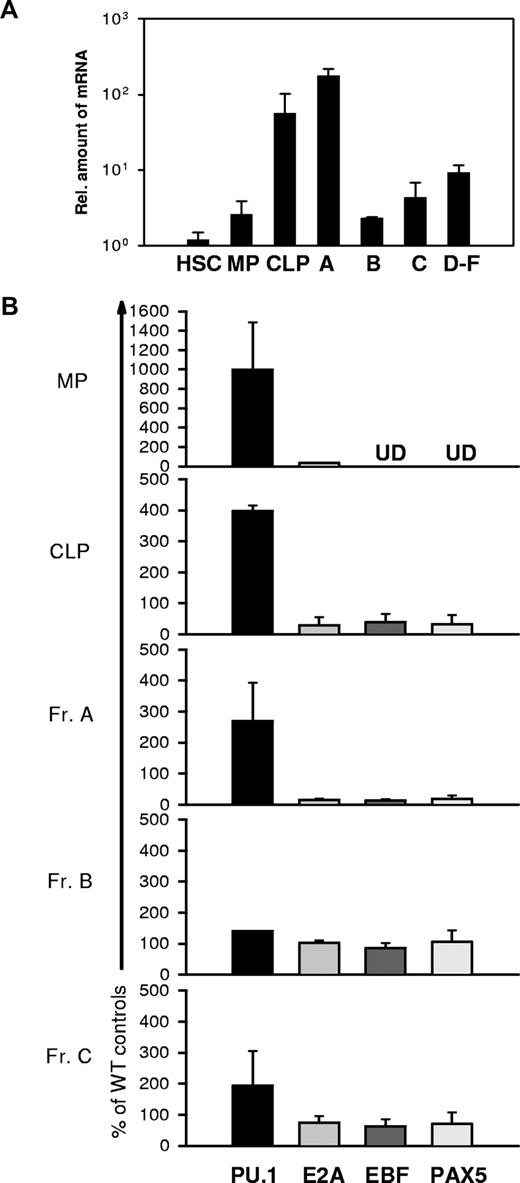
To determine whether the observed regulation of PU.1 and EBF by IRF8 correlates with the function of IRF8 in vivo, we purified HSCs, CLPs, MPs, and Hardy Fr A, B, C, and D-F subpopulations of B cells32 from normal BM and measured levels of IRF8 transcripts by qPCR. Since there is no standardized nomenclature for hematopoietic progenitors, we adopted the scheme defined by the well-documented antibody staining patterns that distinguish HSCs, CLPs, and MPs. HSCs, which include both long-term and short-term repopulating stem cells, are present in the Lin−Sca-1+c-Kit+IL-7R− fraction; CLPs are enriched in Lin−Sca-1loc-KitloIL-7R+ cells; and MPs, comprising common myeloid progenitors (CMPs), granulocyte-monocyte progenitors (GMPs), and megakaryocytic-erythroid progenitors (MEPs), are found in the Lin−Sca-1−c-Kit+IL-7− population.33,34 Cell gating schemes are depicted in Figures 4–5.
As shown in Figure 3A, HSCs and MPs expressed IRF8 transcripts at very low levels. In contrast, the levels expressed by CLPs and Fr A, also termed pre-pro-B, the earliest stage of B-lineage cells, were about 20- to 100-fold higher than those in HSCs and MPs. Strikingly, the transition from Fr A to Fr B and Fr C cells was marked by a precipitous drop of nearly 2 logs10 in IRF8 transcripts to levels approximating those seen in HSCs and MPs. IRF8 transcripts in sorts combining Frs D, E, and F, which contain pre-BII, immature B, and mature B cells, were substantially higher than the levels in cells in Fr B. These data demonstrate that IRF8 is differentially expressed during early B-cell development and may be involved in the development of CLPs and in B-lineage cell commitment.
Expression of Irf8, Pu.1, Ebf, Pax-5, and E2a in HSCs, CLPs, MPs, and various B-cell subsets. Each cell population was sort-purified from a pool of 5 to 6 mice. RNA was extracted and reverse transcribed. qPCR was used to quantitate Irf8 (A), Pu.1, Ebf, E2a, and Pax-5 (B) expression levels. Data are mean plus or minus SEM of 3 independent experiments. UD indicates undetectable.
Expression of Irf8, Pu.1, Ebf, Pax-5, and E2a in HSCs, CLPs, MPs, and various B-cell subsets. Each cell population was sort-purified from a pool of 5 to 6 mice. RNA was extracted and reverse transcribed. qPCR was used to quantitate Irf8 (A), Pu.1, Ebf, E2a, and Pax-5 (B) expression levels. Data are mean plus or minus SEM of 3 independent experiments. UD indicates undetectable.
IRF8 deficiency results in increased expression of PU.1 and diminished expression of EBF in CLPs and early B cells
To determine whether IRF8 deficiency would affect expression of PU.1 and EBF in vivo, we analyzed subpopulations of IRF8−/− BM cells for expression levels of PU.1, EBF, Pax5, and E2A in subsets of BM hematopoietic cells including subsets of B-lineage cells in IRF8−/− mice. It was previously shown that IRF8−/− mice exhibit a progressive increase in the numbers of granulocytes in various lymphoid organs and are deficient in their ability to produce IL-12 and IFNγ.16 To minimize the potential effects of abnormal granulocyte expansion and a biased cytokine environment on B-cell development, we studied 6- to 10-week-old mice.
As shown in Figure 3B, PU.1 was expressed at levels 2.5- to 10-fold higher in MPs, CLPs, and pre-pro-B populations of IRF8−/− mice compared with cells from IRF8+/+ mice. In contrast, the levels of EBF were significantly lower in CLPs and pre-pro-B cells of IRF8−/− mice. These patterns of PU.1 and EBF expression were consistent with the studies described in Figures 1 and 2 indicating that IRF8 mediates the transcriptional repression of PU.1 and activation of EBF in early B-lineage and progenitor cells. The difference in EBF expression between wild-type and IRF8−/− mice was reduced in the later stage B cells in Frs B and C, which was in accord with down-regulated expression of IRF8 in these subsets of normal mice (Figure 3A). Moreover, transcripts levels for E2A and Pax5 in Fr A cells were also reduced in IRF8−/− mice (Figure 3B). Finally, we examined the possibility that the results obtained with our sorted cells were biased by inadvertent contamination with Lin+ myeloid cells. To do this, we tested sorted MP cells, the subset most likely to be contaminated, by measuring the transcripts levels of 2 myeloid restricted genes, Cebpa and Csf2ra, which encode the MCSF receptor, by qPCR. The results indicated minimal if any contamination with myeloid cells in sorted MP population (Figure S1). We therefore conclude that IRF8 regulates expression of both PU.1 and EBF, and thereby contributes to specification of a lymphoid fate and commitment to the B-cell lineage.
IRF8 deficiency impairs development of pre-pro-B cells
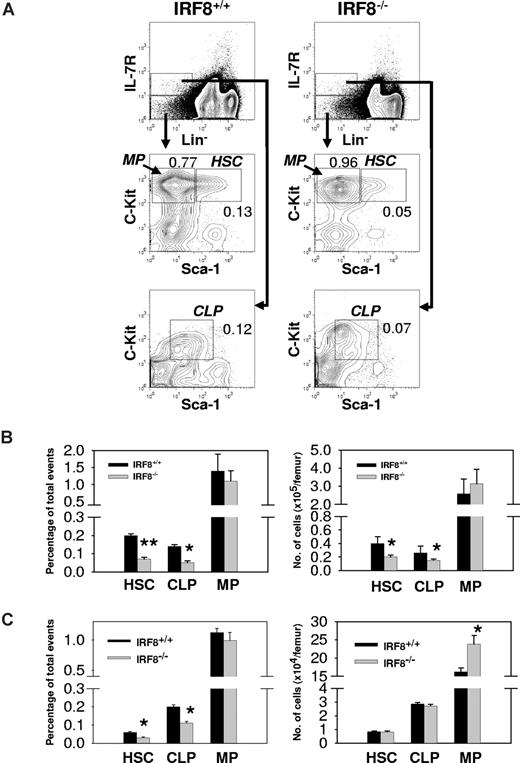
Analyses of BM cells from IRF8−/− mice showed that the frequencies of HSCs and CLPs were reduced 3-fold and 2-fold, respectively, from the levels found in IRF8+/+ littermate controls (Figure 4A,B). Since the BM cellularity of IRF8−/− mice was increased by an average of 44%, the absolute numbers of HSCs and CLPs in the knockouts were reduced by 50% and 42%, respectively (Figure 4A,B; P < .05). In contrast, the total number of MPs in BM of IRF8−/− mice was slightly but not significantly increased (Figure 4A,B). The reduced numbers of HSCs and CLPs in IRF8−/− mice appeared to result from defects of unknown BM microenvironmental factors because adoptive transfer of HSCs sorted from IRF8+/+ and IRF8−/− mice into lethally irradiated normal recipient mice yielded comparable numbers of HSCs and CLPs but not MPs (Figure 4C). The numbers of MPs were higher in mice receiving IRF8-deficient HSCs than in mice receiving IRF8+/+ HSCs (Figure 4C), similar to what was found in unmanipulated IRF8−/− mice (Figure 4B). These results indicated that IRF8 deficiency biased the differentiation of HSCs in favor the myeloid lineages, consistent with previous studies using colony-forming assays35,36 (also see Figure 6).
Bias of MP development in IRF8−/− mice. (A) BM cells from IRF8−/− and IRF8+/+ littermate mice (n = 8) were stained with antibodies against Lineage panel (Lin), IL-7Rα, c-Kit, and Sca-1. The numbers represent percentage of total events. (B) The frequency (left panel) and absolute cell numbers (right panel) of HSCs, CLPs, and MPs in IRF8+/+ and IRF8−/− mice. Data are mean plus or minus SEM of 5 mice per group. *P < .05; **P < .001 compared with controls. (C) Sorted HSCs from IRF8+/+ and IRF8−/− mice were injected into lethally irradiated CD45.2− B6 mice. Two months later, BM cells were analyzed by FACS and the donor cells (CD45.2+) were identified by an anti-CD45.2 antibody. The frequency (left panel) and absolute cell numbers (right panel) of donor HSCs, CLPs, and MPs were shown as mean plus or minus SEM of 6 mice per group. *P < .05 compared with controls.
Bias of MP development in IRF8−/− mice. (A) BM cells from IRF8−/− and IRF8+/+ littermate mice (n = 8) were stained with antibodies against Lineage panel (Lin), IL-7Rα, c-Kit, and Sca-1. The numbers represent percentage of total events. (B) The frequency (left panel) and absolute cell numbers (right panel) of HSCs, CLPs, and MPs in IRF8+/+ and IRF8−/− mice. Data are mean plus or minus SEM of 5 mice per group. *P < .05; **P < .001 compared with controls. (C) Sorted HSCs from IRF8+/+ and IRF8−/− mice were injected into lethally irradiated CD45.2− B6 mice. Two months later, BM cells were analyzed by FACS and the donor cells (CD45.2+) were identified by an anti-CD45.2 antibody. The frequency (left panel) and absolute cell numbers (right panel) of donor HSCs, CLPs, and MPs were shown as mean plus or minus SEM of 6 mice per group. *P < .05 compared with controls.
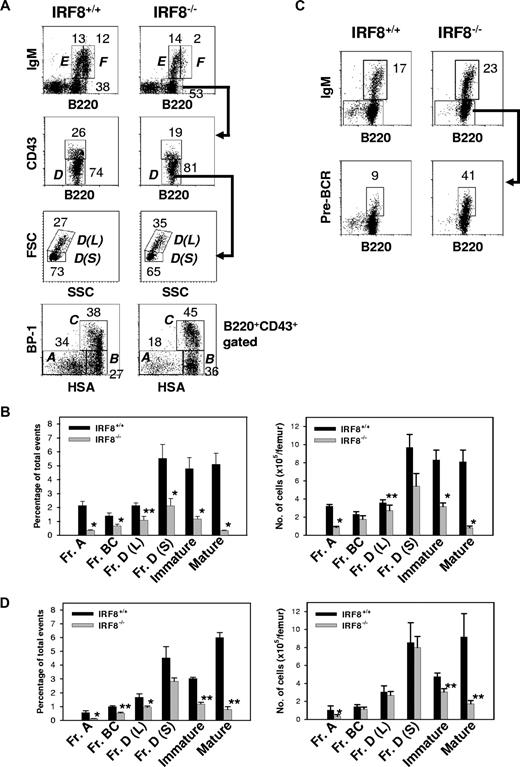
Studies of B-lineage cells in the BM showed that the absolute number of B220+ lymphocytes was decreased approximately 60% in IRF8−/− mice (1.8 ± 0.2 × 106 vs 4.1 ± 0.4 × 106 per femur for IRF8−/− and IRF8+/+ mice, respectively; n = 6; P < .001). BM B-cell lineage subpopulations defined in accordance with the Hardy nomenclature32 are depicted in Figure 5A. In IRF8−/− mice, the frequency and absolute numbers of Fr A pre-pro-B cells were reduced 6-fold and 4-fold, respectively, from the levels in IRF8+/+ mice (P < .001) (Figure 5B). An alternative criteria using the markers B220, CD19, CD43, DX5, and LY6C to characterize pre-pro-B cells37 yielded similar results (Figure S2). Strikingly, the numbers of later stage B-lineage cells in IRF8−/− marrow, including Frs B, C, and D, increased steadily although never reached normal levels (Figure 5B). The ratio of large pre-B II cells [Fr D(L)] to small pre-B II cells [Fr D(S)] was also significantly increased (Figure 5A). The increase of large pre-BII cells in IRF8−/− mice was associated with enhanced pre-BCR expression on pre-B cells (Figure 5C). In addition, both IgM+ immature (Fr E) and mature (Fr F) populations were also significantly reduced in IRF8−/− mice (Figure 2A-B). Taken together, these data indicated that the development of B-lineage cells was significantly impaired in IRF8−/− mice.
Impaired B-cell development in the BM of IRF8−/− mice. (A) BM cells from the indicated mice (n = 8) were stained for B220, IgM, CD43, BP-1, and HSA. The percentage of B cells falling within each gate is given. Hardy Frs A to F are indicated. L indicates large; S, small. (B) The frequency and absolute numbers of each B-cell subset. Data are mean plus or minus SEM of 6 mice. * indicates P value less than .001 compared with control groups; **, P value less than .05 compared with Fr A cells. (C) IL-7 cultured BM cells were stained with antibodies against B220, IgM, and pre-BCR. The anti–pre-BCR antibody (clone SL156) specifically recognizes an epitope composed of the μH chain and the surrogate L chain. The percentage of B cells falling within the gate is given. Data represent 1 of 3 mice with similar results. (D) Sorted HSCs from IRF8+/+ and IRF8−/− mice were injected into lethally irradiated CD45.2− B6 mice. Two months later, BM cells were analyzed by FACS and the donor cells (CD45.2+) were identified by an anti-CD45.2 antibody. The frequency (left panel) and absolute cell numbers (right panel) of donor B cells were shown as mean plus or minus SEM of 6 mice per group. * P < .05; **P < .001 compared with the control group.
Impaired B-cell development in the BM of IRF8−/− mice. (A) BM cells from the indicated mice (n = 8) were stained for B220, IgM, CD43, BP-1, and HSA. The percentage of B cells falling within each gate is given. Hardy Frs A to F are indicated. L indicates large; S, small. (B) The frequency and absolute numbers of each B-cell subset. Data are mean plus or minus SEM of 6 mice. * indicates P value less than .001 compared with control groups; **, P value less than .05 compared with Fr A cells. (C) IL-7 cultured BM cells were stained with antibodies against B220, IgM, and pre-BCR. The anti–pre-BCR antibody (clone SL156) specifically recognizes an epitope composed of the μH chain and the surrogate L chain. The percentage of B cells falling within the gate is given. Data represent 1 of 3 mice with similar results. (D) Sorted HSCs from IRF8+/+ and IRF8−/− mice were injected into lethally irradiated CD45.2− B6 mice. Two months later, BM cells were analyzed by FACS and the donor cells (CD45.2+) were identified by an anti-CD45.2 antibody. The frequency (left panel) and absolute cell numbers (right panel) of donor B cells were shown as mean plus or minus SEM of 6 mice per group. * P < .05; **P < .001 compared with the control group.
The deficiency of pre-pro-B cells in IRF8−/− mice is cell intrinsic
To clarify whether the defective development of pre-pro-B cells in IRF8−/− mice was cell autonomous, we used 3 experimental approaches. First, sorted HSCs from IRF8−/− and IRF8+/+ mice were transferred into lethally irradiated CD45.1 normal mice. Eight weeks after reconstitution, donor cells (CD45.2+) in the BM were analyzed by flow cytometry. As shown in Figure 5D, both the frequency and the absolute number of Fr A cells were significantly lower in IRF8−/− HSC-reconstituted mice than in IRF8+/+ HSC-reconstituted mice. The lower numbers of other B-cell subsets were similar to that in IRF8−/− mice, suggesting that the deficiency of pre-pro-B and other early B cells in IRF8−/− mice may be B-cell intrinsic. Second, sorted HSCs were induced to differentiate in OP9 cocultures in the presence of the B-cell permissive cytokines SCF, IL-7, and Flt3L. Under this condition, IRF8−/− HSCs gave rise to very few B220+CD19+ B cells, which was in sharp contrast to the large numbers recovered from cultured IRF8+/+ HSCs (Figure 6A). Consistent with previous reports,35,36 IRF8−/− HSCs generated a predominant of myeloid cells compared with their recovery from IRF8+/+ HSCs. Finally, we infected HSCs of IRF8−/− and IRF8+/+ mice with constructs expressing GFP alone, GFP-tagged wild-type mouse IRF8, or a GFP-tagged mutant IRF8 in which K at 79 was replaced with Glu (E) (K79E).38 K79E was previously shown to lack DNA-binding and transcriptional activities.38 GFP+ cells were sorted and recultured in conditions conducive to B-cell growth for 6 to 7 days. Exogenous IRF8 rescued B-cell development in IRF8-deficient HSCs, as indicated by the detection of large numbers of B220+ cells (Figure 6B). In contrast, the K79E mutant did not rescue B-cell differentiation, indicating that IRF8 has a cell-intrinsic role in B-cell lineage specification. Real-time qPCR analysis revealed that IRF8 induced expression of EBF and down-regulated expression of PU.1, whereas expression of PAX5 was unchanged (Figure S3). Moreover, enforced expression of IRF8 in HSCs of both IRF8+/+ and IRF8−/− mice resulted in strong suppression on cell growth without affecting the ratio of CD11b+ myeloid to B220+ B-lineage cells (Figure 6C; Table 1). Cell-cycle analysis by BrdU pulse labeling revealed a significant reduction of S phase cells and a modest increase of apoptotic cells in GFP-IRF8–expressing cells (Figure 6C). Introduction of the IRF8 K79E mutant did not result in growth inhibition (Figure 6C; Table 1). These data indicated that IRF8 controls a gene program for proliferation and survival, consistent with a previous report using a myeloid progenitor cell line.38 We conclude that IRF8 modulates cell-cycle progression and B-lineage specification at the myeloid/lymphoid progenitor stage, and that in its absence, the bias is toward myeloid lineage development.
The failure of IRF8-deficient HSCs to differentiate into B cells is cell intrinsic. (A) Sorted HSCs from IRF8+/+ and IRF8−/− mice were induced to differentiate in OP9 cocultures in the presence of SCF, IL-7, and Fl3tL for 7 days. The cells were stained with antibodies recognizing B220, CD19, and CD11b and analyzed by flow cytometry. Cells were gated on 7AAD− living cells. The numbers are percentages of cells falling in each gate. Data represent 1 of 3 independent experiments. (B) Sorted HSCs from each group of mice were cultured in the presence of SCF, Flt3L, IL-3, and IL-6 for 18 hours before they were infected with retroviral vectors encoding an IRF8-GFP fusion protein, an IRF8 mutant K79E-GFP fusion protein, or GFP only for 24 hours in the presence of SCF, Flt3L, IL-3, IL-6, IL-7, GM-CSF, and 4 μg/mL polybrene. GFP+ cells were resorted and plated onto OP9 cell layer in the presence of SCF, IL-7, and Flt3L for 7 days. The cells were analyzed by flow cytometry. The numbers are percentages of cells falling in each gate. Data represent 1 of 4 independent experiments. (C) The sorted GFP+ cells were cultured with OP9 cells at the same conditions as in panel B and were pulsed with BrdU for 40 minutes at day 4 and analyzed by FACS. The numbers are percentages of cells falling in each gate. Data represent 1 of 2 independent experiments.
The failure of IRF8-deficient HSCs to differentiate into B cells is cell intrinsic. (A) Sorted HSCs from IRF8+/+ and IRF8−/− mice were induced to differentiate in OP9 cocultures in the presence of SCF, IL-7, and Fl3tL for 7 days. The cells were stained with antibodies recognizing B220, CD19, and CD11b and analyzed by flow cytometry. Cells were gated on 7AAD− living cells. The numbers are percentages of cells falling in each gate. Data represent 1 of 3 independent experiments. (B) Sorted HSCs from each group of mice were cultured in the presence of SCF, Flt3L, IL-3, and IL-6 for 18 hours before they were infected with retroviral vectors encoding an IRF8-GFP fusion protein, an IRF8 mutant K79E-GFP fusion protein, or GFP only for 24 hours in the presence of SCF, Flt3L, IL-3, IL-6, IL-7, GM-CSF, and 4 μg/mL polybrene. GFP+ cells were resorted and plated onto OP9 cell layer in the presence of SCF, IL-7, and Flt3L for 7 days. The cells were analyzed by flow cytometry. The numbers are percentages of cells falling in each gate. Data represent 1 of 4 independent experiments. (C) The sorted GFP+ cells were cultured with OP9 cells at the same conditions as in panel B and were pulsed with BrdU for 40 minutes at day 4 and analyzed by FACS. The numbers are percentages of cells falling in each gate. Data represent 1 of 2 independent experiments.
The effect of IRF8 expression on cell growth
| . | Fold increase of cell numbers . | ||
|---|---|---|---|
| eGFP . | eGFP-Irf8 . | eGFP-K79E . | |
| Expt 1* | |||
| IRF8+/+ HSCs | 50.6 | 30.9 | 47.2 |
| IRF8−/− HSCs | 190.3 | 69.8 | 174.3 |
| Expt 2† | |||
| IRF8+/+ HSCs | 21.3 | 10.3 | 26.0 |
| IRF8−/− HSCs | 35.2 | 15.8 | 35.2 |
| . | Fold increase of cell numbers . | ||
|---|---|---|---|
| eGFP . | eGFP-Irf8 . | eGFP-K79E . | |
| Expt 1* | |||
| IRF8+/+ HSCs | 50.6 | 30.9 | 47.2 |
| IRF8−/− HSCs | 190.3 | 69.8 | 174.3 |
| Expt 2† | |||
| IRF8+/+ HSCs | 21.3 | 10.3 | 26.0 |
| IRF8−/− HSCs | 35.2 | 15.8 | 35.2 |
HSCs were sorted and transfected with vectors expressing eGFP-IRF8, eGFP-K79E, or eGFP alone and were resorted for GFP+ cells. Cells (1 × 104) were cultured in 24-well plate containing OP9 cells as well as B-cell permissive cytokines SCF, IL-7, and Flt3L for 5 days.
GFP+ cells (3-6 × 104) were cultured at the same conditions as in Expt 1 for 4 days.
Discussion
IRF8 is best known for its involvement in multiple aspects of the development and function of the innate immune system, primarily through studies of mice with a null mutation of the gene.16 More recently, several studies have shown that IRF8 is also involved in the acquisition of acquired immunity, first by promoting the pre-B to B-cell transition20 and second by contributing to transcriptional activation of BCL6 and AICDA, genes critical to the germinal center reaction.17,39 The present study broadens this picture by characterizing IRF8 as a component of the transcriptional complex governing myeloid versus lymphoid lineage specification as well as subsequent differentiation within the B-cell lineage.
Differentiation of myeloid and lymphoid progenitors from short-term HSCs is a constant dynamic process. Lineage specification is regulated, at least in part, by PU.1. The expression level of PU.1 appears to be critical for lineage choice. For example, high levels of PU.1 are required for macrophage development, lower levels are sufficient for B-cell differentiation, whereas intermediate levels of PU.1 are necessary for granulocytic differentiation.40-42 This pattern of graded PU.1 expression in hematopoietic progenitors is consistent with findings using PU.1Gfp reporter mice43,44 and with studies artificially manipulating PU.1 expression levels. For example, overexpression of PU.1 in hematopoietic cells blocks B-cell development and promotes macrophage differentiation but also supports the development of erythroleukemias.45 SiRNA knockdowns of PU.1 expression in ES cells promoted B-cell differentiation.10 However, B-cell commitment would appear to still require a minimal level of PU.1 because PU.1-null mutations result in complete loss of B cells and other hematopoietic lineages3,46 even though prolonged culture of PU.1−/− liver cells can produce CD19+ B cells in vitro.47 What was not established, however, were the mechanisms governing the expression of PU.1 in lymphoid and myeloid lineage cells. Several lines of evidence indicate that the normal balance between generation of MPs and CLPs is dependent, at least in part, on negative regulation of PU.1 by IRF8. First, the ratio of MPs to CLPs was significantly increased in IRF8−/− mice and mice reconstituted with IRF8-deficient HSCs. Second, in the absence of IRF8, the levels of PU.1 transcripts were increased 3-fold or more in MPs, CLPs, and Fr A cells. Third, stable siRNA knockdowns of IRF8 expression in a mouse B lymphoma cell line were associated with greater than 3-fold increases in the expression of PU.1 transcripts and protein. Fourth, overexpression of IRF8 in NFS-203 cells resulted in down-regulation of PU.1 expression at the protein level. Fifth, enforced expression of IRF8 in HSC-derived progenitor cells reduced the levels of PU.1 transcripts (Figure S3). Finally, ChIP analyses of B cells showed that IRF8 occupies a PU.1 promoter sequence containing a recently identified IRF8 target sequence. Previous studies have identified a series of factors that may drive transcriptional activation of PU.1 in hematopoietic cells including those of the B-cell lineage—PU.1 itself, Oct factors, GATA-1, Bob-1, and SpiB.1,48-50 To our knowledge, IRF8 is the first example of a transcriptional repressor for PU.1.
EBF was found to be another important target of the IRF8/PU.1 regulatory complex. EBF is expressed in B cells as well as nonlymphoid cells,51-53 although the mechanisms governing its expression within the B-cell lineage remain poorly understood. Previous studies showed that PU.1 binds a conserved sequence in the promoter of the Ebf1 gene.7 Our studies extend this finding by showing that PU.1 and IRF8 act synergistically to induce transcriptional activation of Ebf1. Two predicted IRF8/PU.1-binding sites in the Ebf1 promoter were validated by ChIP analyses, and mutations of either site reduced the activity of Ebf1 promoter reporter constructs. The conclusion that optimal regulation of Ebf1 is dependent on the expression of both genes is supported by the observation that the levels of Ebf1 transcripts in CLP and Fr A cells of IRF8−/− mice were reduced 80% or more even though the levels of PU.1 transcripts were increased in both cell subsets.
Transcripts of E2a and Pax5, 2 genes that encode additional early B-lineage transcription factors, were also significantly reduced in Fr A cells of IRF8-deficient mice. Similar to EBF, E2A regulates expression of genes activated as part of the early B-cell program, including Igll1 (λ5), Vpreb1 (Vpre-B), Cd79a (mb-1), Cd79b (B29), Rag1, and Cd19, and is absolutely required for early B-cell development.54 How E2A is regulated remains unclear but the data suggest it is not likely to be a direct transcriptional target of IRF8.
PAX5 is known to be induced by EBF and E2A and the effects of IRF8 deficiency on Pax5 could be secondary to reduced expression of EBF and E2A in these cells. However, this assumption could be complicated by the finding that enforced expression of IRF8 in HSC-derived early progenitors failed to induce expression of PAX5 even though IRF8 readily up-regulated expression of EBF and down-regulated expression of PU.1 (Figure 3S). The reason that PAX5 was not expressed under these seemingly favorable circumstances remains unclear. The growth inhibitory effect of IRF8 may change the balance of the transcriptional network required for PAX5 translation. IRF8 is known to directly regulate Prdm1 and Etv3, which encode for BLIMP1 and METS, respectively.55 Both transcription factors suppress expression of Myc, thereby inhibiting cell growth. Whether the expression of PAX5 is correlated with cell-cycle progression warrants further investigation. To attempt to understand how IRF8 controls programs of proliferation as well as lineage commitment, we are currently studying gene expression profiles in IRF8-expressing progenitor cells. Regardless of the mechanisms involved, it is likely that IRF8 acts upstream of the EBF-E2A-PAX5 regulatory complex to modulate B-lineage cell specification, commitment, and differentiation.
Despite the developmental defects at the pre-pro-B-cell stage identified here, total cell numbers in subsequent stages gradually increased even though they never reached normal levels. We attribute this result partially to enhanced pre-BCR expression on IRF8−/− pre-BII cells. The pre-BII cell stage (equivalent to Hardy Fr C′ and Fr D) is a critical checkpoint where productive μH chains are selected based on the ability of the μH chain to form pre-BCRs with the surrogate light chain56 and on the signaling strength of the pre-BCR.57,58 Pre-BCR signals are responsible for a proliferative burst of pre-BII cells. IRF8 and IRF4 have been shown to negatively regulate expression of surrogate L chains and pre-B-cell proliferation.59 Mice deficient for both IRF8 and IRF4 exhibited a complete blockade of B-cell development at the pre-B-cell stage, which was attributed to an inability of pre-B cells to exit cell cycle to initiate L chain gene rearrangement.20 Our data are consistent with these findings and suggest a redundant role of IRF8 and IRF4 in regulating pre-BCR signaling. Moreover, our data also suggested additional functions of IRF8 in development of immature B cells given that IRF8-deficient small pre-BII cells clearly generated fewer IgM+ immature B cells in irradiated recipient mice (Figure 5D). The basis for reductions of mature recirculating B cells in IRF8−/− mice is unknown, but could be due to an altered BM microenvironment that may prevent mature B cells from homing to the BM. Finally, because IRF4 has a redundant role with IRF8 in regulation of pre-B-cell differentiation, it would be interesting to examine whether IRF4 alone also regulates development of pre-pro-B cells in IRF4−/− mice.
Previously, it was shown that IRF8 plays important roles in GC B cells but is apparently dispensable in plasma cells.17,18 Taken in the light of other recent studies,4,7,40 our current results demonstrate that IRF8 is also part of a complex regulatory network governing the specification and commitment to B-cell development that includes PU.1, EBF, E2A, IKAROS, and PAX5.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Keiko Ozato for providing us with IRF8−/− mice and IRF8 constructs, and Dr Jeff X. Zhou for materials. We are grateful to Mehrnoosh Abshari for her assistance in cell sorting and to Nishant Kerrikatte.
This work was supported by the Intramural Research Program of National Institute of Allergy and Infectious Diseases (NIAID) and National Institute of Child Health and Human Development (NICHD), National Institutes of Health.
National Institutes of Health
Authorship
Contribution: H.W. designed, directed, and performed experiments, analyzed data, and wrote the paper; C.H.L. designed and performed experiments, and analyzed data; C.Q., P.T., J.F., S.A., and T.A. performed experiments; and H.C.M. directed experiments and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Herbert C. Morse III, Laboratory of Immunopathology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, MD 20852; e-mail: hmorse@niaid.nih.gov; or Hongsheng Wang, Laboratory of Immunopathology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, MD 20852; e-mail: wanghongs@niaid.nih.gov.
References
Author notes
*H.W. and C.H.L. contributed equally to this work.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal