In this issue of Blood, Purvis and colleagues describe a computational approach, employing 4 interlinked kinetic modules, to model platelet phosphoinositide and calcium regulation in resting platelets and after ADP-mediated P2Y1 purinergic receptor activation.
The model accurately replicates experimental findings, including the broad frequency distribution and asynchronous calcium spiking behavior in single platelets in response to adenosine diphosphate (ADP).
The P2Y1 receptor is a G-protein coupled receptor that signals through Gq and mediates ADP-induced platelet shape change and aggregation. Phospholipase Cβ2 (PLC-β2) is the major signaling molecule downstream of Gq and is responsible for transient increases in inositol 1,4,5-trisphosphate (IP3) and calcium concentration as well as the production of diacylglycerol (DAG) and protein kinase C (PKC) activation.1,2 In resting platelets, the cytoplasmic calcium concentration is actively maintained at approximately 100 nM, primarily through bulk storage of calcium within the dense tubular system (DTS). Calcium homeostasis between the cytoplasm and the DTS is regulated by inositol trisphosphate receptor (IP3R) channels, which release calcium ions from the DTS in response to IP3 and by a sarcoplasmic/endoplasmic reticulum Ca2+ ATPase (SERCA), which pumps calcium ions from the cytoplasmic compartment into the DTS.3
Classically, our understanding of platelet biology has come from a reductionist approach where individual events are studied under a single set of conditions. Here, Purvis et al present the first detailed and integrated “systems biology” approach to platelet signaling, modeling both phosphoinositide and calcium homeostasis in the resting platelet as well as temporal changes post-P2Y1 receptor activation. The model employs 4 interlinked kinetic modules: a calcium module, where the cytosolic and DTS compartments are separated by the DTS membrane, which contains the IP3R channels and SERCA; a phosphatidylinositol (PI) module, in which plasma membrane-bound PIs are cleaved by PLC-β2 to form diffusible inositol phosphates and DAG, which are substrates for resynthesis of PIs; a PKC module, whereby calcium and DAG activate PKC, which migrates to the plasma membrane and phosphorylates PLC-β2; and a P2Y1 module, where extracellular ADP activates P2Y1, accelerating formation of GTP-bound Gq, and subsequent activation of PLC-β2. Each module was initially considered in isolation for optimization. For example, platelet calcium homeostasis was analyzed by fixing the kinetic properties of the IP3R channels and SERCA, the resting calcium concentration, the volume of the platelet from experimental measurement, and assessing combinations of values for the number of IP3R channels per platelet, SERCA pumps per platelet and volume of the dense tubular system, using analog computation. P2Y1 signaling was con-sidered within the constraint that calcium influx was experimentally excluded, obviating the need to model store-operated calcium entry.4
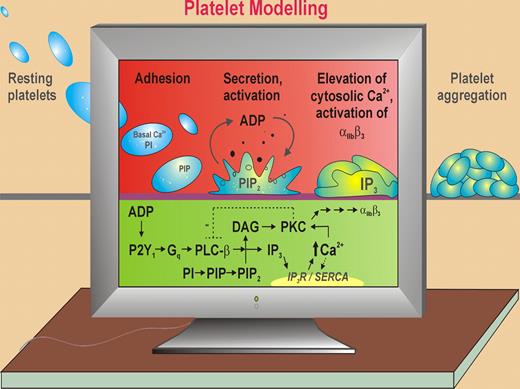
ADP-dependent platelet activation. Purvis et al use a computational model of the human platelet based on published kinetic data, electrochemical calculations, details of platelet ultrastructure, single cell analysis, and new data to predict temporal changes in intracellular Ca2+ levels, PI metabolites, and other ultrastructural parameters in response to ADP-dependent signaling through the purinergic Gq-coupled receptor, P2Y1. In thrombus formation, initial adhesion of circulating resting platelets to the vessel wall triggers platelet activation and secretion of ADP that acts in an autocrine fashion to induce shape change, cytoskeletal rearrangements, elevation of Ca2+, and αIIbβ3-dependent platelet aggregation. ADP binding to P2Y1 leads to down-stream activation of phospholipase C-β2 (PLC-β), generation of DAG (that activates PKC) and inositol trisphosphate (IP3), and elevation of cytosolic Ca2+ mediating platelet shape change and reversible platelet aggregation. Ca2+ levels are regulated by IP3 receptor (IP3R) channels, which release Ca2+ from the DTS and by a sarcoplasmic/endoplasmic reticulum Ca2+ ATPase (SERCA), which uptakes Ca2+. PKC phosphorylates/negatively regulates PLC-β. ADP binding P2Y12 (Gi-coupled) leads to decreased adenyl cyclase/cAMP and consolidates αIIbβ3-dependent platelet aggregation (not shown).
ADP-dependent platelet activation. Purvis et al use a computational model of the human platelet based on published kinetic data, electrochemical calculations, details of platelet ultrastructure, single cell analysis, and new data to predict temporal changes in intracellular Ca2+ levels, PI metabolites, and other ultrastructural parameters in response to ADP-dependent signaling through the purinergic Gq-coupled receptor, P2Y1. In thrombus formation, initial adhesion of circulating resting platelets to the vessel wall triggers platelet activation and secretion of ADP that acts in an autocrine fashion to induce shape change, cytoskeletal rearrangements, elevation of Ca2+, and αIIbβ3-dependent platelet aggregation. ADP binding to P2Y1 leads to down-stream activation of phospholipase C-β2 (PLC-β), generation of DAG (that activates PKC) and inositol trisphosphate (IP3), and elevation of cytosolic Ca2+ mediating platelet shape change and reversible platelet aggregation. Ca2+ levels are regulated by IP3 receptor (IP3R) channels, which release Ca2+ from the DTS and by a sarcoplasmic/endoplasmic reticulum Ca2+ ATPase (SERCA), which uptakes Ca2+. PKC phosphorylates/negatively regulates PLC-β. ADP binding P2Y12 (Gi-coupled) leads to decreased adenyl cyclase/cAMP and consolidates αIIbβ3-dependent platelet aggregation (not shown).
This integrated model developed by Purvis and colleagues accurately accounts for known platelet behavior and replicates experimental data at both averaged platelet and single platelet response. Stochastic simulation of the platelet model generated calcium spiking with peak-to-peak interval times favoring 6 to 8 and 11 to13 s gaps, strikingly similar to calcium responses in video-imaged single platelets. In addition, the model allowed several novel predictions: The calcium spiking in single platelets was a consequence of the small platelet volume, the number of SERCA pumps must signifi-cantly outnumber IP3R channels, and recovery of basal PI levels requires a negative-feedback mechanism in which PKC phosphorylation of PLC-β inhibits its hydrolytic activity. The model further explains that the reason thrombin is a more potent agonist than ADP is primarily due to differences in receptor copy number. The power of the computational approach of Purvis et al is that with continued development and experimental refinement of current variables, the model will not only accurately reflect known platelet behavior, but also predict new experimental findings, allowing a true in silico molecular and kinetic understanding of platelet biology.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal