Abstract
Chemokine-controlled migration plays a critical role in B-cell development, differentiation, and function, as well as in the pathogenesis of B-cell malignancies, including the plasma cell neoplasm multiple myeloma (MM). Here, we demonstrate that stimulation of B cells and MM cells with the chemokine stromal cell–derived factor-1 (SDF-1) induces strong migration and activation of the Ras-like GTPase Ral. Inhibition of Ral, by expression of the dominant negative RalN28 mutant or of RalBPΔGAP, a Ral effector mutant that sequesters active Ral, results in impaired SDF-1–induced migration of B cells and MM cells. Of the 2 Ral isoforms, RalA and RalB, RalB was found to mediate SDF-1–induced migration. We have recently shown that Btk, PLCγ2, and Lyn/Syk mediate SDF-1–controlled B-cell migration; however, SDF-1–induced Ral activation is not affected in B cells deficient in these proteins. In addition, treatment with pharmacological inhibitors against PI3K and PLC or expression of dominant-negative Ras did not impair SDF-1–induced Ral activation. Taken together, these results reveal a novel function for Ral, that is, regulation of SDF-1–induced migration of B cells and MM cells, thereby providing new insights into the control of B-cell homeostasis, trafficking, and function, as well as into the pathogenesis of MM.
Introduction
Depending on their stage of development or differentiation, B cells express specific chemokine receptors enabling them to respond to chemokines that control homing to the bone marrow (BM) and secondary lymphoid organs.1-3 Here they are provided with signals crucial for progression through the consecutive stages of B-cell development and differentiation. The chemokine stromal cell–derived factor-1 (SDF-1, also known as CXCL12) plays an important role in lymphocyte trafficking. SDF-1, originally identified as a growth-stimulatory factor for pre-B cells,4 is constitutively expressed by BM stromal cells.5-7 Its cognate receptor, the 7-transmembrane G-protein–coupled receptor CXCR4, is broadly expressed by cells of the immune system and mediates SDF-1–induced migration of hematopoietic progenitors and lymphocytes.1-3,8,9 In the B-cell lineage, CXCR4 is prominently expressed by pre-B cells and controls localization in cellular niches in the BM obligatory for B-cell development.3,8 Furthermore, CXCR4 expression by mature B cells is required for proper germinal center organization, which is essential for antigen-specific B-cell differentiation.10 In addition, SDF-1 controls plasma cell migration,6 and plasma cells require CXCR4 expression to localize in the BM compartment.11,12
Multiple myeloma (MM), an incurable hematologic malignancy with a median survival of 3 to 4 years, is characterized by the expansion of malignant plasma cells in the BM. These tumor cells also express CXCR4,13 and SDF-1 controls their α4β1-mediated adhesion to VCAM-1, fibronectin, and endothelial cells, as well as their transendothelial migration and homing/retention in the BM tumor microenvironment.13-19 MM cells are critically dependent on the BM microenvironment, where cytokines produced by BM stromal cells provide them with proliferation and survival signals required for their expansion.16 Thus, by controlling homing and retention of MM cells in the BM, SDF-1 plays an important role in the pathogenesis of MM.
The important role of chemokine-induced migration in B-cell development and differentiation, as well as in the pathogenesis of MM, prompted us to explore the underlying signaling pathways. Recently, we have established an important role for Bruton tyrosine kinase (Btk) and phospholipase C-γ2 (PLCγ2) in chemokine-induced integrin-mediated migration and homing of B cells.20 Importantly, however, Btk is not expressed in plasma cells and MM cells.21-23 Several members of the Ras superfamily of small GTPases have also been implicated in chemokine-controlled B-cell migration.24-27 This includes members of the Rho family24,25 and members of the Ras family, such as Ras and Rap1.26,27 Ral, another member of the Ras family of small GTPases, has been shown to mediate a wide variety of cellular responses, including regulation of cytoskeletal rearrangements and cell motility.28 Ral can bind to and regulate the activity of phospholipase D1,29 the Sec5 and Exo84 subunits of the exocyst complex,30,31 the actin binding protein filamin,32 and the Cdc42/Rac GTPase-activating protein Ral binding protein-1 (RalBP1).33,34 Notably, accumulating evidence indicates that Ral is an essential mediator of Ras-induced transformation of human cells.35-38 Interestingly, Ral is involved in invasion and metastasis of transformed cells and in Ras- and growth factor–controlled cell migration39-47 ; however, thus far a role for Ral in chemokine-controlled signaling and migration has not been described. Here we show that Ral is activated in response to SDF-1 and mediates SDF-1–induced chemotaxis of B cells and MM cells.
Methods
Materials
Monoclonal antibodies used were mouse anti-RalA (IgG2a), anti-Rap1 (IgG1), biotinylated rat anti–mouse CXCR4 (2B11/CXCR4) (all Becton Dickinson [BD] Biosciences, San Jose, CA), and Streptavidin-PE (Southern Biotechnology Associates, Birmingham, AL). Polyclonal antibodies used were rabbit anti-RalB (BD Biosciences), rabbit anti–phospho PKB/Akt (Ser 473), rabbit anti–phospho p44/42 MAP kinase (Thr 202/Tyr 204) (both New England Biolabs, Beverly, MA), and HRP-conjugated rabbit antimouse and HRP-conjugated goat antirabbit (both DAKO, Carpinteria, CA). Pharmacological inhibitors used were LY294002, wortmannin, PD98059, and U73122 (Biomol, Plymouth Meeting, PA).
Plasmids
pA-puroII-RasN1748 was kindly provided by Dr T. Kurosaki (Kansai Medical University, Moriguchi, Japan). pRK5-RalBPΔGAP and pMT2-HA-RalAN28 were described previously.49 pRK5-RalBPΔGAP was kindly provided by Dr F. Zwartkruis (Utrecht Medical Center, The Netherlands). pCDNA-HA-RalAN28 was generated by ligating the blunted PstI-digested 952-bp fragment of pMT2-HA-RalAN28 into EcoRV-digested pCDNA3.1+. pA-puroII-HA-RalAN28 was generated by ligating the KpnI-XhoI–digested fragment of pCDNA-HA-RalAN28 in KpnI-SalI–digested pA-puroII. pTER-shRalA and pTER-shRalB were generated by ligating the annealed oligos into BglII-HindIII–digested pTER. The oligonucleotides used were as follows: for RalA, 5′-GATCCCGACAGGTTTCTGTAGAAGATTCAAGAGATCTTCTACAGAAACCTGTCTTTTTGGAAA-3′ and 5′-AG-CTTTTCCAAAAAGACAGGTTTCTGTAGAAGATCTCTTGAATCTTC-TACAGAAACCTTCGG-3′; for RalB, 5′-GATCCCGGTGATCATGGTTG-GCAGCTTCAAGAGAGCTGCCAACCATGATCACCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGGTGATCATGGTTGGCAGCTCTCTTGAAG-CTGCCAACCATGATCACCGG-3′. These target sequences were previously shown to result in efficient knockdown of RalA30 or RalB,50 respectively.
Isolation of primary cells and cell culture
Human tonsillar B cells were isolated essentially as described previously.51 Isolated B cells were maintained in RPMI containing 10% FCS and were used immediately. Human primary MM cells were isolated essentially as described previously.52 The Burkitt lymphoma cell line Ramos was cultured in Iscoves medium (Life Technologies, Breda, The Netherlands) containing 10% fetal clone I serum (HyClone Laboratories, Logan, UT), 100 IU/mL penicillin and 100 IU/mL streptomycin (Life Technologies), 20 mg/mL human recombinant transferrin (Sigma, Bornem, Belgium), and 50 mM β-mercaptoethanol. Isolated primary MM cells and XG-1 MM cells were maintained in the same medium supplemented with 500 pg/mL IL-6 (R&D Systems, Abington, United Kingdom). The Burkitt lymphoma cell line Namalwa, and the MM cell lines NCI-H929 and OPM-1,53,54 were cultured in RPMI 1640 supplemented with 10% fetal clone I serum, 2 mM l-glutamine, 100 IU/mL penicillin, and 100 IU/mL streptomycin (all Life Technologies). The chicken bursal lymphoma cell line DT40 was cultured at 39.5°C as described.48 Stable overexpression of RasN17 in DT40 cells was performed by electroporation, using the pA-puroII-RasN17 expression vector (kindly provided by Dr T. Kurosaki), exactly as described.55
Ral and Rap pull-down assays
Cells were resuspended in RPMI or IMDM to 2.0 × 107 cells/mL and stimulated with SDF-1 (100 ng/mL). Reactions were terminated by adding an equal volume of cold 2 × lysis buffer (100 mM Tris-HCl [pH 7.4], 400 mM NaCl, 5 mM MgCl2, 2% Nonidet P-40, 20% glycerol, 2 × EDTA-free protease inhibitor cocktail tablets [Roche, Indianapolis, IN] per 50 mL). After 10 minutes on ice, cell debris was removed by centrifugation. Cell lysates were used immediately for Ral or Rap1 pull-down assays. For this purpose, glutathione-Sepharose beads (100 μL of a 20% solution per sample) were precoupled with GST-RalBP-RBD or GST-RalGDS-RBD fusion protein by continuous mixing for 30 minutes at 4°C with bacterial cell lysates from Escherichia coli strain AD202 transformed with pGEX4T3-RalBP-RBD or E coli strain BL21 transformed with pGEX4T-RalGDS-RBD96, respectively.56,57 After being washed 3 times with cell lysis buffer, these precoupled beads were added to the cell lysates, and incubated for 30 minutes at 4°C during continuous mixing. Finally, the beads were washed 4 times with lysis buffer, and bound proteins were eluted with sample buffer, separated by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotted with anti-RalA or anti-Rap1.
Generation of stably transfected DT40 cells
Linearized pA-puroII-HA-RalN28 (25 μg) was mixed with 107 DT40 cells in 0.5 mL RPMI 1640 medium in a 0.4-cm electrode gap Gene Pulser Cuvette (BioRad, Hercules, CA) and electroporated using a Gene Pulser Apparatus with Capacitance Extender (BioRad) at 250 V, 960 μF. After 24-hour recovery at 39.5°C in DT40 medium, cells were selected in DT40 medium containing 0.5 μg/mL Puromycin (Sigma). Puromycin-resistant clones were screened for expression of HA-RalN28 by immunoblotting.
Transient transfections
Cells (107) in 0.5 mL RPMI 1640 medium were transfected by electroporation as indicated with 5 μg pEGFP-N3 (for migration assays) or pRL-TK (for adhesion assays), together with the indicated expression plasmids or empty control plasmid up to a total amount of 25 to 30 μg DNA per transfection. Cells were allowed to recover for 16 hours at 37°C in complete medium. Then viable cells were isolated using Ficoll-Paque density gradient centrifugation (Amersham Pharmacia, Uppsala, Sweden), and after 24-hour incubation in complete medium cells were used in migration or adhesion assays. For immunoblot analysis of RalA and RalB knockdown, fluorescence-activated cell sorting (FACS)–sorted GFP-positive cells were used.
Cell migration assay
Migration assays were performed in triplicate using 5-μm (DT40 cells) or 8-μm pore size Transwells (Costar, Cambridge, MA), washed twice with PBS, and blocked for 1 hour at 37°C with 0.5% BSA in RPMI 1640. Unless indicated otherwise, the lower compartment was filled with 600 μL 0.5% BSA/RPMI containing 100 ng/mL SDF-1, and 5 × 105 cells in 100 μL were applied to the upper compartment and allowed to migrate for 5 hours. The amount of viable (GFP-positive) migrated cells was determined by FACS and expressed as a percentage of the input, that is, the number of cells applied directly into the lower compartment in parallel wells. Unless otherwise indicated, the migration of nonpretreated WT or GFP-positive empty control-plasmid transfected cells in the presence of SDF-1 was normalized to 100% (± SD) of triplicates.
Flow cytometry
Quantification of (GFP-positive) migrated cells and analysis of CXCR4 expression was carried out on a FACScalibur flow cytometer (BD Biosciences) with CELLQuest software (BD Biosciences).
Cell adhesion assay
Adhesion assays were performed in triplicate on flat-bottom, high-binding 96-well plates (Costar) coated overnight at 4°C with PBS containing 1 μg/mL sVCAM-1 and 4% BSA, with or without 150 ng/mL SDF-1, and then blocked for 1 hour at 37°C with 4% BSA in RPMI 1640. Cells (1.5 × 105) were plated in 100 μL/well in RPMI containing 1% BSA, spun down briefly (32g, 30 seconds) and incubated at 37°C for 2 minutes. After extensive washing of the plate with RPMI containing 1% BSA to remove nonadhering cells, the adherent cells were lysed and Renilla luciferase activity was determined according to manufacturer's instructions (Dual-Luciferase Reporter Assay System; Promega, Madison, WI). Adhesion was calculated and corrected for transfection efficiency and nonspecific effects of constructs by measuring Renilla luciferase activity of total input cells.
Immunoblotting
Immunoblotting was performed essentially as previously described.51 Quantification was performed using Image-Pro plus (MediaCybernetics, Bethesda, MD) software.
Statistical analysis
The unpaired 2-tailed Student t test was used to determine the significance of differences between means. All relevant comparisons were significantly different (P < .05), unless otherwise indicated.
Results
SDF-1 induces migration and Ral activation in B cells and MM cells
SDF-1 is an important chemoattractant for B cells,3,8-10 as illustrated by its potential to induce migration of the chicken bursal lymphoma cell line DT40, the GC-like B-cell lymphoma cell lines Namalwa and Ramos, and human tonsillar B cells in a Transwell migration assay (Figure 1A). SDF-1 is acting exclusively as a true chemotactic—and not a chemokinetic—factor, since migration occurred only when SDF-1 was present in the lower but not the upper compartment of the Transwells (Figure 1B). To determine whether Ral is involved in chemokine-induced signaling, we performed Ral pull-down assays. We found that SDF-1 stimulation of the chicken B-cell line DT40 resulted in a rapid, transient activation of Ral (Figure 1C). Ral was already maximally activated after 30 seconds of SDF-1 stimulation, and Ral-GTP levels were back to basal levels within 5 minutes. A similar pattern of Ral activation was observed after SDF-1 stimulation of Ramos and Namalwa B-cell lines as well as primary human tonsillar B cells (Figure 1C).
SDF-1 stimulation induces migration and Ral activation in B cells. (A) DT40, Ramos, and Namalwa B-cell lines, and human tonsillar B cells, were allowed to migrate for 4 hours in the absence (c) or presence of 100 ng/mL SDF-1 (SDF-1) in Transwells (shown as the mean ± SD of triplicates). (B) DT40 cells were allowed to migrate for 4 hours in the absence (−) or presence (+) of 100 ng/mL SDF-1 in the upper or lower compartment of Transwells, as indicated (shown as the mean ± SD of triplicates). (C) DT40, Ramos, and Namalwa B-cell lines, and human tonsillar B cells, were stimulated for the indicated periods of time with 100 ng/mL SDF-1 and lysed, and the amount of Ral-GTP in the lysates was determined by pull-down assay (PD) using GST-RalBP-RBD fusion protein. As a control, total lysates (TLs) were immunoblotted and probed using anti-Ral antibodies. (A,B) All relevant comparisons (eg, migration in absence versus migration in presence of SDF-1) were significantly different (P < .05). (A-C) The results are representative of at least 2 independent experiments.
SDF-1 stimulation induces migration and Ral activation in B cells. (A) DT40, Ramos, and Namalwa B-cell lines, and human tonsillar B cells, were allowed to migrate for 4 hours in the absence (c) or presence of 100 ng/mL SDF-1 (SDF-1) in Transwells (shown as the mean ± SD of triplicates). (B) DT40 cells were allowed to migrate for 4 hours in the absence (−) or presence (+) of 100 ng/mL SDF-1 in the upper or lower compartment of Transwells, as indicated (shown as the mean ± SD of triplicates). (C) DT40, Ramos, and Namalwa B-cell lines, and human tonsillar B cells, were stimulated for the indicated periods of time with 100 ng/mL SDF-1 and lysed, and the amount of Ral-GTP in the lysates was determined by pull-down assay (PD) using GST-RalBP-RBD fusion protein. As a control, total lysates (TLs) were immunoblotted and probed using anti-Ral antibodies. (A,B) All relevant comparisons (eg, migration in absence versus migration in presence of SDF-1) were significantly different (P < .05). (A-C) The results are representative of at least 2 independent experiments.
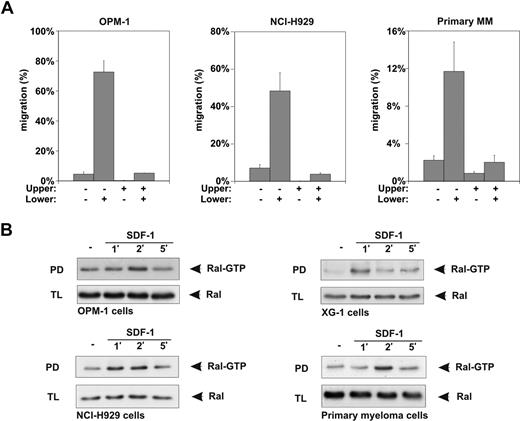
Similar to normal plasma cells,6,11,12 SDF-1 is a potent chemoattractant for MM cell lines13,14,17-19 and the malignant plasma cells from MM patients,13,14,19,58,59 as illustrated by the ability of SDF-1 to induce migration of the MM cell lines NCI-H929 and OPM-1, and of primary MM cells, in the Transwell migration assay (Figure 2A). In addition, for MM cells (cell lines and primary patient material), SDF-1 is acting exclusively as a true chemotactic factor, that is, migration occurred only when SDF-1 was present in the lower but not the upper compartment of the Transwells (Figure 2A). Therefore, a role for Ral in SDF-1 signaling in MM cells was also examined. Similar to B cells, Ral was activated in response to SDF-1 in several MM cell lines and in primary MM cells (Figure 2B). Active Ral levels were maximal after 1 to 2 minutes of SDF-1 stimulation, and returned to basal within 5 minutes. Taken together, these results demonstrate that SDF-1 induces migration and activation of Ral in B cells and MM cells.
SDF-1 stimulation induces migration and Ral activation in MM cells. (A) The MM cell lines OPM-1 and NCI-H929, and the malignant plasma cells from a MM patient, were allowed to migrate for 4 hours in the absence (−) or presence (+) of 100 ng/mL SDF-1 in the upper or lower compartment of Trans-wells, as indicated (shown as the mean ± SD of triplicates). All relevant comparisons (eg, migration in absence versus migration in presence of SDF-1) were significantly different (P < .05), and the results are representative of 3 independent experiments and of freshly isolated malignant plasma cells from 3 MM patients. (B) NCI-H929, OPM-1, and XG-1 MM cell lines, and primary human MM cells, were stimulated for the indicated periods of time with 100 ng/mL SDF-1 and lysed, and the amount of Ral-GTP in the lysates was determined by pull-down assay (PD) using GST-RalBP-RBD fusion protein. As a control, total lysates (TLs) were immunoblotted and probed using anti-Ral antibodies. The results are representative of at least 2 independent experiments.
SDF-1 stimulation induces migration and Ral activation in MM cells. (A) The MM cell lines OPM-1 and NCI-H929, and the malignant plasma cells from a MM patient, were allowed to migrate for 4 hours in the absence (−) or presence (+) of 100 ng/mL SDF-1 in the upper or lower compartment of Trans-wells, as indicated (shown as the mean ± SD of triplicates). All relevant comparisons (eg, migration in absence versus migration in presence of SDF-1) were significantly different (P < .05), and the results are representative of 3 independent experiments and of freshly isolated malignant plasma cells from 3 MM patients. (B) NCI-H929, OPM-1, and XG-1 MM cell lines, and primary human MM cells, were stimulated for the indicated periods of time with 100 ng/mL SDF-1 and lysed, and the amount of Ral-GTP in the lysates was determined by pull-down assay (PD) using GST-RalBP-RBD fusion protein. As a control, total lysates (TLs) were immunoblotted and probed using anti-Ral antibodies. The results are representative of at least 2 independent experiments.
Ral mediates SDF-1–induced migration of B cells and MM cells
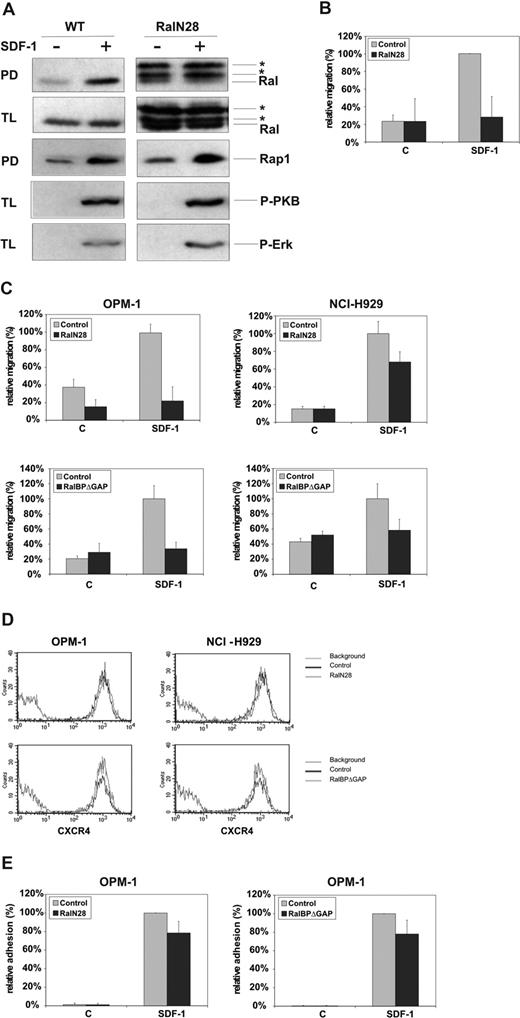
To investigate whether Ral mediates SDF-1–induced B-cell migration, we generated DT40 cells stably expressing a dominant negative Ral mutant, RalN28, which prevents SDF-1–induced activation of endogenous Ral (Figure 3A). Interestingly, in a Transwell migration assay the RalN28-expressing cells showed strongly reduced migration toward SDF-1 in comparison with wild-type (WT) cells (Figure 3B), indicating that Ral mediates SDF-1–induced migration. Expression of RalN28 did not affect SDF-1–induced activation of the small GTPase Rap1, of PI3K (as determined by phosphorylation of PKB), and of the MAP kinase ERK (Figure 3A), which have previously been implicated in SDF-1–induced migration.60-62 Thus, Ral does not mediate migration through activation of these migration-regulatory signaling molecules. In addition, these results demonstrate the specificity of the inhibitory effect of RalN28 on SDF-1–induced activation of Ral and migration.
Ral mediates SDF-1–induced migration of B cells and MM cells. (A) Wild-type (WT) and stably RalN28-transfected DT40 cells were stimulated for 1 minute with 100 ng/mL SDF-1 and lysed. The amount of Ral-GTP and Rap1-GTP in the DT40 lysates was determined by pull-down assay (PD) using GST-RalBP-RBD and GST-RalGDS-RBD fusion protein and immunoblotting with anti-Ral and anti-Rap, respectively. Total lysates (TLs) were immunoblotted and probed using anti-Ral, anti–P-PKB, or anti–P-MAPK (P-ERK) antibodies. The lower (minor) band in the Ral pull-down assays of the RalN28-expressing DT40 cells represents endogenous Ral and the upper 2 (major) bands represent overexpressed HA-tagged RalN28 (*), that is, the full-length and a partial product resulting from instability of RalN28. (B,C) Wild-type (WT) and stably RalN28-transfected DT40 cells (B) or OPM-1 and NCI-H929 MM cells cotransfected with GFP and either RalN28 or RalBPΔGAP (C) were allowed to migrate for 4 hours in the absence (c) or presence of 100 ng/mL SDF-1 (SDF-1) in Transwells. The migration (of GFP-positive cells) in the presence of SDF-1 was normalized to 100%, and is shown as the mean (± SD) of triplicates. All relevant comparisons were significantly different (P < .05). The results are representative of at least 3 independent experiments. (D) OPM-1 and NCI-H929 MM cells cotransfected with GFP and either RalN28 or RalBPΔGAP were analyzed for CXCR4 expression by FACS. The results are representative of at least 3 independent experiments. (E) OPM-1 MM cells cotransfected with Renilla luciferase and either RalN28 or RalBPΔGAP were allowed to adhere to VCAM-1 for 2 minutes in the absence (c) or presence of SDF-1. The Renilla luciferase activity for the cells adhering to VCAM-1 in the presence of SDF-1 (reflecting 52% of input cells) was normalized to 100%, and shown as the mean (± SD) of 3 independent experiments. All relevant comparisons were significantly different (P < .05).
Ral mediates SDF-1–induced migration of B cells and MM cells. (A) Wild-type (WT) and stably RalN28-transfected DT40 cells were stimulated for 1 minute with 100 ng/mL SDF-1 and lysed. The amount of Ral-GTP and Rap1-GTP in the DT40 lysates was determined by pull-down assay (PD) using GST-RalBP-RBD and GST-RalGDS-RBD fusion protein and immunoblotting with anti-Ral and anti-Rap, respectively. Total lysates (TLs) were immunoblotted and probed using anti-Ral, anti–P-PKB, or anti–P-MAPK (P-ERK) antibodies. The lower (minor) band in the Ral pull-down assays of the RalN28-expressing DT40 cells represents endogenous Ral and the upper 2 (major) bands represent overexpressed HA-tagged RalN28 (*), that is, the full-length and a partial product resulting from instability of RalN28. (B,C) Wild-type (WT) and stably RalN28-transfected DT40 cells (B) or OPM-1 and NCI-H929 MM cells cotransfected with GFP and either RalN28 or RalBPΔGAP (C) were allowed to migrate for 4 hours in the absence (c) or presence of 100 ng/mL SDF-1 (SDF-1) in Transwells. The migration (of GFP-positive cells) in the presence of SDF-1 was normalized to 100%, and is shown as the mean (± SD) of triplicates. All relevant comparisons were significantly different (P < .05). The results are representative of at least 3 independent experiments. (D) OPM-1 and NCI-H929 MM cells cotransfected with GFP and either RalN28 or RalBPΔGAP were analyzed for CXCR4 expression by FACS. The results are representative of at least 3 independent experiments. (E) OPM-1 MM cells cotransfected with Renilla luciferase and either RalN28 or RalBPΔGAP were allowed to adhere to VCAM-1 for 2 minutes in the absence (c) or presence of SDF-1. The Renilla luciferase activity for the cells adhering to VCAM-1 in the presence of SDF-1 (reflecting 52% of input cells) was normalized to 100%, and shown as the mean (± SD) of 3 independent experiments. All relevant comparisons were significantly different (P < .05).
Inhibition of Ral by transient transfection of either RalN28 or RalBPΔ GAP, a Ral effector mutant that sequesters active Ral, also resulted in reduced SDF-1–induced migration of the MM cell lines OPM-1 and NCI-H929 (Figure 3C). Since Ral has been implicated in the regulation of gene transcription and vesicle transport,30,31,55,63 the impaired responsiveness to SDF-1 upon inhibition of Ral could have been the consequence of reduced surface expression of CXCR4; however, CXCR4 expression of OPM-1 and NCI-H929 cells expressing either dominant negative Ral or RalBPΔGAP was similar to untransfected cells (Figure 3D), and inhibition of Ral did not affect SDF-1–induced internalization or recycling of CXCR4 either (data not shown). In line with this, SDF-1–induced phosphorylation of PKB and ERK was not affected by expression of RalN28 in OPM-1 MM cells (data not shown): please note that a GTPase pull-down assay for Rap1 (or Ral) cannot be performed in these transiently transfected MM cells due to limited transfection efficiency and assay sensitivity.
Integrin-mediated adhesion is an important aspect of chemokine-controlled migration and of the pathogenesis of MM. To investigate whether Ral regulates SDF-1–induced integrin-mediated adhesion of MM cells, OPM-1 cells were transiently transfected with RalN28 or RalBPΔ GAP and allowed to adhere to VCAM-1 in the presence of SDF-1. SDF-1 strongly induced adhesion, and inhibition of Ral due to expression of RalN28 or RalBPΔ GAP resulted in a modest reduction of SDF-1–induced adhesion (Figure 3E). Notably, inhibition of Rap1, another mediator of chemokine-induced lymphocyte migration and adhesion,62 resulted in a similarly modest inhibition of adhesion (data not shown). Taken together, our results show that Ral mediates SDF-1–induced migration, which may, at least in part, involve the control of integrin activation.
RalB rather than RalA mediates SDF-1–induced MM migration
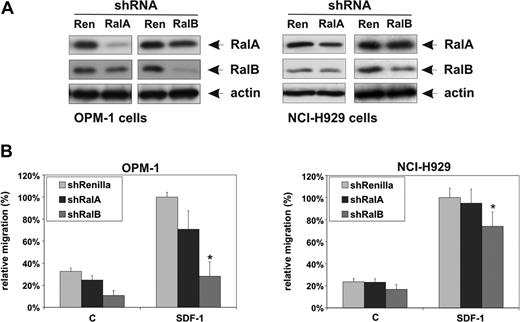
Mammalian cells can express 2 isoforms of Ral, RalA and RalB, which show 85% identity. Although these proteins are highly similar, they appear to have different functions. Whereas RalA is an essential mediator of Ras-controlled tumorigenesis, RalB has shown to be involved in tumor cell motility.37,46 To determine which Ral isoform is involved in SDF-1–induced MM migration, OPM-1 and NCI-H929 were transiently transfected with a GFP construct and constructs expressing short-hairpin RNA (shRNA) directed against either RalA, RalB, or, as a control, Renilla. In OPM-1 cells, and to a lesser extent NCI-H929 cells, these constructs were found to efficiently knock down expression of RalA or RalB, respectively (Figure 4A). Notably, however, expression of RalA shRNA also resulted in decreased anti-RalB signal, and vice versa (Figure 4A), indicating that although these antibodies display isoform preference, they are not strictly specific. This was confirmed by the observation that the anti-RalB antibody also detects ectopically expressed RalA (data not shown). Interestingly, MM cells transfected with shRNA against RalB showed impaired migration toward SDF-1, whereas only a modest or no decrease in migration was observed in cells with reduced RalA ex-pression (Figure 4B). Furthermore, among the MM cell lines there appears to be a correlation between the efficiency of RalB silencing and the suppression of migration. Taken together, our data suggest that SDF-1–induced MM cell migration is predominantly mediated by RalB.
RalB rather than RalA mediates SDF-1–induced MM cell migration. (A) Lysates of OPM-1 and NCI-H929 MM cells co-transfected with GFP and shRenilla, shRalA, or shRalB were immunoblotted to determine RalA and RalB expression. Actin served as a loading control. (B) OPM-1 and NCI-H929 MM cells co-transfected with GFP and shRenilla, shRalA, or shRalB were allowed to migrate for 4 hours in the absence (c) or presence of 100 ng/mL SDF-1 (SDF-1) in Transwells. The migration of the shRenilla-transfected GFP-positive cells in the presence of SDF-1 was normalized to 100%, and shown as the mean ± SD of triplicates (*P < .05). The results are representative of at least 2 independent experiments.
RalB rather than RalA mediates SDF-1–induced MM cell migration. (A) Lysates of OPM-1 and NCI-H929 MM cells co-transfected with GFP and shRenilla, shRalA, or shRalB were immunoblotted to determine RalA and RalB expression. Actin served as a loading control. (B) OPM-1 and NCI-H929 MM cells co-transfected with GFP and shRenilla, shRalA, or shRalB were allowed to migrate for 4 hours in the absence (c) or presence of 100 ng/mL SDF-1 (SDF-1) in Transwells. The migration of the shRenilla-transfected GFP-positive cells in the presence of SDF-1 was normalized to 100%, and shown as the mean ± SD of triplicates (*P < .05). The results are representative of at least 2 independent experiments.
SDF-1–induced Ral activation is independent of Lyn/Syk, Btk, PLC, PI3K, and Ras activity
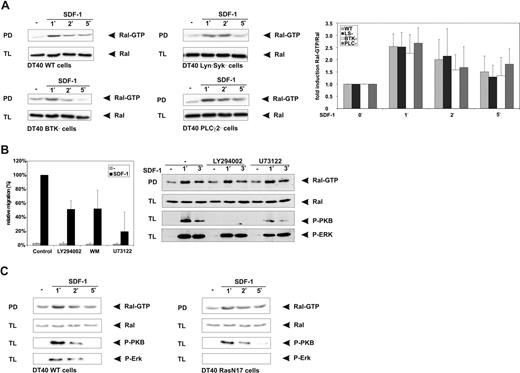
Recently, we have established a prominent role for the cytoplasmic kinases Lyn, Syk, and Btk, and for Btk-mediated activation of PLCγ2, in chemokine-controlled B-cell migration.20 Therefore, we used cell lines deficient in Lyn and Syk, Btk, or PLCγ2 to investigate if Ral may act downstream of these signaling molecules. Interestingly, however, SDF-1–induced activation of Ral in these gene-deficient cells was similar to the wild-type (WT) cells (Figure 5A). In addition, the PLC isoform PLCβ and PI3K have been shown to mediate SDF-1–induced migration60,61 ; for PI3K, this includes studies in B cells20,64-67 and myeloma cells.14,19,68,69 In line with this, pretreatment with either the pharmacological PI3K inhibitors LY294002 and wortmannin or the general PLC inhibitor U73122 inhibited migration of DT40 cells (Figure 5B); however, these pharmacological inhibitors did not affect SDF-induced Ral activation either (Figure 5B).
SDF-1–induced activation of Ral is independent of Lyn/Syk, Btk, PLC, PI3K, or Ras. DT40 cells, either wild type (WT) or deficient in Lyn/Syk, Btk, or PLCγ2 (A); DT40 cells pretreated for 30 minutes with either 20 μM LY294002 or 10 μM U73122 (B); or DT40 cells stably transfected with RasN17 (C) were stimulated for the indicated periods of time with 100 ng/mL SDF-1 and lysed, and the amount of Ral-GTP in the lysates was determined by pull-down assay (PD) using GST-RalBP-RBD fusion protein. As a control, total lysates (TLs) were immunoblotted and probed using anti-Ral, anti–P-PKB, or anti–P-MAPK (P-ERK) antibodies. (A) The right panel displays quantification of the Ral activation; the fold induction of the Ral-GTP/Ral ratio is shown as the mean (± SD) of at least 2 independent experiments. (B) DT40 cells pretreated for 30 minutes with either 20 μM LY294002, 100 nM wortmannin (WM), or 10 μM U73122 were allowed to migrate for 4 hours in the presence of the inhibitors, and in the absence (c) or presence of 100 ng/mL SDF-1 (SDF-1) in Transwells. The migration was normalized to 100% for the cells allowed to migrate in the presence of SDF-1, and shown as the mean (± SD) of triplicates. All relevant comparisons were significantly different (P < .05). (B,C) The results are representative of at least 2 independent experiments.
SDF-1–induced activation of Ral is independent of Lyn/Syk, Btk, PLC, PI3K, or Ras. DT40 cells, either wild type (WT) or deficient in Lyn/Syk, Btk, or PLCγ2 (A); DT40 cells pretreated for 30 minutes with either 20 μM LY294002 or 10 μM U73122 (B); or DT40 cells stably transfected with RasN17 (C) were stimulated for the indicated periods of time with 100 ng/mL SDF-1 and lysed, and the amount of Ral-GTP in the lysates was determined by pull-down assay (PD) using GST-RalBP-RBD fusion protein. As a control, total lysates (TLs) were immunoblotted and probed using anti-Ral, anti–P-PKB, or anti–P-MAPK (P-ERK) antibodies. (A) The right panel displays quantification of the Ral activation; the fold induction of the Ral-GTP/Ral ratio is shown as the mean (± SD) of at least 2 independent experiments. (B) DT40 cells pretreated for 30 minutes with either 20 μM LY294002, 100 nM wortmannin (WM), or 10 μM U73122 were allowed to migrate for 4 hours in the presence of the inhibitors, and in the absence (c) or presence of 100 ng/mL SDF-1 (SDF-1) in Transwells. The migration was normalized to 100% for the cells allowed to migrate in the presence of SDF-1, and shown as the mean (± SD) of triplicates. All relevant comparisons were significantly different (P < .05). (B,C) The results are representative of at least 2 independent experiments.
Ral can be activated by Ras-independent mechanisms via Ca2+56 ; and calmodulin70 or β-arrestins71 ; however, most cytokines have been found to activate Ral in a Ras-dependent manner.72,73 Therefore, we used DT40 cells stably expressing RasN17, a dominant negative Ras mutant, to examine whether SDF-1–induced Ral activation requires Ras activity. However, whereas BCR-controlled activation of Ral55 and SDF-1–induced phosphorylation of the downstream Ras target Erk were completely abolished in these RasN17-expressing cells, activation of Ral by SDF-1 was not affected (Figure 5C).
Taken together, these data demonstrate that activation of Ral is required but not sufficient for full SDF-1–induced migration, and SDF-1–induced activation of Ral and Ral-mediated migration occurs independent of the established migration-regulatory pathways involving Lyn/Syk, Btk, PLC, PI3K, or Ras.
Discussion
We found that SDF-1 stimulation of B cells and MM cells results in activation of the small GTPase Ral (Figures 1C and 2B). Expression of RalN28 or RalBPΔGAP led to reduced migration of B cells and MM cells toward SDF-1 (Figure 3), indicating that Ral mediates SDF-1–induced B-cell and MM cell migration. Notably, inhibition of Ral in the Jurkat T-cell line also resulted in diminished SDF-1–induced migration (data not shown), suggesting that Ral is involved in SDF-1–induced lymphocyte migration in general and not only in migration of B cells. The recently identified mediators of chemokine-induced B-cell migration and homing, Btk and PLCγ2,20 are not involved in GTP loading of Ral upon SDF-1 stimulation (Figure 5A). In addition, PI3K and other PLC isoforms that are known to mediate chemokine-induced responses (Figure 5B)60,61 are not involved in SDF-1–induced activation of Ral (Figure 5B). Interestingly, whereas Ras mediates Ral activation in response to several stimuli, inhibition of Ras activity by stable expression of a dominant negative Ras mutant did not affect SDF-1–induced Ral activation (Figure 5C). Thus, activation of Ral and Ral-mediated migration appears to occur independent of these established migration-regulatory pathways. DOCK2 is another important mediator of lymphocyte chemotaxis, which controls activation of the migration-regulatory GTPases Rac and Rap1.74,75 Therefore, it would be interesting to be determine whether chemokine-induced Ral activation may occur in a DOCK2-dependent manner.
Members of the Rho family of small GTPases have been identified as important mediators of lymphocyte migration. The C3 exoenzyme from the bacterial pathogen Clostridium botilinum, which ADP ribosylates and thereby inactivates RhoA, RhoB, and RhoC, is widely used as tool to elucidate the cellular functions of Rho GTPases. For example, C3 treatment impairs SDF-1–induced MM cell adhesion to VCAM-1 and fibronectin.15 Interestingly, C3 was also found to bind with high affinity to GDP-bound Ral GTPases.76,77 C3 binding to Ral stabilizes the GDP-bound, inactive conformation, and as a consequence C3 interferes with Ral signaling.77 Moreover, by binding of C3 to Ral, the ribosylation and inactivation of Rho is blocked.76,77 This can be overcome by increasing the C3 concentration,76 suggesting that C3-mediated inactivation of Rho occurs only after all Ral proteins have been inhibited. Therefore effects of C3 that are ascribed to Rho inactivation could also be due to inhibition of Ral.
Ral might regulate chemokine-controlled migration in various manners. Ral GTPases may control chemotaxis by modulating the actin cytoskeleton through interaction with the Cdc42/Rac GTPase-activating protein RalBP1.33,34 In addition, RalA can control filopodia formation by recruitment of the Ral-effector complex the exocyst78 and the actin filament cross-linking protein filamin,32 which can affect migration. However, RalB rather than RalA was found to mediate MM cell chemotaxis (Figure 5B). Similarly, specifically RalB was recently shown to be involved in cancer cell motility.46,47 By recruiting the exocyst, RalB is believed to control the coordinated delivery of secretory vesicles to the sites of dynamic plasma membrane expansion that specify directional movement.47 The differential requirement for Ral isoforms in cell migration might be the consequence of differences in the C-terminal variable regions of RalA and RalB. Due to their different C-termini, RalA and RalB localize to different subcellular compartments, which determines their specific functions.37,79 In addition, despite the high degree of effector-binding domain sequence homology, the affinity for effectors varies between the 2 Ral isoforms.79
Previously, Bhattacharya et al found that Ral is involved in fMLP-induced cytoskeletal reorganization.71 In this study, the RalGEF RalGDS was found to be activated by β-arrestins in a Ras-independent manner. Interestingly, β-arrestins are important mediators of G-protein–coupled receptor signaling,80 and are required for CXCR4-mediated lymphocyte chemotaxis.81 β-Arrestin-mediated SDF-1–induced chemotaxis was found to be mediated by the p38 MAPK,82 a signaling molecule activated by Ral in growth factor signaling.83 Combined with our observation that SDF-1 induces Ral activation independent of Ras activity (Figure 5C), it is tempting to speculate that SDF-1 activates Ral via β-arrestins, which may lead to p38-mediated chemotactic responses. JNK, another MAPK, was also found to be involved in B-cell chemotaxis,84 and in mediating several Ral-dependent processes, including tissue development,85 protection against oxidative stress,86 and survival of tumor cells.38 Thus, p38 as well as JNK may act downstream of Ral in SDF-1–induced chemotaxis of B cells and MM cells.
We have shown that Ral is activated upon stimulation with SDF-1 and mediates SDF-1–induced migration of MM cells (Figures 2B and 3B). Several chemokines, including SDF-1, are produced by BM stromal cells and act as chemoattractants for MM cells.13,14,17-19,58,59 As a consequence, MM cells reside in the BM, where various cytokines produced in the microenvironment induce MM growth and survival. Hence, chemoattractants controlling homing of MM cells to the BM can be considered equally important for MM growth as the cytokines that provide the actual proliferative and antiapoptotic signals. Consequently, Ral may play an important role in the pathogenesis of MM. In addition, Ral appears to be an important mediator in Ras-induced transformation of solid tumors,35-38 and may also directly control the survival and proliferation of MM cells. Thus, by inhibition of Ral both migration to the BM and direct MM growth might be affected. This putative property of Ral, being involved in both MM cell migration and MM growth and survival, could make this protein a very attractive target for therapeutic intervention.
In conclusion, we have shown that Ral mediates SDF-1–controlled migration of B cells and MM cells and may therefore play an important role in B-cell development and function, as well as in the pathogenesis of MM.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr F. Zwartkruis for kindly providing pRK5-RalBPΔ GAP and Dr T. Kurosaki for pA-puroII-RasN17. We thank Berend Hooibrink for sorting the GFP-positive MM cells; Richard Bende for providing patient myeloma cells; and M. Klitsie, P. van Rijn, N. de Vries, and coworkers of the Department of Otolaryngology of the Sint Lucas and Andreas hospital Amsterdam for providing the tonsils.
This work was supported by grants from the Dutch Cancer Society (KWF).
National Institutes of Health
Authorship
Contribution: D.J.J.G. performed research, analyzed data, and wrote the paper; R.M.R., E.A.B., H.P.H.N., and A.K. performed research; M.J.K. provided materials; S.T.P. analyzed data; M.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marcel Spaargaren, Department of Pathology, Academic Medical Center, University of Amsterdam, PO Box 22700, 1100 DE Amsterdam, The Netherlands; e-mail: marcel.spaargaren@amc.uva.nl.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal