Abstract
These days it has been increasingly recognized that mast cells (MCs) are critical components of host defense against pathogens. In this study, we have provided the first evidence that MCs can kill bacteria by entrapping them in extracellular structures similar to the extracellular traps described for neutrophils (NETs). We took advantage of the ability of MCs to kill the human pathogen Streptococcus pyogenes by a phagocytosis-independent mechanism in order to characterize the extracellular antimicrobial activity of MCs. Close contact of bacteria and MCs was required for full antimicrobial activity. Immunofluorescence and electron microscopy revealed that S pyogenes was entrapped by extracellular structures produced by MCs (MCETs), which are composed of DNA, histones, tryptase, and the antimicrobial peptide LL-37. Disruption of MCETs significantly reduced the antimicrobial effect of MCs, suggesting that intact extracellular webs are critical for effective inhibition of bacterial growth. Similar to NETs, production of MCETs was mediated by a reactive oxygen species (ROS)–dependent cell death mechanism accompanied by disruption of the nuclear envelope, which can be induced after stimulation of MCs with phorbol-12-myristate-13-acetate (PMA), H2O2, or bacterial pathogens. Our study provides the first experimental evidence of antimicrobial extracellular traps formation by an immune cell population other than neutrophils.
Introduction
Mast cells (MCs) are multifunctional and highly effective tissue-dwelling cells that play a prominent role in a wide variety of important biological processes. They are well known for their role in the initiation of allergic diseases and their activation during certain parasitic infections (reviewed by Puxeddu et al1 ). MCs derived from bone-marrow progenitor cells circulate in the peripheral blood and migrate into vascularized tissue before undergoing final maturation under the influence of local factors. Maturated MCs are commonly found in tissues that interface with the external environment such as the skin and mucosa of the respiratory and gastrointestinal tract (reviewed by Mekori and Metcalf2 ). Because these sites are also common portals of infection, MCs are likely to be among the first inflammatory cells to interact with invading pathogens.
Several recent reports in the literature indicate that MCs can mediate a variety of antimicrobial activities following activation upon contact with pathogens. First, MCs have been shown to release preformed and newly synthesized inflammatory mediators, proteases, cytokines, and chemokines that recruit neutrophils to the site of infection.3,4 They are the only cell type known to prestore TNF-α in their secretory granules, which can be released immediately upon activation by pathogens to initiate the early phase of the inflammatory response.5,6 Secondly, there is increasing experimental evidence that MCs themselves can directly kill various Gram-negative and Gram-positive bacteria.7-10 Finally, experiments using MC-deficient mice have clearly demonstrated that MCs are essential for mounting an effective immune response against bacterial infections such as Citrobacter rodentium,9 Pseudomonas aeruginosa,11 Klebsiella pneumoniae,6 or enteropathogenic Escherichia coli.12-15 Based on these observations, it has been proposed that MCs play a central role in the host defense against infectious pathogens (recently reviewed by Dawicki and Marshall16 ).
Regarding the direct antimicrobial activity of MCs, several studies have shown that MCs are capable of bacterial recognition and intracellular uptake. Bacteria endocytosed after opsonin-mediated binding are internalized via a route involving the endosome-lysosome pathway, in which the bacteria are killed through a combination of oxidative and nonoxidative killing systems (reviewed in Féger et al8 ). These observations suggest that MCs are able to eliminate bacteria through an intracellular bactericidal mechanism similar to that of professional phagocytes.
However, recent investigations have reported that various potent broad-spectrum antimicrobial peptides such as the cathelicidin LL-37,17 β-defensins,9 or piscidins18 are present in MC granules and are secreted upon stimulation with various pathogens,17 indicating that an extracellular bactericidal mechanism may also be operating in these cells. In contrast to the numerous reports addressing the intracellular bactericidal activity of MCs, limited data exist regarding their possible extracellular killing activities. Recently, it has been shown that murine bone marrow–derived MCs significantly suppressed the growth rate of the Gram-positive bacterium Streptococcus pyogenes in cocultures, and that this growth inhibition was mediated, at least in part, by the MC's antimicrobial cathelicidins.17 Addition of cathelicidins to the culture medium resulted in complete suppression of S pyogenes proliferation. Moreover, Wei et al9 showed that MCs exhibit an extracellular antimicrobial activity against the Gram-negative bacterium C rodentium, but the underlying mechanism was not described.
Taken together, these observations clearly indicate that the release of cathelicidins like LL-37, and probably of other antimicrobial compounds stored in the MCs granules, may provide a potent mechanism that enables MCs to participate in host defense against invading pathogens. Although several lines of evidence support the hypothesis of an extracellular antimicrobial mechanism of MCs, the exact mechanism remains to be clarified.
In this study, we have characterized the mechanism underlying the extracellular antimicrobial activity exerted by human and murine MCs. S pyogenes was chosen for these studies because in preliminary experiments we found that, in contrast to other microorganisms, MCs can kill S pyogenes by an extracellular, phagocytosis-independent mechanism. Therefore, this infection model provides an excellent system to study the extracellular antimicrobial mechanisms of MCs without the influence of phagocytic uptake.
Our results demonstrated that MCs exert their extracellular antimicrobial activity through the production of extracellular structures similar to the recently reported neutrophil extracellular traps (NETs).19,20 The major components of these MC extracellular traps (MCETs) are DNA, histones, and granule proteins such as tryptase and the antimicrobial peptide LL-37. Our study provides the first evidence that besides neutrophils, other innate immune cells can exert antimicrobial activity by formation of extracellular traps.
Methods
The animal experiments were approved by the appropriate national ethical board (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, Oldenburg, Germany). The human study was approved by the Human Subjects Review Committee of the University of Toronto, and informed consent was obtained from all patients in accordance with the Declaration of Helsinki.
Bacterial strains
The bacterial strains used in this study were S pyogenes strain A20 (M-type 23), a human isolate obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ 2071 Braunschweig, Germany), the P aeruginosa strain PAO, and the S aureus strain S6.21 S pyogenes was cultured at 37°C in Todd Hewitt broth (Oxoid, Wesel, Germany) supplemented with 1% yeast extract, P aeruginosa in Luria-Bertani broth (Carl Roth, Karlsruhe, Germany) and S aureus in brain heart infusion broth (Carl Roth). Bacteria were collected at the midpoint of log-growth phase, washed twice with sterile phosphate-buffered saline (PBS), and diluted to the required concentration. The number of viable bacteria (colony-forming units [CFUs]) was determined after serial diluting and plating on blood agar containing 5% sheep blood (Invitrogen, Karlsruhe, Germany).
For some experiments, bacteria were labeled with carboxyfluorescein (Invitrogen) by incubating a suspension of 5 × 108 bacteria with 0.2 mg/mL carboxyfluorescein for 30 minutes at 4°C in the dark. After incubation, bacteria were washed several times to remove unbound dye and used for the corresponding assay.
Mice
Female C3H/HeN mice (8-10 weeks old) used for isolation of bone marrow–derived MCs (BMMCs) were purchased from Harlan-Winkelmann (Borchen, Germany). The experiments were approved by the appropriate national ethical board (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, Oldenburg, Germany).
Culture of MCs
The human MC line HMC-1 was obtained from Joseph H. Butterfield (Mayo Foundation for Medical Education and Research, Rochester, MN).22 HMC-1 cells were cultured in Iscove modified Dulbecco medium (IMDM) supplemented with 10% fetal calf serum (FCS), 1.2 mM α-thioglycerol, and 100 U/mL penicillin/100 μg/mL streptomycin (Invitrogen).
Murine BMMCs were isolated from C3H/HeN mice as previously described.23 The resulting cell population consisted of more than 97% BMMCs as determined by flow cytometry analysis using anti-mouse CD117 antibody (c-Kit; Caltag Laboratories, Karlsruhe, Germany), toluidine blue staining, and scanning electron microscopy.
In vitro coculture of MCs and bacteria
MCs were washed twice with medium without antibiotics, plated in 24-well tissue-culture plates at a density of 106 cells/mL (BMMCs) or 5 × 105 cells/mL (HMC-1). Bacteria were added directly to cultured MCs at a multiplicity of infection (MOI) of 25 bacteria per MC.
The bacterial viability in the presence of MCs was monitored at different time points by the determination of CFUs in the cell culture supernatants. Total bacterial growth was determined in samples containing medium alone. For preparation of the inoculum, S pyogenes was subjected to extensive washing to ensure the elimination of exotoxins and other bacterial products released during culturing. This treatment generates high levels of stress for the microorganisms, which often require an adaptation phase after being seeded in medium alone or in coculture with MCs. The length of this lag phase can vary from experiment to experiment, resulting in different bacterial growth kinetics.
In experiments using transwell systems, MCs were seeded in the lower chamber of a transwell polystyrene plate (polycarbonate membrane with 0.4-μm pore size, 6.5-mm diameter; Costar Corning, Schiphol-Rijk, The Netherlands), and bacteria were added to the top chamber of the transwell plate.
In some assays, MCs were treated with cytochalasin D (Sigma-Aldrich, Taufkirchen, Germany) at a final concentration of 10 μg/mL to inhibit phagocytosis. MCs were stimulated with 25 nM phorbol-12-myristate-13-acetate (PMA) or 100 mU/mL of the H2O2-producing enzyme glucose oxidase (GO; Sigma-Aldrich) to induce formation of MCETs. In some experiments, H2O2 was neutralized by addition of 100 U/mL of catalase (Sigma-Aldrich). To dismantle MCETs, stimulated MCs were treated with 40 U/mL DNase to degrade DNA and 1 μmol myeloperoxidase (Sigma-Aldrich) to degrade tryptase as previously described.24 To inhibit the production of reactive oxygen species (ROS), MCs were treated with 10 μM of the NADPH oxidase inhibitor diphenylene iodonium (DPI; Sigma-Aldrich).
Double immunofluorescence microscopy
S pyogenes– or S aureus–infected MCs were fixed with paraformaldehyde (4%), and double immunofluorescence analysis was performed as previously described.25
Examination of MCETs
MCs were seeded on poly-L-lysine–covered coverslides, treated with 25 nM PMA for 10 minutes, and infected with carboxyfluorescein-labeled (for confocal microscopy) or unlabeled (for electron microscopy) S pyogenes at a MOI of 1:25. After different times of infection, in-fected cells were centrifuged for 5 minutes at 52g and fixed with 4% paraformaldehyde.
For fluorescence microscopy, fixed cells were washed 3 times in PBS before staining with specific antibodies. For LL-37 and tryptase staining, coverslides were incubated with goat anti–mouse tryptase β-1 antibodies (1:60 diluted; R&D Systems, Minneapolis, MN), mouse anti–human MC tryptase (clone A41, 1:200 diluted; Dako, Hamburg, Germany), or mouse anti–human LL37 (clone 1-1C12, 1:50 diluted; HyCult Biotechnology, Uden, The Netherlands), followed by rabbit anti–goat/mouse IgG 488–Alexa–labeled antibodies (1:300 diluted; Invitrogen). For histone staining, MCs were incubated with mouse anti–bovine histone antibodies (1:500 diluted; United States Biological, Swampscott, MA) with a species cross-reactivity to human and mouse followed by rabbit-anti–mouse IgG 488–Alexa–labeled antibodies (1:300 diluted; Invitrogen). Murine BMMCs were blocked with a goat anti–mouse IgG (1:200; Southern Biotechnology Associates, Birmingham, AL) before incubation with the secondary anti-mouse IgG. All incubations with antibodies were performed for 45 minutes at room temperature. Finally, coverslides were washed and mounted onto glass slides using Prolong Gold (Invitrogen), a mounting medium that contains the DNA-staining dye DAPI (4,6-diamino-2-phenylindole: blue).
The LIVE/DEAD BacLight Bacterial Viability Kit was used to determine viability of S pyogenes entrapped in the MCETs by fluorescence microscopy following the recommendations of the manufacturer (Invitrogen).
Mounted samples were examined using a confocal laser-scanning microscope (MRC1024UV; Bio-Rad, Hemel Hempstead, United Kingdom) with an inverted Zeiss Axiovert 100TV microscope (with a 63×/1.25 oil objective; Carl Zeiss, Oberkochen, Germany). Images were obtained using a T1/E2 filter combination and a krypton/argon laser. Images were collected and processed with Lasersharp software 3.2 (Bio Rad). Alternatively, images were recorded using a Zeiss Axiophot microscope (20×/0.50, 40×/1.30 oil, 63×/1.25 oil, or 100×/1.25 oil objective) with an attached Zeiss Axiocam HRc digital camera and Axiovision software 4.5 (Carl Zeiss) at calibrated magnifications.
Field emission scanning electron microscopy
MCs were fixed with 4% paraformaldehyde, washed with TE-buffer (20 mM Tris, 1 mM EDTA [pH 6.9]), dehydrated by incubating with a graded series of ethanol (10%, 30%, 50%, 70%, 90%, and 100%) on ice for 15 minutes, critical-point dried with liquid CO2 (CPD 30; BAL-TEC, Balzers, Liechtenstein), and covered with a gold film by sputter coating (SCD 40; Balzers Union) before being examined in a field emission scanning electron microscope (Zeiss DSM 982 Gemini; Carl Zeiss) using the Everhart Thornley SE detector and the inlens detector (Carl Zeiss) in a 50:50 ratio at an acceleration voltage of 5 kV.
Transmission electron microscopy
Samples were fixed with a solution containing 5% formaldehyde and 2% glutaraldyhde in cacodylate buffer (0.1 M cacodylate, 0.01 M CaCl2, 0.01 M MgCl2, 0.09 M sucrose [pH 6.9]) for 1 hour on ice, washed with cacodylate buffer, and osmificated with 1% aqueous osmium for 1 hour at room temperature. Samples were then dehydrated with a graded series of acetone (10%, 30%, 50%, 70%, 90%, and 100%) for 30 minutes at each step. Dehydration in the 70% acetone step was done with 2% uranyl acetate overnight. Samples were infiltrated with an epoxy resin according to the Spurr formula.26 Ultrathin sections were cut with a diamond knife, counterstained with uranyl acetate and lead citrate, and examined in a TEM910 transmission electron microscope (Carl Zeiss) at an acceleration voltage of 80 kV. Images were taken at calibrated magnifications using a line replica.
Determination of ROS
For determination of ROS, MCs were seeded on poly-L-lysine–coated coverslides, infected with S pyogenes at a MOI of 1:25, and incubated for 15 minutes at 37°C, 5% CO2. After washing to remove nonadherent cells, MCs were incubated with 1 mg/mL Nitro Blue Tetrazolium (NBT) dissolved in Krebs-Ringer phosphate glucose (KRPG) buffer for 45 minutes at 37°C. After incubation, the cells were washed twice with KRPG buffer, fixed in 4% paraformaldehyde, and counterstained with safranin. Samples were examined by light microscopy for the presence of blue-black formazan precipitate.
Mitochondrial ROS generation was measured by fluorescence microscopy and by flow cytometry using a FACScalibur (Becton Dickinson, Heidelberg, Germany) after loading the treated and untreated HMC-1 cells with 5 μm dihydrorhodamine 123 (DHR 123; Sigma-Aldrich) for 30 minutes at 37°C.
Quantification and visualization of cell death
MCs were stained with 10 μg/mL propidium iodide (Sigma-Aldrich) and dead cells as well as extracellular DNA were quantified by flow cytometry. For microscopic examination of cell death, infected MCs were cultured on poly-L-lysine–coated glass coverslides and analyzed for viability using the LIVE/DEAD viability/cytotoxicity kit for mammalian cells (Invitrogen) following the recommendations of the manufacturer.
Quantification of DNA released by dying MCs
To quantify DNA release, MCs were seeded and stimulated as described. Released DNA was stained by adding 5 μM of Sytox Orange (Invitrogen), a non–cell-permeable DNA-binding dye that has been previously used for the quantification of DNA release by neutrophils as a marker for NETs.19,27 The intensity of fluorescence as direct quantification of the amount of DNA release was determined using the Xenogen Vivo Vision IVIS 200 System (Xenogen, Hopkinton, MA) with the excitation passband of 500 to 550 nm and the emission passband of 575 to 650 nm; analysis was done using the Igor Pro 4.09A software (Wavemetrics, Lake Oswego, OR).
Statistical analysis
Data were analyzed by using Excel 2000 (Microsoft, Redmond, WA) or GraphPad Prism 4.0 (GraphPad Software, San Diego, CA). Each in vitro experiment was performed at least 3 times at independent occasions, and within each experiment all samples were processed in duplicate. Comparison between groups was made by use of a variance analysis (F-test). P values of .05 or less were considered significant.
Results
MCs kill S pyogenes via an extracellular mechanism
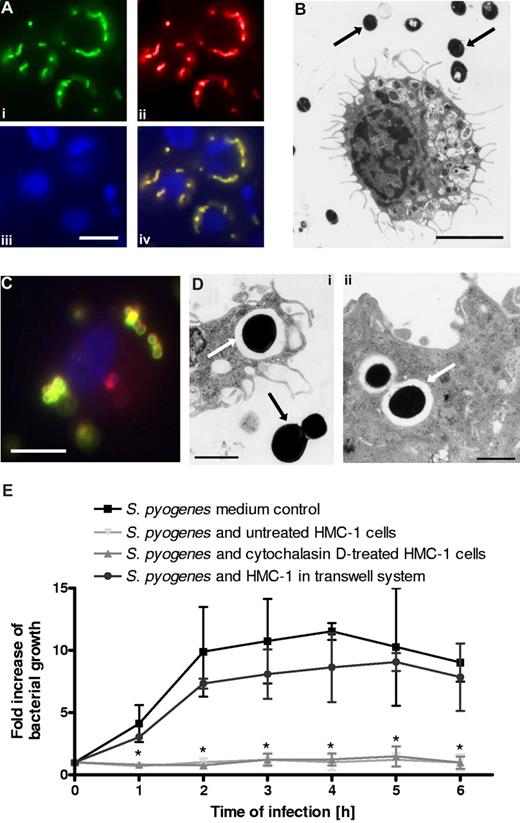
It has been previously reported that MCs can inhibit growth of S pyogenes.17 However, whether growth inhibition was mediated by phagocytic uptake and subsequent intracellular bacterial killing or by an extracellular killing mechanism mediated by the release of antimicrobial peptides remained to be determined. To examine if cultured human MCs (HMC-1 cells) are able to uptake S pyogenes, HMC-1 cells were incubated with S pyogenes for 3 hours, and the bacterial location (extracellular versus intracellular) was determined by fluorescence microscopy. Photographs displayed in Figure 1A provide clear evidence of the failure of HMC-1 cells to uptake S pyogenes, since only green/yellow-labeled bacteria (extracellular) but not red-labeled S pyogenes (intracellular) were detected. These observations were further confirmed by transmission electron microscopic examination where only extracellular bacteria were seen (Figure 1B black arrows). The failure of HMC-1 cells to phagocytose S pyogenes was not due to the assay conditions used in this study. This could be demonstrated by the efficient uptake of S aureus, a pathogen previously shown to be phagocytosed by MCs,7 under similar experimental settings (double immunofluorescence in Figure 1C; scanning electron microscopy in Figure 1Di,ii).
In vitro interactions of bacteria with human MCs (HMC-1). (A) Double immunofluorescence staining for determination of extracellular/intracellular location of S pyogenes associated with MCs (bar, 3.5 μm). Extracellular bacteria were stained with polyclonal rabbit anti–S pyogenes antibodies, followed by Alexa green–conjugated goat antirabbit antibodies (Sigma-Aldrich). After several washes, the cells were permeabilized by 0.025% Triton X-100 in PBS and washed again, and intracellular (as well as extracellular) bacteria were stained by anti–S pyogenes antibodies, followed by Alexa red–conjugated goat antirabbit antibodies (Sigma-Aldrich). Exclusively extracellular bacteria are labeled in green (i), extracellular plus intracellular bacteria are labeled in red (ii), and MC nuclei are labeled in blue (iii). An overlaid merged image where extracellular bacteria are labeled yellow and intracellular bacteria in red is shown (iv). (B) Transmission electron microscopic examination of cross-sections of HMC-1 cells cocultured with S pyogenes (bar, 2.5 μm). Bacteria are indicated by black arrows. Notice that all streptococcal microorganisms are extracellularly located. (C) Immunofluorescence staining for determination of extracellular (yellow) and intracellular (red) location of S aureus associated with MCs (bar, 3.5 μm). (D) Transmissionelectron microscopic examination of cross-sections of HMC-1 cells showing internalized S aureus (i,ii; white arrows). Extracellular bacteria (i) are indicated by a black arrow (bars, 1 μm). (E) Growth of S pyogenes in medium alone, in coculture with MCs, in coculture with cytochalasin D–treated MCs, or in coculture with MCs separated by a transwell system. Data are expressed as x-fold increase in bacterial growth with respect to the original inoculum. Each point represents the mean plus or minus SD of 3 independent experiments. *P < .05 by F-test for S pyogenes growth in medium control versus S pyogenes growth in the presence of either untreated or cytochalasin D–treated HMC-1 cells.
In vitro interactions of bacteria with human MCs (HMC-1). (A) Double immunofluorescence staining for determination of extracellular/intracellular location of S pyogenes associated with MCs (bar, 3.5 μm). Extracellular bacteria were stained with polyclonal rabbit anti–S pyogenes antibodies, followed by Alexa green–conjugated goat antirabbit antibodies (Sigma-Aldrich). After several washes, the cells were permeabilized by 0.025% Triton X-100 in PBS and washed again, and intracellular (as well as extracellular) bacteria were stained by anti–S pyogenes antibodies, followed by Alexa red–conjugated goat antirabbit antibodies (Sigma-Aldrich). Exclusively extracellular bacteria are labeled in green (i), extracellular plus intracellular bacteria are labeled in red (ii), and MC nuclei are labeled in blue (iii). An overlaid merged image where extracellular bacteria are labeled yellow and intracellular bacteria in red is shown (iv). (B) Transmission electron microscopic examination of cross-sections of HMC-1 cells cocultured with S pyogenes (bar, 2.5 μm). Bacteria are indicated by black arrows. Notice that all streptococcal microorganisms are extracellularly located. (C) Immunofluorescence staining for determination of extracellular (yellow) and intracellular (red) location of S aureus associated with MCs (bar, 3.5 μm). (D) Transmissionelectron microscopic examination of cross-sections of HMC-1 cells showing internalized S aureus (i,ii; white arrows). Extracellular bacteria (i) are indicated by a black arrow (bars, 1 μm). (E) Growth of S pyogenes in medium alone, in coculture with MCs, in coculture with cytochalasin D–treated MCs, or in coculture with MCs separated by a transwell system. Data are expressed as x-fold increase in bacterial growth with respect to the original inoculum. Each point represents the mean plus or minus SD of 3 independent experiments. *P < .05 by F-test for S pyogenes growth in medium control versus S pyogenes growth in the presence of either untreated or cytochalasin D–treated HMC-1 cells.
We next determined if, despite their inability to phagocytose S pyogenes, HMC-1 cells could still inhibit the growth of this pathogen under our in vitro experimental conditions. Bacterial growth was quantified by counting the number of surviving bacteria (CFUs) at increasing time points of coculture and was expressed as fold increase of bacterial growth over the initial inoculum. As shown in Figure 1E, complete inhibition of bacterial growth was observed after incubation with HMC-1 cells compared with time-matched growth in medium alone. As expected, the growth inhibition was unaffected by treatment with the phagocytosis inhibitor cytochalasin D, further demonstrating that growth inhibition is independent of phagocytic uptake of S pyogenes by HMC-1 cells (Figure 1E). Interestingly, close proximity between the bacteria and the MCs was required for efficient growth inhibition as shown by the significantly lower levels of growth inhibition observed when bacteria and HMC-1 cells were separated by a 0.4-μm pore-size membrane in a transwell system (Figure 1E).
S pyogenes is entrapped by extracellular structures consisting of DNA, histones, tryptase, and the cathelicidin LL-37 produced by activated MCs
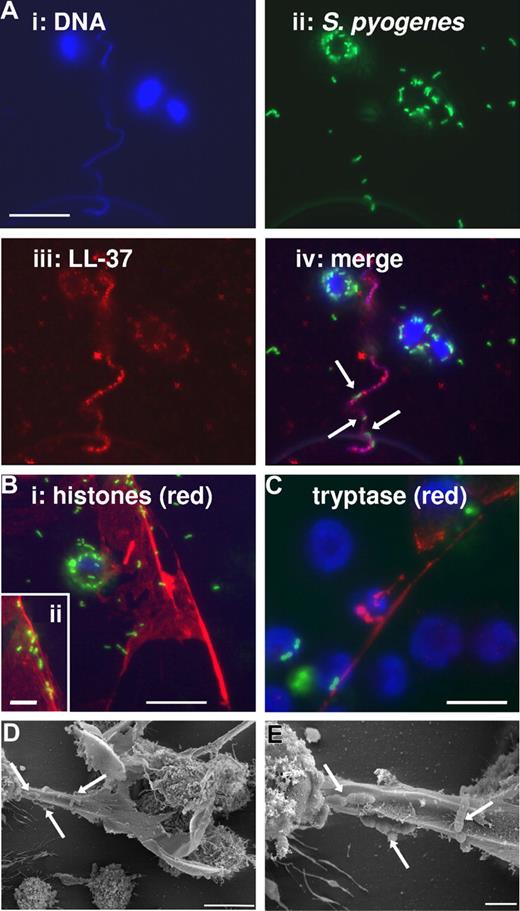
It has been recently shown that antimicrobial activity of MCs against S pyogenes involves the production and/or the release of cathelicidins by MCs.17 To determine whether cathelicidin LL-37 produced by HMC-1 cells was released after incubation with S pyogenes, MCs were seeded on poly-L-lysine–coated coverslides and infected with green-labeled S pyogenes at a MOI of 1:25. At different times after infection, infected cells were fixed with 4% paraformaldehyde and stained with Alexa-red antibodies against LL-37. HMC-1 cells are visualized in Figure 2 by the blue staining of the nuclear DNA (Figure 2Ai), S pyogenes by the green fluorescence (Figure 2Aii), and LL-37 by the red fluorescence (Figure 2Aiii). Unexpectedly, the overlaid image depicted in Figure 2Aiv shows that extracellular LL-37 (red) was associated with highly defined structures containing DNA and, even more interesting, S pyogenes can be found entrapped in these structures (arrows). These formations were similar to the recently described extracellular traps produced by neutrophils (NETs),19 and therefore we designated them MCETs. The formation of MCETs can be greatly increased after stimulation of MCs with 25 nM PMA for 10 minutes prior to infection, similar to NETs.19 Further microscopic examination of the composition of these MCETs revealed that, in addition to DNA and cathelicidins, other structural components are histones (red; Figure 2Bi,ii) and tryptase (red; Figure 2C). The morphology of the MCETs with entrapped S pyogenes (arrows Figure 2D,F) was visualized by field emission scanning electron microscopy (FESEM).
Immunofluorescence and FESEM examination of human MCETs. MCs were seeded on poly-L-lysine–coated glass slides, stimulated with 25 nM PMA for 10 minutes, then infected with FITC-labeled S pyogenes (MOI 25:1) for 1 hour and fixed with 4% paraformaldehyde. (A) Colocalization of DNA, S pyogenes, and LL-37 in MCETs (bars, 10 μm): (i) Dapi-stained DNA; (ii) FITC-labeled S pyogenes; (iii) immunostaining of LL-37 with Alexa-red–labeled antibodies against human LL-37; and (iv) overlay of A1-3. (B) Immunostaining of MCETs with Alexa-red-labeled antibodies against histones (bar, 10 μm). Insert in the lower-left corner (ii) shows a higher magnification of S pyogenes (green) entrapped in the MCETs (red; bar, 3.5 μm). (C) Immunostaining of MCETs with Alexa-red–labeled antibodies against tryptase. S pyogenes are labeled green and DNA are labeled blue (bar, 8 μm). (D) FESEM image of MCETs produced by human MCs during coculture with S pyogenes. Microorganisms entrapped in the MCETs structures are indicated by white arrows (bar, 5 μm). (E) Higher magnification showing S pyogenes entrapped in the fibers of the MCETs (white arrows; bar, 1 μm).
Immunofluorescence and FESEM examination of human MCETs. MCs were seeded on poly-L-lysine–coated glass slides, stimulated with 25 nM PMA for 10 minutes, then infected with FITC-labeled S pyogenes (MOI 25:1) for 1 hour and fixed with 4% paraformaldehyde. (A) Colocalization of DNA, S pyogenes, and LL-37 in MCETs (bars, 10 μm): (i) Dapi-stained DNA; (ii) FITC-labeled S pyogenes; (iii) immunostaining of LL-37 with Alexa-red–labeled antibodies against human LL-37; and (iv) overlay of A1-3. (B) Immunostaining of MCETs with Alexa-red-labeled antibodies against histones (bar, 10 μm). Insert in the lower-left corner (ii) shows a higher magnification of S pyogenes (green) entrapped in the MCETs (red; bar, 3.5 μm). (C) Immunostaining of MCETs with Alexa-red–labeled antibodies against tryptase. S pyogenes are labeled green and DNA are labeled blue (bar, 8 μm). (D) FESEM image of MCETs produced by human MCs during coculture with S pyogenes. Microorganisms entrapped in the MCETs structures are indicated by white arrows (bar, 5 μm). (E) Higher magnification showing S pyogenes entrapped in the fibers of the MCETs (white arrows; bar, 1 μm).
Murine BMMCs are also able to produce MCETs in response to S pyogenes
In order to demonstrate that the MCETs formation was not just an artifact of the human MC line used for these studies, we examined the ability of murine BMMCs to produce MCETs in response to S pyogenes.
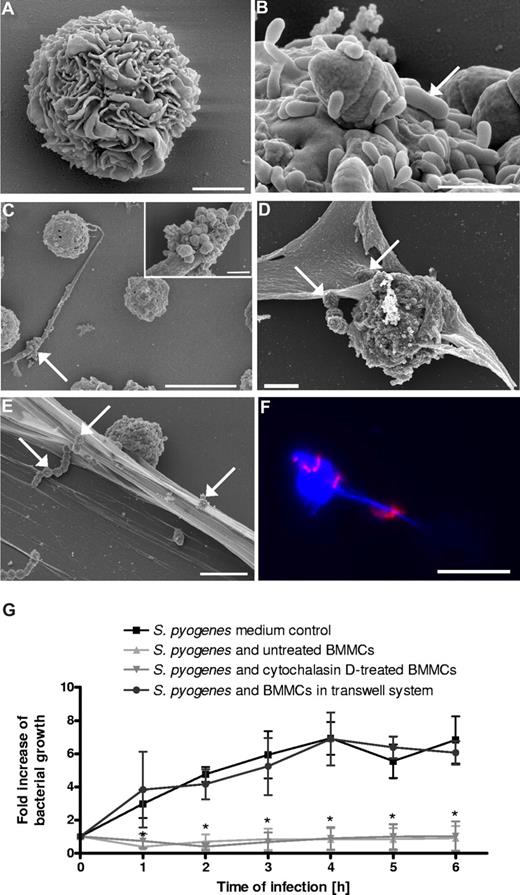
BMMCs were isolated from murine bone marrow and differentiated with recombinant murine IL-3 over 3 weeks in in vitro culture. The cells were determined to be more than 97% BMMCs by immunostaining with anti-CD117 antibodies, as well as toluidine blue staining (data not shown), and by their morphologic appearance using scanning electron microscopy (Figure 3A). BMMCs were infected for 3 hours with S pyogenes, fixed, and processed for either electron or immunofluorescence microscopy. As shown in Figure 3B, S pyogenes was also capable of attaching to the surface of the BMMCs but were not internalized by these cells (data not shown). However, in addition to the cell-surface–attached bacteria, S pyogenes can also be found trapped by extracellular structures produced by the BMMCs similar to those observed in HMC-1 cells (Figure 3C-F). The composition of the MCETs produced by BMMCs was determined by immunofluorescence microscopy. As with the MCETs produced by the human MCs, the MCETs produced by BMMCs were also composed of DNA (Figure 3F; bacteria in red, DNA in blue) and proteins such as tryptase and histones (data not shown). Similar to the human MCs, these BMMCs were able to inhibit bacterial growth in a contact-dependent manner (Figure 3G).
In vitro interactions of S pyogenes with murine BMMCs. (A) FESEM images of uninfected murine BMMCs after 21 days in culture medium supplemented with recombinant mouse IL-3 (bar, 2 μm). (B) S pyogenes (arrow) attached to the surface of BMMCs (bar, 1 μm). (C) A clump of S pyogenes (arrow) trapped by extracellular fibers produced by BMMCs (bar, 10 μm). Insert in top-right corner shows a higher magnification of an entrapped bacterial clump (bar, 2 μm). (D) MCs in the process of producing MCETs. Some streptococci trapped by incipient MCETs are indicated by white arrows (bar, 5 μm). (E) S pyogenes captured in MCETs (white arrows; bar, 5 μm). (F) Immunofluorescence photograph showing blue labeled DNA (BMMC nuclei and extracellular MCET fibers) and associated red-labeled S pyogenes (bar, 10 μm). (G) Growth of S pyogenes in medium alone, in coculture with MCs, in coculture with cytochalasin D–treated MCs, or in coculture with MCs separated by a transwell system. Data are expressed as x-fold increase in bacterial growth with respect to the original inoculum. Each point represents the mean plus or minus SD of 3 independent experiments. *P < .05 by F-test for S pyogenes growth in medium control versus S pyogenes growth in the presence of either untreated or cytochalasin D–treated BMMCs.
In vitro interactions of S pyogenes with murine BMMCs. (A) FESEM images of uninfected murine BMMCs after 21 days in culture medium supplemented with recombinant mouse IL-3 (bar, 2 μm). (B) S pyogenes (arrow) attached to the surface of BMMCs (bar, 1 μm). (C) A clump of S pyogenes (arrow) trapped by extracellular fibers produced by BMMCs (bar, 10 μm). Insert in top-right corner shows a higher magnification of an entrapped bacterial clump (bar, 2 μm). (D) MCs in the process of producing MCETs. Some streptococci trapped by incipient MCETs are indicated by white arrows (bar, 5 μm). (E) S pyogenes captured in MCETs (white arrows; bar, 5 μm). (F) Immunofluorescence photograph showing blue labeled DNA (BMMC nuclei and extracellular MCET fibers) and associated red-labeled S pyogenes (bar, 10 μm). (G) Growth of S pyogenes in medium alone, in coculture with MCs, in coculture with cytochalasin D–treated MCs, or in coculture with MCs separated by a transwell system. Data are expressed as x-fold increase in bacterial growth with respect to the original inoculum. Each point represents the mean plus or minus SD of 3 independent experiments. *P < .05 by F-test for S pyogenes growth in medium control versus S pyogenes growth in the presence of either untreated or cytochalasin D–treated BMMCs.
Extracellular killing of S pyogenes by MCs requires intact MCETs
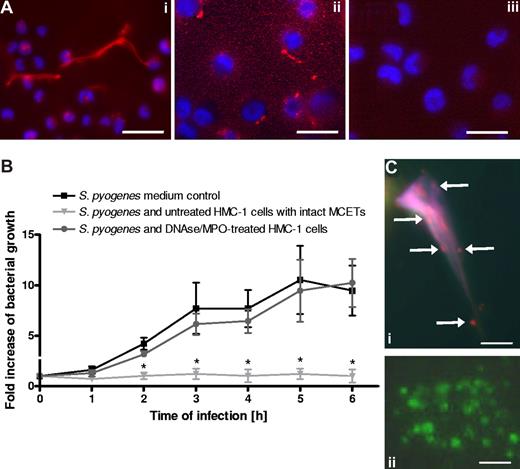
MCETs can be disrupted after treatment with DNase (to target DNA) plus myeloperoxidase (to target tryptase24 ). Shown in Figure 4Ai are the MCETs produced by PMA-stimulated HMC-1 cells labeled red with antihistone antibodies and blue for DNA. Treatment of MCETs with DNase and myeloperoxidase resulted in a complete disruption of these structures (Figure 4Aii). The respective isotype control is shown in Figure 4Aiii. As shown in Figure 4B, disruption of MCETs abolished the antimicrobial effect of HMC-1 cells against S pyogenes.
Intact MCs extracellular traps are required for effective growth inhibition of S pyogenes. (A) Immunostaining with Alexa-red–labeled antibodies against histones of (i) intact MCETs or (ii) disrupted MCETs after treatment with DNAse and myeloperoxidase (bars, 10 μm). (iii) Isotype control antibody (bar, 10 μm). The nucleus of MCs was stained with Dapi (blue). (B) Growth of S pyogenes after coculture with MCs treated with DNAse and MPO to dismantle extracellular trap structures, untreated MCs, or in medium without MCs supplemented with DNAse and MPO. Data are expressed as x-fold increase in bacterial growth with respect to the original inoculum. Each point represents the mean plus or minus SD of 3 independent experiments. *P < .05 by F-test for S pyogenes growth in the presence of untreated HMC-1 cells versus growth of S pyogenes in the presence of DNAse/MPO-treated HMC-1 cells. (C) Analysis of viable (green) versus dead (red) bacteria entrapped in MCETs (i) or grown in medium control (ii) as determined by the LIVE/DEAD BacLight Bacterial Viability assay (bars, 3 μm).
Intact MCs extracellular traps are required for effective growth inhibition of S pyogenes. (A) Immunostaining with Alexa-red–labeled antibodies against histones of (i) intact MCETs or (ii) disrupted MCETs after treatment with DNAse and myeloperoxidase (bars, 10 μm). (iii) Isotype control antibody (bar, 10 μm). The nucleus of MCs was stained with Dapi (blue). (B) Growth of S pyogenes after coculture with MCs treated with DNAse and MPO to dismantle extracellular trap structures, untreated MCs, or in medium without MCs supplemented with DNAse and MPO. Data are expressed as x-fold increase in bacterial growth with respect to the original inoculum. Each point represents the mean plus or minus SD of 3 independent experiments. *P < .05 by F-test for S pyogenes growth in the presence of untreated HMC-1 cells versus growth of S pyogenes in the presence of DNAse/MPO-treated HMC-1 cells. (C) Analysis of viable (green) versus dead (red) bacteria entrapped in MCETs (i) or grown in medium control (ii) as determined by the LIVE/DEAD BacLight Bacterial Viability assay (bars, 3 μm).
These results clearly indicate that intact MCETs structures are required for an effective growth inhibition of S pyogenes by MCs. Determination of bacterial viability by the LIVE/DEAD viability assay shows that most bacteria trapped in the MCETS were nonviable as indicated by the red staining (Figure 4Ci), while mainly viable green bacteria were detected in the medium control (Figure 4Cii) or after disruption of the MCETs (data not shown).
MCETs are formed by MCs undergoing NADPH oxidase–dependent cell death
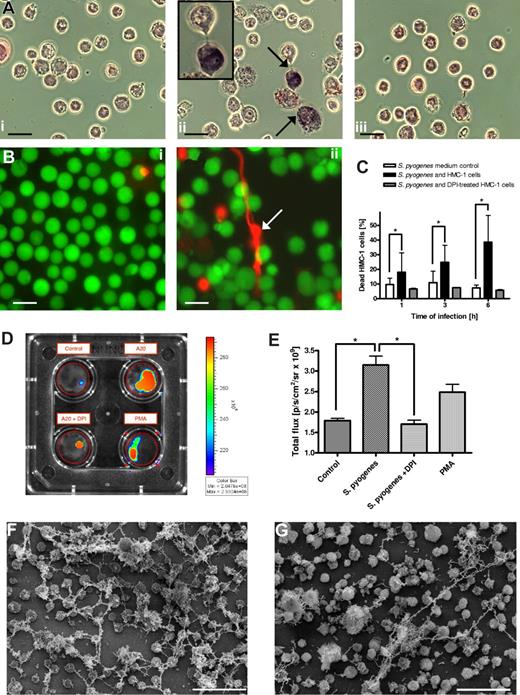
A recent report by Fuchs et al20 has shown that NETs are released by dying neutrophils, and that this process is dependent on the generation of ROS by activated NADPH oxidases. To test whether a similar mechanism underlies the formation of MCETs, we first determined the NADPH oxidase–dependent production of ROS by MCs after incubation with S pyogenes. HMC-1 cells were cocultured with S pyogenes for 15 minutes, and the production of ROS was determined by the NBT reaction. Uninfected HMC-1 cells were used as a control (Figure 5Ai). As shown in Figure 5A, high levels of NBT-reducing ROS activity (dark blue precipitate) were detected in HMC-1 cells, which exhibited an incipient formation of extracellular traps (Figure 5Aii; arrows and insert). That the production of ROS by HMC-1 cells after stimulation with S pyogenes was triggered by the assembly of NADPH oxidase was demonstrated by the lack of NBT stain of S pyogenes–stimulated MCs after adding the NADPH oxidase inhibitor DPI to the culture medium (Figure 5Aiii). Experiments using dihydrorhodamine 123 demonstrated that ROS production was dependent of the cytosolic rather than mitochondrial oxidase activity (data not shown).
Formation of MCETs is dependent of reactive oxygen species (ROS)-induced MCs death. (A) Production of ROS by S pyogenes–infected MCs in the presence or absence of NADPH oxidase inhibitor DPI. Uninfected MCs (i) or S pyogenes–infected MCs cultured in the absence (ii) or presence of DPI (iii) were incubated with NBT for 45 minutes and examined by light microscopy. Precipitation of formazan indicating active production of ROS is indicated by arrows in panel ii (bars, 10 μm). (B) Immunofluorescence photograph showing viable (green) versus dead (red) MCs of uninfected cells (i) or cells after coculture with S pyogenes (ii). Note the release of DNA by dying MCs (arrow) in panel ii. All bars in panel B represent 10 μm. (C) Quantification of MC death in control medium (□), coculture with S pyogenes (■), or coculture with S pyogenes in the presence of DPI (▩). The data are presented as percentage of PI-stained dead cells determined by flow cytometry. Bars represent the means plus or minus SD of 3 independent experiments. *P < .05 by F-test. (D) Quantification of DNA release by MCs after 1 hour of culture in the presence of PMA or S pyogenes and with or without DPI. The amount of DNA was measured as intensity/total flux of red fluorescence (Sytox orange) using the Xenogen Vivo Vision IVIS 200 System with the filter setting of 532 nm excitation/580 nm emission and Igor Pro 4.09A software. A diagram showing the quantitative data are displayed in panel E. (F) FESEM image of MCETs produced by human MCs after 6 hours of either stimulation with 100 mU/mL of glucose oxidase (F) or with S pyogenes (G). Bar, 50 μm.
Formation of MCETs is dependent of reactive oxygen species (ROS)-induced MCs death. (A) Production of ROS by S pyogenes–infected MCs in the presence or absence of NADPH oxidase inhibitor DPI. Uninfected MCs (i) or S pyogenes–infected MCs cultured in the absence (ii) or presence of DPI (iii) were incubated with NBT for 45 minutes and examined by light microscopy. Precipitation of formazan indicating active production of ROS is indicated by arrows in panel ii (bars, 10 μm). (B) Immunofluorescence photograph showing viable (green) versus dead (red) MCs of uninfected cells (i) or cells after coculture with S pyogenes (ii). Note the release of DNA by dying MCs (arrow) in panel ii. All bars in panel B represent 10 μm. (C) Quantification of MC death in control medium (□), coculture with S pyogenes (■), or coculture with S pyogenes in the presence of DPI (▩). The data are presented as percentage of PI-stained dead cells determined by flow cytometry. Bars represent the means plus or minus SD of 3 independent experiments. *P < .05 by F-test. (D) Quantification of DNA release by MCs after 1 hour of culture in the presence of PMA or S pyogenes and with or without DPI. The amount of DNA was measured as intensity/total flux of red fluorescence (Sytox orange) using the Xenogen Vivo Vision IVIS 200 System with the filter setting of 532 nm excitation/580 nm emission and Igor Pro 4.09A software. A diagram showing the quantitative data are displayed in panel E. (F) FESEM image of MCETs produced by human MCs after 6 hours of either stimulation with 100 mU/mL of glucose oxidase (F) or with S pyogenes (G). Bar, 50 μm.
We next determined whether ROS formation by S pyogenes–stimulated HMC-1 cells leads to MC death as a final step for MCET formation. For this purpose, HMC-1 cells were incubated with S pyogenes for 1 hour and then analyzed microscopically for cell viability versus death using the LIVE/DEAD viability/cytotoxicity kit for mammalian cells (Invitrogen). Uninfected HMC-1 cells were used as a control. As shown in the photographs displayed in Figure 5Bii, significantly higher numbers of dead MCs (red staining) were observed on HMC-1 cells cocultured with S pyogenes compared with the uninfected control (Figure 5Bi). Quantification of dead HMC-1 cells cultured in medium alone or with S pyogenes, either in the presence or absence of DPI inhibitor shown in Figure 5C revealed that S pyogenes–induced HMC-1 cell death can be inhibited by the addition of DPI to the culture medium. Also evidenced in Figure 5Bii is that dying HMC-1 cells release DNA in the extracellular milieu for the initiation of the MCETs formation (arrows). The amount of DNA released by infected HMC-1 cells was quantified by adding the DNA dye Sytox orange, which stains only extracellular DNA, to the cultures and measuring the fluorescence intensity using the Xenogen Vivo Vision IVIS 200 System. PMA-stimulated MCs served as a positive control for formation of MCETs. As depicted in Figure 5D (top right), incubation of HMC-1 cells with S pyogenes resulted in a significantly greater DNA release compared with that of the untreated control (Figure 5D; top left) and even higher than that seen by PMA-stimulated HMC-1 cells (Figure 5D bottom right). Again, the NADPH oxidase inhibitor DPI prevented the DNA release by HMC-1 cells upon activation with S pyogenes (Figure 5D bottom left). The quantification data of DNA release are shown in Figure 5E.
Addition of glucose oxidase and the subsequent generation of H2O2 resulted in strong induction of MCETs (Figure 5F) that was superior to the induction of MCETS after exposure of MCs to S pyogenes (Figure 5G). The presence of catalase completely inhibited the formation of MCETs after treatment with glucose oxidase, indicating that the triggering agent was indeed the generated H2O2 (data not shown).
Taken together, these results show that production of MCETs in response to stimulation with S pyogenes is mediated by a ROS-dependent cell death mechanism.
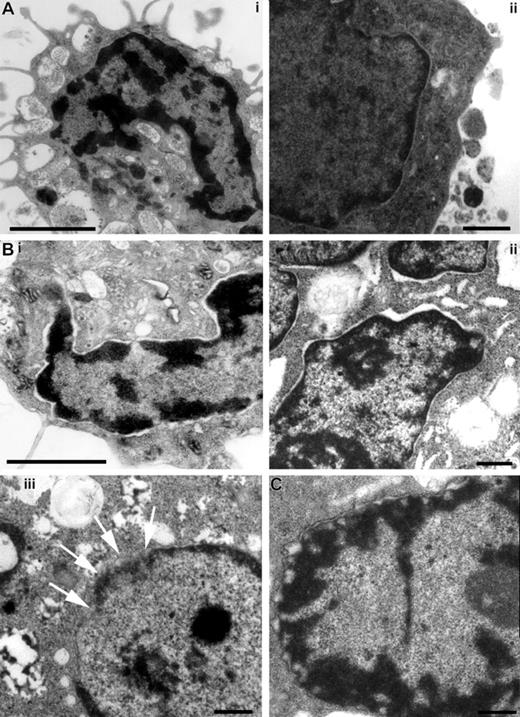
Since the process of MCET formation exhibits a high degree of similarity to the recently described NETs, we determined whether MCs undergo similar morphologic changes leading to the cell death as described for neutrophils.20 Transmission electron photographs displayed in Figure 6 show that the external and internal sheets of the nuclear membrane of MCs stimulated either with S pyogenes (Figure 6B) or H2O2 after adding glucose oxidase (Figure 6C) started to separate (Figure 6Bi,ii), leading to a final disintegration into vesicles surrounding the DNA (Figure 6Biii,C).
Structural examination of MCs stimulated with either S pyogenes or glucose oxidase by transmission electron microscopy. MCs were incubated with medium alone (A), with S pyogenes for 6 hours (B), or with 100 mU/mL of glucose oxidase (C). Unstimulated MCs show intact nuclear membrane (A), whereas the external and internal sheets of the nuclear membrane start to separate in S pyogenes–stimulated (B) and glucose oxidase–stimulated (C) MCs. Desintegration of the nuclear membrane can be seen in panels Biii (arrows) and C. Bars are 2 μm (Ai,Bi) and 0.5 μm (Aii,Bii,iii,C).
Structural examination of MCs stimulated with either S pyogenes or glucose oxidase by transmission electron microscopy. MCs were incubated with medium alone (A), with S pyogenes for 6 hours (B), or with 100 mU/mL of glucose oxidase (C). Unstimulated MCs show intact nuclear membrane (A), whereas the external and internal sheets of the nuclear membrane start to separate in S pyogenes–stimulated (B) and glucose oxidase–stimulated (C) MCs. Desintegration of the nuclear membrane can be seen in panels Biii (arrows) and C. Bars are 2 μm (Ai,Bi) and 0.5 μm (Aii,Bii,iii,C).
Induction of MCET formation by other bacterial pathogens
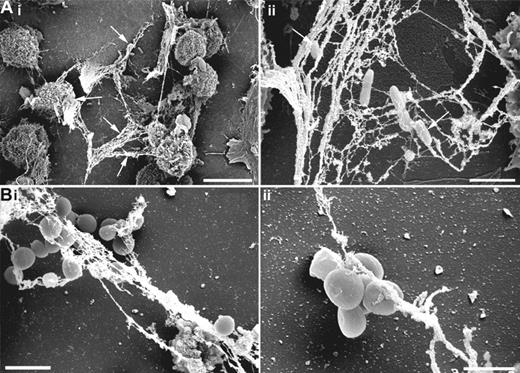
Finally, we determined whether other bacterial pathogens such as the Gram-negative P aeruginosa or the Gram-positive S aureus were able to induce formation of MCETs. Scanning electron microscopy photographs displayed in Figure 7 clearly demonstrated that the induction of MCETs is not an exclusive ability of S pyogenes and that P aeruginosa (Figure 7Ai,ii) and S aureus (Figure 7Bi,ii) are able to induce and subsequently be entrapped by MCETs.
Induction of MCETs by S aureus and P aeruginosa. HMC-1 cells were seeded on poly-L-lysine–coated glass slides, then infected with S aureus or P aeruginosa (MOI 1:25) for 6 hours and fixed with 4% paraformaldehyde and examined by FESEM. (A) P aeruginosa entrapped by MCETs (white arrows; bars are 10 μm for panel Ai and 2 μm for panel Aii). (B) S aureus entrapped by extracellular fibers produced by MCs (bars are 2 μm for panel Bi and 1 μm for panel Bii).
Induction of MCETs by S aureus and P aeruginosa. HMC-1 cells were seeded on poly-L-lysine–coated glass slides, then infected with S aureus or P aeruginosa (MOI 1:25) for 6 hours and fixed with 4% paraformaldehyde and examined by FESEM. (A) P aeruginosa entrapped by MCETs (white arrows; bars are 10 μm for panel Ai and 2 μm for panel Aii). (B) S aureus entrapped by extracellular fibers produced by MCs (bars are 2 μm for panel Bi and 1 μm for panel Bii).
Discussion
The contribution of MCs to many aspects of host defense has become increasingly recognized over recent years (reviewed by Marshall28 and Dawicki and Marshall16 ). Rapidly activated by a variety of mechanisms in response to bacterial infection, MCs can be crucial for the early recruitment of effector cells such as neutrophils through the release of chemoattractant compounds. However, MCs also have the ability to recognize and phagocytose infectious agents through specific receptors present on their surface.16,28 Similar to other immune cells, MCs possess complement receptor 3 (CR3) and FcγR and therefore have the capacity to recognize pathogens that have been opsonized by either complement or IgG.29,30 Nevertheless, MCs also have the innate capacity to recognize pathogens even in the absence of opsonins. This attribute is mediated by the expression of cell-surface receptors for a particular bacterial component, such as toll-like receptors (TLRs) or the mannose receptor (reviewed by Marshall28 ). Indeed, MCs have long been known to possess phagocytic properties; however, since the bacterial phagocytic efficiency of MCs is much less than professional phagocytic cells, the physiologic relevance of this phenomenon is unclear.
We have shown here that MCs can also exert an antimicrobial effect in the absence of phagocytosis. S pyogenes was used to investigate the phagocytosis-independent extracellular antimicrobial mechanism of MCs, since MCs are unable to phagocytose but can still efficiently inhibit the growth of this important human pathogen. That growth inhibition of S pyogenes was not the result of a simple release of antimicrobial peptides by the MCs into the culture supernatant was demonstrated by the requirement of direct contact or close proximity between S pyogenes and MCs for full antimicrobial effect. This was shown by the disappearance of this antimicrobial activity after separation of S pyogenes from MCs in a transwell system that allows the diffusion of released molecules but not direct contact of bacteria with MCs. The most prominent finding of the present study is that the extracellular killing of S pyogenes by MCs was mediated by formation of extracellular structures that strongly resembled the recently described NETs.19 These structures, referred to here as MCETs, are composed of DNA, histones, the MCs-specific protease tryptase,31,32 antimicrobial peptides, and maybe other MC granule components not yet identified. These granule components are critical for MCET formation since the formation of MCETs by MCs was greatly impaired by treatment with compound 48/80 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). However, similar to the NETs, the structure of the MCETs is maintained by DNA. Close examination of the MCETs by fluorescence and electron microscopy corroborated the presence of streptococcal microorganisms entrapped in these structures. Most of the entrapped bacteria were nonviable, indicating that these microorganisms are probably killed within the MCETs by being exposed to a high local concentration of MCET-associated antimicrobial compounds such as LL-3717 or histones33 similar to NETs (reviewed in Brinkmann and Zychlinsky34 ). Bacterial entrapment was required for the antimicrobial activity, since dismantling the MCET structure by treatment with DNase plus myeloperoxidase strongly reduced bacterial growth inhibition.
Furthermore, some microbes such as S pyogenes can form clumps (as shown in Figure 3C), and this has been suggested to be a mechanism to escape phagocytic killing, since the clumps are too big to be phagocytosed.35 The MCETs could support killing of clumps that are too large to be efficiently phagocytosed. Therefore, formation of MCETs represents a novel mechanism by which MCs contribute to host defenses.
Notably, accumulation of MCs has often been observed at sites of infection.36,37 MCs identified by the tryptase-positive staining were observed in soft-tissue biopsies of S pyogenes–infected patients (Figure S2). Interestingly, a diffuse gradient of extracellular tryptase staining was often observed in areas with large numbers of bacteria, which may indicate a massive release of this enzyme and possibly the formation of MCETs at the site of infection (Figure S2B compared with the isotype control Figure S2A). In addition, immunofluorescence staining and confocal microscopical analysis of infected tissue showed colocalization of tryptase with S pyogenes in the vicinity of MCs (Figure S2C-E). These observations indicate that MCs can target extracellular S pyogenes during in vivo infections, possibly by the formation of MCETs.
The rapid degranulation of MCs after exposure to pathogens provides important signals for the initiation of vascular changes, as well as for the mobilization and recruitment of effector cells. On the other hand, degranulation of MCs can also have an enormous impact on the tissue environment since an excessive release of mediators such as potent proteolytic enzymes with tryptase-, chymase-, or carboxypeptidase-like activities, metalloproteases, cytokines, chemokines, or arachidonic acid metabolites may produce an adverse effect in the surrounding tissue and exacerbate the inflammatory response and tissue damage.38 Therefore, in addition to killing microbes, the formation of MCETs may also contribute to minimizing tissue damage by sequestering these harmful compounds at the site of infection and not allowing them to diffuse away in surrounding noninfected tissue.
Regarding the mechanism triggering the MCET formation, the fact that MCs are extremely long-lived cells8 clearly indicates that MCET formation is not the result of passive release of DNA and granule proteins during cellular disintegration, but rather an active and controlled process in response to specific stimulation. Recent work from Fuchs et al20 has implicated the production of reactive oxygen radicals (ROS) and induction of cell death in the production of NETs. Similar to these observations, our results also show that MCETs are formed by dying MCs, and that this process is strongly dependent on the production of ROS since inhibition of ROS by DPI resulted in significant reduction of MC death and DNA release. Although ROS had been previously associated with the induction of neutrophil apoptosis, Fuchs et al20 showed that the process accounting for NET formation is neither typical apoptosis nor necrosis, but rather a new form of ROS-dependent cell death recently termed “NETosis.”39 During this process, disintegration of the nuclear membrane occurs concomitant with cytoplasmic granule dissolution, allowing the NET components to mix in the cytoplasm. Examination of stimulated MCs by electron microscopy confirmed that MCs undergo a similar mechanism of cell death accompanied by a disruption of the nuclear membrane leading to the formation of MCETs.
In summary, the findings of our study provide the first experimental evidence of antimicrobial extracellular trap formation by an immune cell population other than neutrophils.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors wish to thank D. E. Low (Mount Sinai Hospital, Toronto, ON) for providing soft tissue biopsy samples, Joseph H. Butterfield (Mayo Foundation for Medical Education and Research, Rochester, MN) for providing HMC-1 cells, and Ina Schleicher and Claudia Höltje for excellent technical assistance.
This work was supported in part by the Nationales Genomforschungsnetz II (Grant 01GS0404), Impuls und Vernetzungsfond, HGF Präsidentenfonds, Hemholtz Association, Germany, and by grants from the Swedish Research Council, the Swedish Foundation for Strategic Research, and the Söderbergs Foundation.
Authorship
Contribution: M.v.K.-B. designed and performed research, analyzed and interpreted data, and wrote the manuscript; M.R. designed, performed, analyzed, and interpreted electron microscopic investigations, and provided critical reading of the manuscript; A.N.-T. and P.T. performed analysis of human soft-tissue biopsies; K.H. performed research; P.T., A.N.-T., and O.G. provided critical design of the research, interpretation of data, and reading of the manuscript; and E.M. designed research, interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eva Medina, Infection Immunology Research Group, Helmholtz Center for Infection Research, Inhoffenstrasse 7, 38124 Braunschweig, Germany; e-mail: eva.medina@helmholtz-hzi.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal