Dyskerin gene is mutated in patients with X-linked dyskeratosis congenita (X-DC), which results in greatly reduced levels of telomerase activity. A genetic suppressor element (GSE) termed GSE24-2 has been isolated in a screening for cisplatin resistance. GSE24-2–expressing cells presented impaired telomerase inhibition following in vitro exposure to chemotherapies, such as cisplatin, or telomerase inhibitors. The promoter of the telomerase component hTERT was constitutively activated in GSE24-2 cells in a c-myc expression–dependent manner. Deletion analyses and mutagenesis of the human c-myc promoter demonstrated that the target sequence for activation was the nuclease hypersensitive element-III (NHEIII) site located upstream to the P1 region of the promoter. Further, expression of GSE24-2 in cell lines derived from patients with X-DC and in VA13 cells induced increased hTERT RNA and hTR levels and recovery of telomerase activity. Finally, expression of GSE24-2 was able to rescue X-DC fibroblasts from premature senescence. These data demonstrate that this domain of dyskerin plays an important role in telomerase maintenance following cell insults such as cisplatin treatment, and in telomerase-defective cells in patients with X-DC. The expression of this dyskerin fragment has a dominant function in X-DC cells and could provide the basis for a therapeutic approach to this disease.
Introduction
Telomeres consist of TTAGGG repeats,1 which shorten in each cell cycle due to the inability of DNA polymerases to replicate the ends of linear DNA molecules. This telomere erosion is counteracted by telomerase, a protein-RNA molecule that mediates the synthesis of the telomeric repeats from a template encoded by telomere RNA (TR).2,3
Telomere shortening, or loss of secondary structure, causes cells to enter senescence and/or apoptosis. These 2 processes act as biologic checkpoints to prevent uncontrolled cell division and genetic instability.4,5 Defects in telomere length have been implicated in the pathology of several diseases, such as premature aging syndromes and cancer.6 Active telomerase complexes comprise hTERT, hTR, and Dyskerin.7 Telomerase expression is repressed in most postnatal somatic cells but is maintained in the majority of human cancers, thus contributing to the immortal phenotype by ensuring telomere integrity.6 Telomerase activity is regulated, mainly, at hTERT transcription level8 and, in humans, is controlled by a 5′ regulatory region of the hTERT gene, which is required for maximal activity and contains a myc binding site (E-box).8,9 c-MYC transcription factor stimulates hTERT expression, whereas enforced expression of mad1 represses hTERT.10,11 Transcriptional regulation of c-myc involves multiple promoters, the predominant ones being P1 and P2.12 The nuclease hypersensitive element-III (NHEIII) of the c-MYC promoter controls 85% to 90% of c-MYC transcription and was originally identified as a major DNAseI hypersensitivity site.13 NHEIII contains a purine-rich encoding strand capable of establishing an equilibrium between a duplex-helix structure and non–B-forms of DNA.14 The DNA-purine–rich strand can form 2 different intramolecular G-quadruplex structures,15 which, since they function as transcriptional repressor elements, have been targeted for different anticancer therapies.
One of the premature aging syndromes associated with telomere erosion is dyskeratosis congenita (DC). It is a rare, inherited disorder characterized by bone marrow failure and increased susceptibility to cancer,16,–18 the main cause of mortality in these patients. The X-linked form of DC (X-DC) is caused, mainly, by point mutations in the DKC gene.19,20 This gene codes for dyskerin, a 58 kDa protein with a putative pseudouridine synthase domain, a component of the telomerase complex. Dyskerin binds to the H/ACA box present in SnoRNAs (small nucleolar RNAs) and TR in vertebrates.21 SnoRNAs act as a guide for dyskerin-mediated rRNA pseudouridylation. H/ACA RNAs contain 2 hairpins of variable length separated by a single-stranded region that includes a conserved H box, and followed by a short tail containing an ACA trinucleotide.22 Three different proteins (Nap2, Gar1, and NOP10) associate with DKC at H/ACA SnoRNAs.3,22,–24 DKC, Nap2, and NOP10 are tightly associated and are involved in maintaining the stability of the ribonucleoprotein complex, whereas interaction with Gar 1 is labile and transient.25,,–28 Therefore, dyskerin is required for telomerase function and is essential for the nuclear accumulation of hTR7,24,,,,–29 and rRNA pseudouridylation.30 Chromosomal instability in patients with DC increases with age as a consequence of decreased telomerase activity and incorrect maintenance of telomere structure. In X-DC lymphoblasts and fibroblasts, telomerase activity and hTR levels are reduced21 and telomeres are shorter than in nonaffected cells. The telomerase defect can be overcome by forced expression of hTERT and hTR.21,31 Other forms of DC result from mutations or deletions in hTR, resulting in accelerated telomere shortening and premature death. This defect can only be overcome by hTR re-expression.31
In the present study we present data on a genetic suppressor element (GSE) named 24-2, coding for the pseudouridine synthase domain of DKC. Expression of this domain can increase survival against cisplatin and telomerase inhibitors in human cells. GSE24-2 activates c-myc promoter through NHEIII and, consequently, activates the hTERT promoter. There is a concomitant increase in telomerase activity in VA13 human cells, and in cells derived from patients with X-DC. Furthermore, expression of GSE24-2 rescues X-DC cells from premature senescence. This suggests that expression of GSE24-2 could be a new therapeutic approach for telomerase-deficient syndromes such as X-DC.
Methods
All mice were housed under similar humane conditions in accordance with the regulations of the Spanish Government concerning experimental animal research.
Constructs and cell lines
GSE24-2, DKC, DKC5′, motif I, motif II, and the CBF5 region, homologous to GSE24-2, were cloned in pLNCX (BD Biosciences, San Jose, CA). The hTERT promoter construct was a gift from Dr T. Kim (Institute for Molecular Biology and Genetics, Seoul National University, Seoul, Korea)32 ; the hTR promoter construct was a gift from Dr N. Keith (Centre for Oncology and Applied Pharmacology, University of Glasgow, United Kingdom)9 ; those of the c-MYC (px3.2) promoter were from Dr A. Aranda (Instituto de Investigaciones Biomedicas, Madrid, Spain); and the Myc/mad hybrid construct, pECL and pECLc-myc, were from Dr J. León (Departamento de Biologia Molecular, Universidad de Cantabria, Santander, Spain). BABE-TERT plasmid was obtained from K. Collins (Department of Molecular and Cell Biology, University of California at Berkeley, CA).31 PGATEV plasmid33 was obtained from Dr G. Montoya (Centro Nacional de Investigaciones Oncológicas, Madrid, Spain). PGATEV-24-2 was obtained by subcloning the 24-2 fragment into the NdeI/XhoI sites of the pGATEV plasmid. The PCDNA3-9E10-24-2 sequence was obtained by subcloning the 24-4 fragment between the EcoR1/XbaI sites of the vector containing the 9E10-myc–recognized epitope. 293T (American Type Culture Collection, Manassas, VA) and Phoenix cells were maintained in Dulbecco modified Eagle medium (DMEM) 10% fetal bovine serum (FBS). The VA13 cell line was obtained from Dr M. Serrano. X-DC cells were obtained from the Coriell Cell Repository and maintained in RPMI 20% FBS. The X-DC lymphoblast family (DC, DC1, DC2, and DC3) has been clinically described34,35 and the dyskerin gene has the single amino-acid substitution T66A. X-DC GMO1787 dermal fibroblasts coding for a leucine 3–lacking dyskerin protein31 were cultured in DMEM supplemented with 10% fetal calf serum (FCS). Murine embryonic fibroblasts (MEFs) were cultured in DMEM supplemented with 10% FBS and nonessential amino acids (Invitrogen, Carlsbad, CA). For stable transfection, 293T cells were cotransfected with 10 μg/million cells GSE24-2 or pLNCX and 1 μg p-BabePUR plasmid (Clontech, Mountain View, CA) and selected for puromycin resistance 24 hours after transfection. Pam 212 cells were transfected with pECL vector, pECLc-myc, or GSE24-2 and were selected for G418 resistance. For infection of X-DC fibroblasts, Phoenix cells were transfected with the pLNCX-derived vectors using calcium phosphate. The viruses were collected 48 hours after transfection and used to infect X-DC fibroblasts over 48 hours. Stable cells were selected with G-418 (Invitrogen).
GSE library and cisplatin selection
The GSE library was constructed essentially as described by Roninson et al.36
Drugs
Cisplatin, telomerase inhibitor I, and SP60125 were purchased from Calbiochem (San Diego, CA). Tumor necrosis factor α (TNFα) was purchased from Upstate Biotechnology (Charlottesville, VA).
GSE24-2 peptide production and purification
E coli DH5α cells were transformed with pGATEV GSE24-2 and lysates were prepared as described.33 The fusion protein was purified with glutation-sepharose and purity was analyzed by gel electrophoresis. GSE24-2 was obtained by TEV protease digestion according to the manufacturer's recommendations. Typically, over 90% of the fusion protein was cleaved, as determined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The protein was passed twice over a 5 mL Hi-Trap Ni-NTA column to remove the polyhistidine tags, uncleaved protein, tobacco etch virus (TEV) protease, and impurities.
Site-directed mutagenesis
Mutation of single guanines in the NHEIII region of the c-MYC promoter (px3.2) was performed using the Quickchange X-L site-directed mutagenesis kit (Stratagene, Santa Clara, CA) according to the manufacturer's protocol.
Telomeric repeat amplification protocol assay
Telomerase activity was measured using the TRAPeze37 telomerase detection kit (Serologicals, Norcross, GA) according to the manufacturer's recommendations. The telomeric repeat amplification protocol (TRAP) assay was performed following a titration of each extract and normalized for total protein concentration.36 Where indicated, a telomere-specific probe was used for the TRAP assay.38 The in vitro platinum-exposure assays had cisplatin added directly to the TRAP reaction.
Western blotting and antibodies
Cells were lysed as previously described39 and 20 μg of protein analyzed in 10% SDS-PAGE. Antibodies used were: anti-pJNK (V7391; Promega, Madison, WI); anti-JNK1 (C-17; Santa Cruz Biotechnologies, Santa Cruz, CA); anti p-P38 (Cell Signaling Technology, Danvers, MA); 9-E10 (A14; Santa Cruz Biotechnologies); and anti-Flag (Invitrogen, Carlsbad, CA).
cDNA preparation and reverse transcription–polymerase chain reaction
Total cellular RNA was extracted using Trizol (Invitrogen) according to the manufacturer's instructions. In each reaction, 2 μg total RNA was reverse transcribed into cDNA using M-Mlv reverse transcriptase (Promega). For a list of oligonucleotides used, see Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Transfection and analysis of gene expression
293T cells were transiently transfected using the calcium phosphate method as previously described.39 The total amount of DNA was kept constant at 10 μg per million cells. X-DC cells were transiently transfected by electroporation of 3 μg pLNCX-GSE24-2 per million cells. VA13 cells and MEFs were transfected with 16 μg DNA/million cells using lipofectamine plus (Invitrogen), according to the manufacturer's instructions. Protein extracts were prepared and luciferase determined using a commercial kit (Promega). Each assay was performed in triplicate, and repeated in 3 different experiments. Transfection efficiencies were corrected by cotransfection of p-CMV-Renilla.
Electrophoretic mobility gel shift assay
293T cells were transfected with PCDNA3-E1024-2 or an empty vector and stable cells were obtained by G-418 selection. These cells expressed an E10-myc epitope fusion GSE24-2 protein. Nuclear protein extraction40 and binding reactions were performed essentially as described previously.41 Oligonucleotides TEL1, containing the telomeric sequence, and CXext, which hybridized to the TEL1 overhang (a negative control),41 were labeled at the 5′ end with T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) and γ32P ATP (Amersham Bioscences, Orsay, France). Specific competition of GSE24-2 binding was performed with the tag-antibody 9E10. DNA-bound complexes were separated in nondenaturing polyacrylamide gels in 0.5% tris borate EDTA buffer.
Cell viability and PLD assays
Cell growth and viability were determined using a crystal violet staining method, as previously described.39 DKC fibroblasts were pooled after infection and selection for G-418 resistance, and grown on bulk culture with the population doubling (PLD) count initiated at the first split of postselection culture.
Results
GSE24-2 protects telomerase activity against cisplatin and telomerase inhibitors
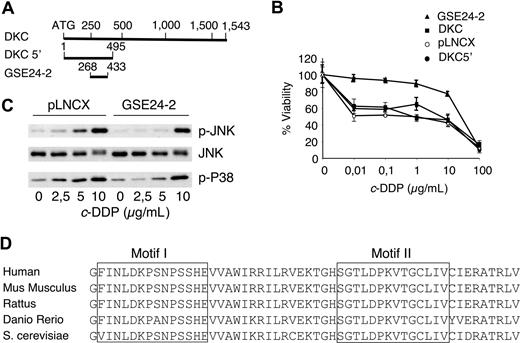
A GSE approach was designed to isolate cDNA sequences conferring survival capacity to cells treated with cisplatin. A cDNA library normalized for low-abundance mRNAs36 was prepared from human placenta, cloned into a retroviral vector, and used to infect Pam212 cells. Cells growing at a cisplatin concentration of 3 μg/mL were selected; 100 resistant clones were isolated and 5 containing a 165-bp fragment of dyskerin were termed GSE24-2. The DNA sequence of GSE24-2 was localized at the N-terminal sequence of dyskerin (Figure 1A). Since the fragment isolated from the library contained 3 different reading frames,36 we subcloned the open reading frame corresponding to dyskerin in the GSE24-2 into the pLNCX vector. In human 293T cells expressing GSE24-2, the 50% inhibitory concentration (IC50) to cisplatin was close to one order of magnitude higher than control cells (Figure 1B). The activity of this plasmid was equivalent to the DNA fragment isolated in the original screening, and it strongly indicated that cisplatin resistance was indeed induced by an internal sequence of dyskerin. Hence, we used this construct for the experiments presented in this report. We had previously shown that JNK/p38 signaling pathway activation is required for apoptosis in cisplatin-treated cells.39 We studied the response in GSE24-2 and control cell lines and found that GSE24-2 cells required higher doses of cisplatin for JNK and p38 activation (Figure 1C). As activation of both kinases is related to DNA damage response,39 the results indicated that GSE24-2–expressing cells were less responsive to DNA damage induced by cisplatin, and that the increased survival against cisplatin correlated with biochemical parameters related to cell death.39 These results are specific for cisplatin since the survival of GSE24-2 cells against Taxol (Sigma-Aldrich, St Louis, MO) was equivalent (data not shown) and, with adriamycin, GSE24-2–expressing cells were more sensitive than control cells (data not shown). This correlated with an increased phosphorylation of p38 in GSE24-2–treated cells (data not shown).
The genetic suppressor element GSE24-2 increases survival against cisplatin and alters the kinetics of JNK and p38 activation. (A) Diagram of the dyskerin cDNA that shows the localization of GSE24-2 and the construct DKC 5′. (B) pLNCX, DKC, DKC 5′, and GSE24-2 cell lines were treated with increasing amounts of cisplatin. Survival was measured 72 hours after treatment. Data represent the means and standard deviations of 2 experiments performed in quadruplicate. (C) Cells were cultured in serum-free medium for 16 hours and then treated with several doses of cisplatin for 6 hours. p-JNK, p-p38, and JNK1 were detected by immunoblot using specific antibodies. The experiments were repeated 3 times, with similar results. (D) Comparison of amino-acid sequences from the pseudouridine synthase domain of GSE24-2 and the corresponding regions from other organisms.
The genetic suppressor element GSE24-2 increases survival against cisplatin and alters the kinetics of JNK and p38 activation. (A) Diagram of the dyskerin cDNA that shows the localization of GSE24-2 and the construct DKC 5′. (B) pLNCX, DKC, DKC 5′, and GSE24-2 cell lines were treated with increasing amounts of cisplatin. Survival was measured 72 hours after treatment. Data represent the means and standard deviations of 2 experiments performed in quadruplicate. (C) Cells were cultured in serum-free medium for 16 hours and then treated with several doses of cisplatin for 6 hours. p-JNK, p-p38, and JNK1 were detected by immunoblot using specific antibodies. The experiments were repeated 3 times, with similar results. (D) Comparison of amino-acid sequences from the pseudouridine synthase domain of GSE24-2 and the corresponding regions from other organisms.
To determine whether cisplatin resistance was due specifically to expression of the 165-bp fragment, we cloned the full-length cDNA for dkc(DKC) as well as the first 495 nucleotides that included the GSE24-2 sequence (DKC 5′), into pLNCX (Figure 1A). Cells expressing GSE24-2 showed less sensitivity to cisplatin than the other cell lines (Figure 1B). Thus, the activity that induced resistance to cisplatin was limited to the GSE24-2 fragment, which includes the pseudouridine synthase domain of DKC,19,,–22 a domain that is highly conserved in all eukaryotes (Figure 1D).
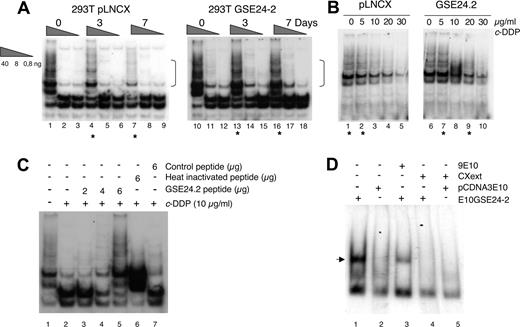
The TTAGGG telomeric sequence is a good target for cisplatin. Intramolecular quadruplexes formed at the telomere consist of 4 consecutive human telomeric DNA repeats.6 These structures are cross-linked by cisplatin. The result is inhibition of telomerase and shortening of the telomeres.42 Therefore, since DKC is one of the components of the telomerase complex,7 we studied the effect of cisplatin and the expression of GSE24-2 on telomerase activity using the TRAP assay.37 Control cells (pLNCX) as well as GSE24-2 cells were pretreated for 3 days with a low cisplatin concentration of 0.5 μg/mL. At this dose, replication takes place without telomere formation.42,43 Cells were then treated with the concentration used for GSE selection (3 μg/mL) for 3 or 7 days. At 7 days (Figure 2A), we observed inhibition of telomerase activity in pLNCX cells, as has been described in the literature.42,43 However, GSE24-2 cells showed higher telomerase activity even after 7 days of treatment (Figure 2A dots). Indeed, the level of activity in control cell extracts (lane 1) matched the level of activity in GSE24-2 cell extracts treated for 3 or 7 days (Figure 2A lanes 13-16). In an in vitro platinum dose-response assay, the level of telomerase activity in GSE24-2 cell extracts, following incubation with 5 μg/mL or 20 μg/mL cisplatin (Figure 2B lanes 7-9), matched the level of activity in control cell extracts (drug-free or with 5 μg/mL of the drug; Figure 2B lanes 1,2). Therefore, cells expressing GSE24-2 required higher doses of cisplatin and longer exposure times to produce an inhibition of telomerase activity. Moreover, since the TRAP probe used in the in vitro platinum exposure assay was telomeric,3,37 the activity of GSE24-2 was also able to protect telomeres from cisplatin crosslinking. Telomerase activity inhibition by cisplatin was also assayed in vitro using cell extracts from 293T cells and a telomeric probe, treated with cisplatin (Figure 2C). In this assay, if GSE24-2 peptide was added, a dose-dependent protection of telomerase activity of cell extracts (Figure 2C lanes 3-5) against cisplatin inhibition was achieved. In contrast, heat-inactivated GSE24-2 peptide or a nonrelated peptide was not able to protect against cisplatin-induced telomerase inhibition (Figure 2C lanes 6,7). This is a further indication that inhibition was, indeed, on telomerase activity and that GSE24-2 peptide was able to inhibit cisplatin-induced telomere crosslinking in vitro. We analyzed the possible binding of GSE24-2 to the telomeric probe using gel mobility shift assays and a short hairpin oligonucleotide (TEL1), which reconstitutes the double-stranded telomere with a short 3′ overhang.41 One specific GSE24-2/TEL1 complex was detected (Figure 2D). CXext, which hybridizes to the single-stranded extension of TEL1, inhibits complex formation. Altogether these results indicate that GSE24-2 was able to bind telomeric sequences; the lack of inhibition of telomerase activity by cisplatin could be due in part to a “protection effect” of GSE24-2 on the telomeric probe.
Effect of expression of GSE24-2 on telomerase activity in response to cisplatin. (A) pLNCX and GSE24-2 cells were treated with 3 μg/mL c-DDP for the indicated time periods. Telomerase activity was detected with the TRAP assay kit. Different extract dilutions are presented for each TRAP assay (total protein amount indicated in triangles). Asterisks indicate the samples that showed important differences and square brackets indicate the important bands. The experiments were repeated 3 times, with similar results. (B) Telomerase activity following in vitro exposure to platinum. GSE24-2 and pLNCX cell extracts were exposed in vitro to platinum with the indicated doses of cisplatin (0-30 μg/mL). Telomerase activity was detected as in panel A, using a telomere-specific template. The experiments were repeated 3 times, with similar results. (C) Telomerase activity following in vitro treatment of 293T extracts with cisplatin and the 24-2 peptide. After in vitro treatment of 293T extracts with cisplatin, different amounts of the 24-2 purified peptide (lanes 3-5), heat-inactivated 24-2 peptide (lane 6), or control peptide (lane 7) were added to the TRAP assay mix. A telomere-specific template was used for the telomerase assay. The experiment was repeated 3 times, with similar results. (D) Band-shift assay of cell extracts expressing GSE24-2. Nuclear cell extracts of cells expressing the plasmid pcDNA3–9E1024–2 or pcDNA3–9E10 were subjected to a band-shift retardation assay. A TEL1 probe containing a telomere-specific sequence was used. Binding specificity was established using 9E10 specific antibody (lane 3) and the CXext oligonucleotide that hybridized to the TEL1 overhang as a competitor (lanes 4-5). The experiment was repeated 3 times, with similar results.
Effect of expression of GSE24-2 on telomerase activity in response to cisplatin. (A) pLNCX and GSE24-2 cells were treated with 3 μg/mL c-DDP for the indicated time periods. Telomerase activity was detected with the TRAP assay kit. Different extract dilutions are presented for each TRAP assay (total protein amount indicated in triangles). Asterisks indicate the samples that showed important differences and square brackets indicate the important bands. The experiments were repeated 3 times, with similar results. (B) Telomerase activity following in vitro exposure to platinum. GSE24-2 and pLNCX cell extracts were exposed in vitro to platinum with the indicated doses of cisplatin (0-30 μg/mL). Telomerase activity was detected as in panel A, using a telomere-specific template. The experiments were repeated 3 times, with similar results. (C) Telomerase activity following in vitro treatment of 293T extracts with cisplatin and the 24-2 peptide. After in vitro treatment of 293T extracts with cisplatin, different amounts of the 24-2 purified peptide (lanes 3-5), heat-inactivated 24-2 peptide (lane 6), or control peptide (lane 7) were added to the TRAP assay mix. A telomere-specific template was used for the telomerase assay. The experiment was repeated 3 times, with similar results. (D) Band-shift assay of cell extracts expressing GSE24-2. Nuclear cell extracts of cells expressing the plasmid pcDNA3–9E1024–2 or pcDNA3–9E10 were subjected to a band-shift retardation assay. A TEL1 probe containing a telomere-specific sequence was used. Binding specificity was established using 9E10 specific antibody (lane 3) and the CXext oligonucleotide that hybridized to the TEL1 overhang as a competitor (lanes 4-5). The experiment was repeated 3 times, with similar results.
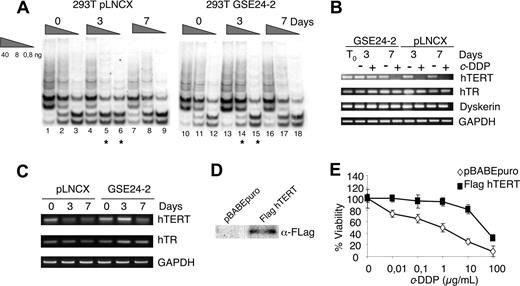
Other compounds that stabilize telomeric G-rich single-strand overhangs and block telomere replication are considered antitumor agents.42,,,–46 The telomerase inhibitor I (TI1; 2,6-diaminoanthraquinone),47 a compound which stabilizes the G-quadruplex structures, was less toxic in GSE24-2 cells than in control cells (data not shown), indicating that the product of GSE24-2 expression would interfere with the capacity of TI1 to stabilize the G-quadruplexes.48 Accordingly, cells expressing GSE24-2 required longer periods of TI1 treatment to inhibit telomerase activity (Figure 3A lanes 5,6 and 14,15).
hTERT expression in response to telomerase inhibitor 1 (TI1) and cell viability of hTERT-expressing cells to cisplatin. (A) Telomerase activity following treatment with TI1. pLNCX and GSE24-2 cells were treated with TI1 (5 μM) for the indicated time periods (0, 3, and 7 days). Telomerase activity was detected with the TRAP assay kit. Different extract dilutions are presented for each TRAP assay (total protein amount indicated in triangles). Asterisks indicate the samples that showed important differences. The experiments were repeated 3 times, with similar results. (B) pLNCX and GSE24-2 cells were untreated (−) or treated (+) with c-DDP (3 μg/mL) for the indicated times (0, 3, and 7 days). Cells were harvested and mRNA levels for hTERT, hTR, and DKC were determined by RT-PCR. GAPDH mRNA was used as control. The experiments were repeated 3 times, with similar results. (C) pLNCX and GSE24-2 cells untreated (0) or treated with TI1 for the indicated times (0, 3, and 7 days) were harvested and mRNA levels for hTERT, hTR, and GAPDH were determined as in panel B. (D) 293T cells were transfected with pBABE or pBABE-flag-hTERT and stable cell lines. hTERT expression was detected by immunoblot using a specific antibody against the FLAG tag. (E) pBABE and pBABEhTERT cell lines were treated with increasing amounts of cisplatin. Survival was measured 72 hours after treatment. Data represent the means and standard deviations of 2 experiments performed in quadruplicate.
hTERT expression in response to telomerase inhibitor 1 (TI1) and cell viability of hTERT-expressing cells to cisplatin. (A) Telomerase activity following treatment with TI1. pLNCX and GSE24-2 cells were treated with TI1 (5 μM) for the indicated time periods (0, 3, and 7 days). Telomerase activity was detected with the TRAP assay kit. Different extract dilutions are presented for each TRAP assay (total protein amount indicated in triangles). Asterisks indicate the samples that showed important differences. The experiments were repeated 3 times, with similar results. (B) pLNCX and GSE24-2 cells were untreated (−) or treated (+) with c-DDP (3 μg/mL) for the indicated times (0, 3, and 7 days). Cells were harvested and mRNA levels for hTERT, hTR, and DKC were determined by RT-PCR. GAPDH mRNA was used as control. The experiments were repeated 3 times, with similar results. (C) pLNCX and GSE24-2 cells untreated (0) or treated with TI1 for the indicated times (0, 3, and 7 days) were harvested and mRNA levels for hTERT, hTR, and GAPDH were determined as in panel B. (D) 293T cells were transfected with pBABE or pBABE-flag-hTERT and stable cell lines. hTERT expression was detected by immunoblot using a specific antibody against the FLAG tag. (E) pBABE and pBABEhTERT cell lines were treated with increasing amounts of cisplatin. Survival was measured 72 hours after treatment. Data represent the means and standard deviations of 2 experiments performed in quadruplicate.
hTERT but not hTR promoter is activated by GSE24-2
Resistance to G-quadruplex ligands has been associated with telomerase overexpression10 in human cells. We investigated whether the resistance to cisplatin and TI1 induced by GSE24-2 could be due to increased expression of some of the telomerase complex components, resulting in higher levels of telomerase activity. The effect of cisplatin on the expression of hTR, hTERT, and dyskerin was determined by semiquantitative PCR. Cisplatin induced a decrease in hTERT mRNA at 3 days of treatment in control cells as previously described.49,–51 However, 7 days of treatment were required for a comparable decrease in GSE24-2 cells (Figure 3B). We did not observe a significant effect of cisplatin on hTR RNA or dkc mRNA levels (Figure 3B). The effect of TI1 treatment on hTERT and hTR RNA levels was also tested (Figure 3C). hTERT mRNA decreases in pLNCX cells after 3 days of treatment whereas in GSE24-2 cells this decrease was observed only after 7 days of treatment. No significant differences were observed in hTR levels (Figure 3C). These results suggest that the differences observed in telomerase activity following cisplatin or TI1 treatment were due, at least in part, to the ability of GSE24-2 cells to maintain higher hTERT RNA levels than control cells. In agreement with this hypothesis, 293T cells that overexpressed hTERT (Figure 3D) were more resistant to cisplatin (Figure 3E) than control cells.
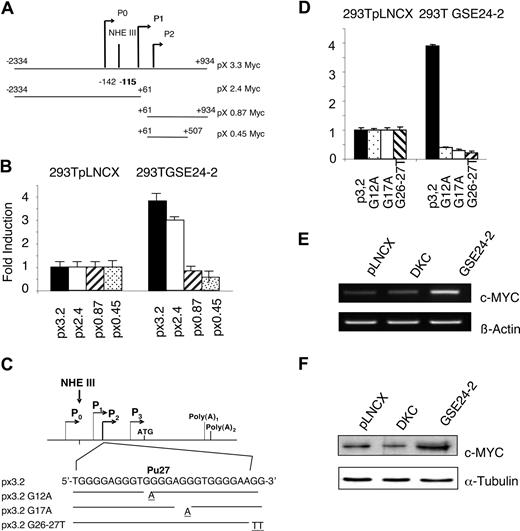
Since cisplatin induces a decrease in hTERT mRNA and the expression of GSE24-2 appears to compensate for this effect, we investigated whether this could be due to changes in hTERT transcription.52 We assessed the activity of the hTERT promoter in GSE24-2 cells using a luciferase reporter construct containing 3402 bp of the promoter.32 Expression of GSE24-2, but not DKC, was able to stimulate hTERT promoter activity in transient transfection assays (Figure 4A). Further, cisplatin stimulated higher promoter activity in GSE24-2 than in control cells (Figure 4B). These results indicate that expression of GSE24-2 was able to stimulate transcription even in the presence of cisplatin, resulting in increased levels of hTERT mRNA (Figure 3B). Further, we were able to detect stimulation of hTERT promoter in MEF cells transfected with the GSE24-2 plasmid (data not shown). We did not observe any effect of GSE24-2 expression on hTR promoter activity9 using either DKC or GSE24-2 cells (Figure 4C). Conversely, treatment with the JNK inhibitor SP60125 was able to activate this promoter (data not shown), as has been previously described.9 These results indicate that the activity of GSE24-2 was specific for hTERT expression.
GSE24-2 increases hTERT promoter activity. (A) 293T cells were cotransfected with pLNCX, DKC, GSE24-2, Cbf5, motif I, or motif II (10 μg DNA per million cells) and the hTERT-luc reporter (1 μg per million cells). Luciferase activity was measured following 24 hours of transfection. Asterisk indicates values with statistical significance of P <.05. (B) 293T (■) and 293T24-2 (□) cells were cotransfected with the hTERT-luc reporter (1 μg per million cells). After 24 hours, the cells were treated with cisplatin (3 μg/mL) for 8 hours and the luciferase activity was determined. (C) 293T cells were cotransfected with the indicated constructs (10 μg DNA per million cells) and the hTR-luc reporter (1 μg per million cells). Luciferase activity was measured following 24 hours of transfection. (D) Cell lines were cotransfected with the hTERT-luc reporter and different amounts of the Mad/myc expression vector. Luciferase activity was measured following 24 hours of transfection. (E) Cell lines were cotransfected with the HIV-luc reporter and increasing amounts of the mad/myc expression vector. Following 24 hours of transfection, cells were stimulated with 50 ng/mL TNFα for 6 hours and luciferase activity was measured. CMV-Renilla (0.1 μg/mL per million cells) was used as a control for transfection efficiency. Data points represent the mean and standard deviations of 2 experiments performed in quadruplicate.
GSE24-2 increases hTERT promoter activity. (A) 293T cells were cotransfected with pLNCX, DKC, GSE24-2, Cbf5, motif I, or motif II (10 μg DNA per million cells) and the hTERT-luc reporter (1 μg per million cells). Luciferase activity was measured following 24 hours of transfection. Asterisk indicates values with statistical significance of P <.05. (B) 293T (■) and 293T24-2 (□) cells were cotransfected with the hTERT-luc reporter (1 μg per million cells). After 24 hours, the cells were treated with cisplatin (3 μg/mL) for 8 hours and the luciferase activity was determined. (C) 293T cells were cotransfected with the indicated constructs (10 μg DNA per million cells) and the hTR-luc reporter (1 μg per million cells). Luciferase activity was measured following 24 hours of transfection. (D) Cell lines were cotransfected with the hTERT-luc reporter and different amounts of the Mad/myc expression vector. Luciferase activity was measured following 24 hours of transfection. (E) Cell lines were cotransfected with the HIV-luc reporter and increasing amounts of the mad/myc expression vector. Following 24 hours of transfection, cells were stimulated with 50 ng/mL TNFα for 6 hours and luciferase activity was measured. CMV-Renilla (0.1 μg/mL per million cells) was used as a control for transfection efficiency. Data points represent the mean and standard deviations of 2 experiments performed in quadruplicate.
Dyskerin was originally identified as the human cbf5 homologue53 showing 85% identity to S cerevisiae Cbf5 gene at the protein level.54 Therefore, we cloned the yeast S cerevisiae cbf5 gene sequence equivalent to GSE24-2 (Figure 1D) into the pLNCX vector. Expression of this construct also increased hTERT(Figure 4A), but not the hTR promoter activity (Figure 4C). This indicates a high degree of functional conservation in this DKC region.
The pseudouridine synthase TRUB domain of DKC comprises 2 structural subdomains, motif I and motif II,53 which exactly match the GSE24-2 encoded region (Figure 1D). These 2 domains are critical for maintaining the global protein structure of DKC. Additionally, motif II contains at least one residue (asp125) essential for enzymatic activity. We assayed the effect of both individual motifs on hTERT promoter activation. Only motif I significantly increased the activity of the hTERT promoter, indicating that it contains the minimal sequence required for maximal activation (Figure 4A).
The NHEIII site at the c-MYC promoter is the target of GSE24-2
The hTERT core promoter contains 2 E-boxes, which can bind myc/max heterodimers.32,55 A hybrid molecule containing the DNA-binding domain of c-myc and the transactivation domain of mad is able to bind E-boxes, thus inhibiting c-myc–dependent transcription. Expression of mad/myc was able to inhibit the activity of the hTERT promoter in a dose-dependent manner in control and DKC cells (Figure 4D). Moreover, the expression of myc/mad was able to abolish transcription activation mediated via the GSE in GSE24-2 cells. This indicated that activation of hTERT transcription induced by GSE24-2 was c-myc dependent (Figure 4D). Inhibition was specific since transfection of the myc/mad construct together with an NFκB-dependent promoter (HIVLuc) did not affect transcription following stimulation with TNFα (Figure 4E).
A reporter vector containing 3.2 kb of c-myc gene (px3.2myc), including the P0, P1, and P2 promoters linked to the luciferase gene (Figure 5A), was transfected in control and GSE24-2 cells. A 3-fold increase in activity was observed in the GSE24-2 cells (Figure 5B). A similar result was obtained in 293T cells transfected with either pLNCX vector or the plasmid expressing GSE24-2 together with the px3.2myc reporter (data not shown). Three deletion mutants of the c-myc promoter were used to define the region necessary for GSE24-2 activation (Figure 5A). Only those regions containing the distal P1 and P0 promoters, including NHEIII, were able to support transcription in GSE24-2, but not in control cells (Figure 5B). This indicated that the proximal P2 promoter was not involved in GSE activation.
c-MYC promoter activity induced by GSE24-2. (A) Schematic representation of the c-MYC promoter indicating the different constructs used in the experiments. (B) 293T pLNCX and 292T GSE24-2 cells were transfected with the different constructs of the c-MYC-luc reporter (1 μg per million cells). Luciferase activity was measured following 24 hours of transfection. CMV-Renilla (0.1 μg/mL per million cells) was used as a control for transfection efficiency. Data points represent the mean and standard deviations of 2 experiments performed in quadruplicate. (C) Schematic representation of the mutations generated in NHEIII. (D) The indicated cell lines were transfected with the mutants of the px3.2 reporter (1 μg per million cells). Luciferase activity was measured following 24 hours of transfection. CMV-Renilla (0.1 μg/mL per million cells) was used as a control for transfection efficiency. Data points represent the mean and standard deviations of 2 experiments performed in quadruplicate. (E) MEFs were transfected with pLNCX-, DKC-, or GSE24-2–expressing vectors. Following 24 hours of transfection, total RNA was isolated and expression of mouse c-myc and β-actin RNA was estimated by RT-PCR. The experiment was repeated 3 times, with similar results. (F) MEFs were transfected with pLNCX-, DKC-, or GSE24-2–expressing vectors. Following 24 hours of transfection, total protein extracts were obtained and expression of mouse c-myc and α-tubulin were detected by immunoblot using specific antibodies. The experiments were repeated 3 times, with similar results.
c-MYC promoter activity induced by GSE24-2. (A) Schematic representation of the c-MYC promoter indicating the different constructs used in the experiments. (B) 293T pLNCX and 292T GSE24-2 cells were transfected with the different constructs of the c-MYC-luc reporter (1 μg per million cells). Luciferase activity was measured following 24 hours of transfection. CMV-Renilla (0.1 μg/mL per million cells) was used as a control for transfection efficiency. Data points represent the mean and standard deviations of 2 experiments performed in quadruplicate. (C) Schematic representation of the mutations generated in NHEIII. (D) The indicated cell lines were transfected with the mutants of the px3.2 reporter (1 μg per million cells). Luciferase activity was measured following 24 hours of transfection. CMV-Renilla (0.1 μg/mL per million cells) was used as a control for transfection efficiency. Data points represent the mean and standard deviations of 2 experiments performed in quadruplicate. (E) MEFs were transfected with pLNCX-, DKC-, or GSE24-2–expressing vectors. Following 24 hours of transfection, total RNA was isolated and expression of mouse c-myc and β-actin RNA was estimated by RT-PCR. The experiment was repeated 3 times, with similar results. (F) MEFs were transfected with pLNCX-, DKC-, or GSE24-2–expressing vectors. Following 24 hours of transfection, total protein extracts were obtained and expression of mouse c-myc and α-tubulin were detected by immunoblot using specific antibodies. The experiments were repeated 3 times, with similar results.
Several NHEIII mutants were obtained by substituting guanine residues at the purine-rich region involved in maintaining the secondary structure of G-quadruplex (Pu27). We mutated G12A at the second G-quartet group, G17A at the second G-triplet and, finally, 2 consecutive guanines (G26A/G27A) at the end of the purine-rich region (Figure 5C). The results indicated that only the wild-type (WT) px3.2 myc promoter region, but none of the mutants of the Pu27 region, is activated by GSE24-2 (Figure 5D). Taken together, these results suggest that GSE24-2 is able to induce a modification of the secondary structure of the Pu27 region into an active conformation enabling c-myc transcription.
We further explored whether these changes in promoter activity resulted in c-myc expression. MEF cells were transiently transfected with the empty vector, DKC, or GSE24-2–expressing vectors, and protein and mRNA levels of c-myc were analyzed (Figure 5E,F). As expected, only expression of GSE24-2 was able to cause increased c-MYC mRNA and protein expression. One of the undesirable effects of GSE24-2 expression could be that an increase in c-MYC expression would induce cell transformation. To evaluate this possibility, nude mice were inoculated with Pam212 cells expressing empty vector, human c-myc, or GSE24-2. Only animals inoculated with c-myc developed tumors during the first 10 days, whereas GSE24-2 cells did not develop tumors even after 3 months (data not shown), indicating that expression of GSE24-2 is not tumorigenic.
Telomerase activity in X-DC and VA13 cells is stimulated by GSE24-2
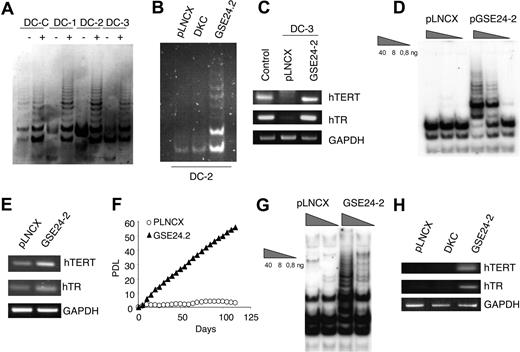
X-DC cells have low levels of telomerase RNA, reduced levels of telomerase activity, and shorter telomeres than normal cells.21 Hence, we evaluated the effect of GSE24-2 expression on telomerase activity of cells derived from patients with X-DC. Commercially available lymphoblasts from a carrier mother (DC, normal dkc allele) and 3 of her clinically affected sons (DC1, DC2, and DC3, mutant dkc allele) were used. These cells are not immortalized and eventually enter senescence in culture. Expression of GSE24-2 increased telomerase activity in all these cell lines (Figure 6A). Interestingly, we did not observe an increase in telomerase activity upon expression of full-length DKC (Figure 6B), as previously reported.21 An increase in hTERT promoter activity was also observed upon GSE24-2 transfection (data not shown). Compared with the healthy control (carrier) mother, hTR levels are low in DC cells expressing a mutant dkc gene21,56 (Figure 6C). RNA levels of hTERT and hTR increased in cells transfected with GSE24-2 (Figure 6C), suggesting that recovery of telomerase activity upon GSE24-2 transfection is due to an increase in both molecules. Primary dermal fibroblasts from patients with X-DC became completely senescent within 3 to 4 weeks of continuous culture, whereas normal cells proliferate up to 2 months before reaching senescence.21 X-DC cells expressing hTERT acquired extended life span and grew continuously for several months.21 Expression of GSE24-2 in GMO1787 X-DC cells was able to induce an increase in telomerase activity (Figure 6D). We also found an increase in hTR and hTERT RNA levels in X-DC fibroblasts expressing GSE24-2 (Figure 6E), and these cells were able to grow continuously (Figure 6F); the results resembled those obtained after hTERT expression.31 Interestingly, in the telomerase-deficient cell line VA13 (wild-type dkc), which lacks hTERT and hTR57 and maintain telomeres by ALT mechanism, expression of GSE24-2 was able to recover telomerase activity (Figure 6G), hTERT RNA, and hTR (Figure 6H).
GSE24-2 reactivates telomerase in X-linked dyskeratosis congenita and VA13 cells. (A) Telomerase activity in X-linked dyskeratosis congenita patient lymphoblast cells obtained from a carrier mother (DC-C) and her affected children (DC-1, DC-2, and DC-3). Cells were transiently transfected with 3 μg of the empty vector (−) or GSE24-2 (+) per million cells. Telomerase activity was measured 24 hours later. (B) DC2 cells were transiently transfected with 3 μg pLNCX-, GSE24-2–, or DKC-expressing vectors, per million cells. Telomerase activity was measured 24 hours later. (C) Expression levels of hTERT and hTR in cells from one of the patients (DC-3) transfected with 3 μg of the empty vector or GSE2-2 per million cells. RNA levels were detected by RT-PCR. GAPDH was used as a control. (D) GMO1787 X-DC patient-derived fibroblasts were infected with pLNCX- or GSE24-2–derived viruses and stable cell lines were used to determine telomerase activity (total protein amount indicated in triangles). (E) Expression levels of hTERT and hTR in GMO1787 transfected with the pLNCX of pGSE24-2 plasmids were determined by RT-PCR. GAPDH was used as a control. (F) Growth rates of the GMO1787-transfected cells described in panel D are compared by cumulative PDLs with time. All the experiments were repeated 3 times, with similar results. (G) Telomerase activity in VA13 cells. Cells were transiently transfected with 16 μg pLNCX control vector or GSE24-2 per million cells. Telomerase activity was measured 24 hours later, using serial dilutions of protein extracts (total protein amount indicated in triangles). (H) Expression levels of hTERT and hTR in VA13 cells transfected with 16 μg pLNCX, DKC, or GSE24-2 plasmid per per million cells. RNA levels were determined by RT-PCR. GAPDH was used as a control.
GSE24-2 reactivates telomerase in X-linked dyskeratosis congenita and VA13 cells. (A) Telomerase activity in X-linked dyskeratosis congenita patient lymphoblast cells obtained from a carrier mother (DC-C) and her affected children (DC-1, DC-2, and DC-3). Cells were transiently transfected with 3 μg of the empty vector (−) or GSE24-2 (+) per million cells. Telomerase activity was measured 24 hours later. (B) DC2 cells were transiently transfected with 3 μg pLNCX-, GSE24-2–, or DKC-expressing vectors, per million cells. Telomerase activity was measured 24 hours later. (C) Expression levels of hTERT and hTR in cells from one of the patients (DC-3) transfected with 3 μg of the empty vector or GSE2-2 per million cells. RNA levels were detected by RT-PCR. GAPDH was used as a control. (D) GMO1787 X-DC patient-derived fibroblasts were infected with pLNCX- or GSE24-2–derived viruses and stable cell lines were used to determine telomerase activity (total protein amount indicated in triangles). (E) Expression levels of hTERT and hTR in GMO1787 transfected with the pLNCX of pGSE24-2 plasmids were determined by RT-PCR. GAPDH was used as a control. (F) Growth rates of the GMO1787-transfected cells described in panel D are compared by cumulative PDLs with time. All the experiments were repeated 3 times, with similar results. (G) Telomerase activity in VA13 cells. Cells were transiently transfected with 16 μg pLNCX control vector or GSE24-2 per million cells. Telomerase activity was measured 24 hours later, using serial dilutions of protein extracts (total protein amount indicated in triangles). (H) Expression levels of hTERT and hTR in VA13 cells transfected with 16 μg pLNCX, DKC, or GSE24-2 plasmid per per million cells. RNA levels were determined by RT-PCR. GAPDH was used as a control.
Taken together, these results indicated that expression of GSE24-2 was able to trigger a recovery of telomerase activity by up-regulating hTERT and hTR RNA levels in both types of cell lines; those expressing a mutant DKC and those with a wild-type DKC allele and low telomerase activity. This increase in telomerase activity resulted in rescue of senescence in X-DC fibroblasts.
Discussion
Controlling telomere length is of considerable relevance in cancer development and aging.6 The findings reported in this study provide the basis for a therapeutic tool that can be used for reactivation of telomerase activity in telomerase-defective syndromes characterized by a faster rate of telomere loss and genomic instability. The therapeutic application can also be for premature aging diseases, inducing a rescue from senescence.
GSEs are useful tools for functional screening of proteins and protein domains.36 We have shown that expression of a dyskerin internal domain is able to induce cisplatin resistance. This is specific for lesions induced by cisplatin since it does not confer resistance to other DNA-damaging agents such as adriamycin. This difference in behavior might be related to the damage that cisplatin induces to the telomeres, given the reactivity of cisplatin with guanines in the DNA. The nonrepaired cisplatin adducts on the telomere repeats will block DNA replication,42 thus inducing apoptosis. A significant shortening of the telomeres has been reported at cisplatin doses used for the in vitro selection of the GSEs.42 Our results indicate that GSE24-2 is able to protect telomeres from cisplatin crosslinks in vitro and up-regulate telomerase activity in vivo. Besides, GSE24-2 cells are also more resistant to TI1 and this would support the concept that GSE could have some effect on G-triplets at telomeres.58 Support for this hypothesis is the down-regulation of telomerase activity observed in response to cisplatin in the cisplatin-sensitive ovarian carcinoma cell line A2780, but not in the cisplatin-resistant cell line A2780CP.59 Conversely, telomerase activities in both cell lines were reduced when cells were treated with adriamycin. This indicates different effects on telomerase activity of the 2 anticancer drugs.59 In addition, GSE24-2 rescues the cisplatin stabilization of the G-quadruplexes formed at the NHEIII site in the c-myc promoter.
Regulation of TERT mRNA expression appears to be the most important limiting step for telomerase activation6 since hTERT is not expressed in cells approaching senescence. GSE24-2 is involved in transcriptional control of hTERT. There is a good correlation between the ability of GSE24-2 to induce cell survival (and to protect telomerase activity against cisplatin) and hTERT promoter activation. This suggests that an increase in telomerase expression would be, at least in part, responsible for increased survival against cisplatin of GSE24-2–expressing cells. Consequently, overexpression of hTERT in 293T cells induced survival against cisplatin. This is also supported by the observation that activation of the TERT promoter in GSE24-2 cells is higher in the presence of cisplatin than under basal conditions (2.5-fold vs 5-fold). hTERT promoter is also activated in cells transfected with the GSE24-2 equivalent fragment from the yeast cbf5 gene,53,54 which implies an evolutionary conserved function of this domain, relevant for hTERT expression.
It is very likely that GSE24-2 activates hTERT transcription in modulating c-myc expression since the myc/mad hybrid molecule totally abolished hTERT transcriptional activation. The GSE24-2 target region appears to be a polypurine sequence (Pu27) also found in other promoters such as CCR5 and PDGFA, and is often associated with unusual DNA structures.60 Mutational analysis suggests that GSE24-2 requires an intact Pu27 to stimulate c-myc transcription. Therefore, alteration of the secondary structure at the G-repeats of the NHEIII site will impair GSE24-2 activity. There are a few known proteins that interact and activate NHE sites. One of them is NM23-H2/NDP kinase B, a sequence-specific DNA-binding protein with affinity for the NHE site at the c-myc promoter. It also activates the expression of other genes such as myeloperoxidase, CD11b, and CCR5, and represses the expression of others such as PDGFA.60,61 Interestingly, this protein can bind single-strand G-enriched sequences in vitro,60,61 including those at the NHE-c-myc promoter and human telomeres. Therefore, a possible mechanism by which GSE24-2 can activate transcription through the NHE is by association with proteins such as NM23H2, which facilitate their interaction with DNA and thereby increase transcription.
Our results obtained in X-DC and VA13 cells indicated that GSE24-2 could increase telomerase activity through a combination of 2 different mechanisms: (1) activating transcription of hTERT, and (2) increasing hTR levels by stabilization of the RNA, in the absence of promoter activation. In addition, GSE24-2 might facilitate the assembly of other proteins such as hNaf1 or Nop10 within the H/ACA box, even in the presence of defective DKC.28 We had not observed any increase in hTR in 293T cells, probably because active telomerase complexes are already present in these cells. Conversely, expression of full-length dyskerin does not result in hTR accumulation in cells from patients with X-DC, as has been described previously,31 probably because the increase in DKC levels is potentially very disruptive for the biogenesis and function of H/ACA motif RNPs.
Independently of the possible mechanism involved, the present results open up the possibility of treatment of X-DC cells with the small GSE24-2 gene fragment, or the encoded peptide. The outcome would be to recover telomerase activity in X-DC cells and maintain hematopoietic renewal, the progressive loss of which is fatal for patients with X-DC. Our results indicate that GSE24-2 expression does not induce tumorigenicity and would not result in aberrant cell growth. Indeed, Wong and Collins31 recently reported that the telomerase defect in patients with X-DC suffering premature cell senescence arise from reduced accumulation of hTR. Our results demonstrate that expression of GSE24-2 is able to increase hTR and hTERT levels in X-DC cells, and recover the capacity to divide. Further, GSE24-2 increased hTERT and hTR expression in VA13 cells. Therefore, the possibility arises that it could be used to rescue telomerase activity in different cell types that have telomerase defects, including cells expressing normal DKC such as VA13, and those from other human syndromes such as autosomal dominant DC or aplastic anemia. Temporary telomerase reactivation could prevent critical telomere shortening and associated pathologies in premature aging syndromes characterized by an increased rate of telomere loss and genomic instability. Telomerase reactivation could be used therapeutically to target those dividing-cell types that maintain organ homeostasis. Telomerase activation therapy could also be used to increase survival of stem-cell populations in patients receiving human stem cell (HSC) transplantation treatment since, even if these cells express telomerase, they are not able to maintain telomere length over a protracted period of time.6
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We gratefully acknowledge M. Blasco and M. Serrano for helpful suggestions; M. Bessler and P. Mason for suggesting some experiments; and N. Keith, A. Aranda, J. León, K. Collins, T. K. Kim, and G. Montoya for providing plasmids used in this work. We also acknowledge J. Perez for the artwork.
This work was supported by grants 04/1164 and 05/1305 from Fondo de Investigación Sauitaire (FIS) and Biologia Fundamental BFU-05-0138. R.M.-P. is supported by Fundación Genoma and grant C03/10 provided by Red Temàtica de Investigación de Centros de Cáncer (RTICC). I.S.-P. and J.-R.M. are Ramón y Cajal program researchers funded by Ministerio de Educación y Ciencia (MEC).
Authorship
Contribution: I.S.-P. and R.M.-P. performed the greater part of the experimental work and contributed equally to the manuscript preparation; J.-R.M. implemented the initial experiments; L.S. provided assistance on specific techniques and experimental design; R.P. was responsible for experimental design and drafting the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rosario Perona, Instituto de Investigaciones Biomédicas, C/Arturo Duperier, 4, 28029 Madrid, Spain; e-mail: rperona@iib.uam.es.
References
Author notes
I.S.-P. and R.M.-P. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal