To the editor:
Virtually all cases of mantle cell lymphoma (MCL) carry the t(11;14)(q13;q32) translocation, leading to the juxtaposition of the CCND1/CYCLIND1 gene to the immunoglobulin heavy chain (IGH) joining region, resulting in cyclin D1 mRNA and protein overexpression.1-3 The existence of “true” MCL negative for cyclin D1 has been controversial but was recently substantiated by gene expression profiling.4 Fu et al reported 6 cases of cyclin D1-negative lymphomas with pathologic and clinical features otherwise typical of MCL, and a molecular signature similar to that of cyclin D1-positive MCL. In these cyclin D1-negative MCL, the tumor cells overexpressed instead either cyclin D2 or cyclin D3, but had no evidence of chromosomal aberration involving the corresponding CCND2 and CCND3 genetic loci. Subsequently, 2 additional cases of cyclin D1-negative/cyclin D2-positive MCL were reported to harbor a t(2;12)(p11;p13) fusing the CCND2 gene to the IGK locus.5
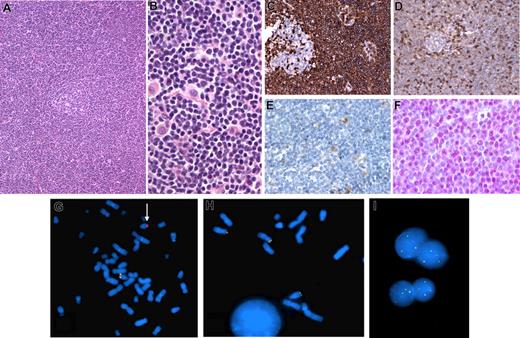
Here, we report a case of cyclin D1-negative MCL with strong nuclear expression of cyclin D2 and a cryptic t(12;14)(p13;q32), juxtaposing the CCND2 gene next to the IGH locus. This lymphoma occurred in a 52-year-old man with stage IV disease involving the bone marrow and gastrointestinal tract. The diagnostic lymph node biopsy showed morphologic and immunopheno-typic features typical of MCL (CD20+, CD5+, CD10−, CD23−, CD43+, BCL-2+, BCL-6−), except for lack of cyclin D1 expression (Figure 1A-E). Conversely, most nuclei were positive for cyclin D2 (Figure 1F) and by competitive real-time–polymerase chain reaction (RT-PCR) cyclin D2 mRNA was overexpressed. Conventional cytogenetic analysis yielded 4 mitoses with a normal karyotype. Interphase FISH performed with LSI IGH/CCND1 Dual Color, Dual Fusion Translocation Probes (Vysis, Downer's Grove, IL) was negative for the t(11;14)(q13;q32), but demonstrated rearrangement of the IGH locus. Q-banded karyotype established from a bone marrow sample was 43,XY,−1,der(8)t(8;8)(p23;q13),−11,−13,add(15)(p11),der(17)tdic(1;17)p22;p12),add(21)(q22),-22, +mar[cp2]/46,XY[17]. FISH performed on bone marrow cells with the LSI IGH Dual Color, Break Apart Rearrangement Probe (Vysis), showed a rearrangement of the 14q32 region (Figure 1G). Because of apparent cyclin D2 overexpression, the tumor cells were investigated for a CCND2 rearrangement, using a break apart FISH assay as previously described.4 A break in the CCND2 locus was clearly demonstrated, with the telomeric probe mapping to 14q32 (Figure 1H). The short arms of both chromosomes 12 were normal, demonstrating a derivative 14 through a cryptic t(12;14)(p13;q32) translocation. Interphase FISH confirmed a CCND2 rearrangement in the lymph node (Figure 1I).
Histologic, immunohistologic, and cytogenetic features in a case of cyclin D1-negative lymphoma with CCND2-IGH fusion. (A,B) Hematoxylin and eosin–stained lymph node biopsy showing a vaguely nodular lymphoid infiltrate around an atrophic residual germinal center (A), composed of small cells with irregular nuclei admixed with scattered histiocytes (B). (C-F) Immunohistochemical findings: strong CD5 expression in the tumor cells around a negative residual germinal center (C); CD43 immunostaining of the lymphoma cells, with a lesser intensity than the reactive T cells (D); lack of cyclin D1 expression in the tumor cells; note that reactive histiocytes exhibit moderate nuclear staining (E); cyclin D2 nuclear expression in the tumor cells detected by a polyclonal anti-cyclin D2 antibody (Cell Signaling) (F). (G-I) FISH results on the bone marrow (G,H) and the lymph node (I) samples. Hybridization of spread metaphases with LSI IGH Dual Color, Break Apart Rearrangement probe (Vysis) shows one chromosome 14 with a normal hybridization pattern (juxtaposed centromeric orange and telomeric green probes) and one chromosome 14 hybridizing only with the centromeric probe, indicating an IGH break (arrow); loss of the telomeric part of the probe precluded identification of the partner chromosome (G). A break apart FISH assay for CCND2 locus rearrangement (telomeric BAC clone: RP11-578L13; centromeric BAC clone: RP11-388F6) shows 2 chromosomes 12 with a normal hybridization pattern and hybridization of the telomeric green probe to 14q, indicating a cryptic t(12;14)(p13:q32) (H). The CCND2 rearrangement is confirmed in the lymph node sample by interphase FISH as the nuclei show 2 yellow (normal chromosomes 12) and one or 2 green (derivative 14) signals with the 2 BAC clones (I). H&E and immunostains images were visualized under a Nikon Eclipse 80i microscope (Nikon, Tokyo, Japan) equipped with Nikon Plan Fluor 10×, 20×/0.50 NA, 40×/ objective lenses and a CFW-1310C camera (Scion, Frederick, MD); images were acquired using Histolab 5.131.1 (Alphelys, Plaisir, France) and processed using Adobe Photoshop v7.0. FISH images were acquired with a 100× immersion objective with an Olympus BX51 fluorescence microscope equipped with the appropriate filter sets, and were documented and processed using the FISH cytovision software.
Histologic, immunohistologic, and cytogenetic features in a case of cyclin D1-negative lymphoma with CCND2-IGH fusion. (A,B) Hematoxylin and eosin–stained lymph node biopsy showing a vaguely nodular lymphoid infiltrate around an atrophic residual germinal center (A), composed of small cells with irregular nuclei admixed with scattered histiocytes (B). (C-F) Immunohistochemical findings: strong CD5 expression in the tumor cells around a negative residual germinal center (C); CD43 immunostaining of the lymphoma cells, with a lesser intensity than the reactive T cells (D); lack of cyclin D1 expression in the tumor cells; note that reactive histiocytes exhibit moderate nuclear staining (E); cyclin D2 nuclear expression in the tumor cells detected by a polyclonal anti-cyclin D2 antibody (Cell Signaling) (F). (G-I) FISH results on the bone marrow (G,H) and the lymph node (I) samples. Hybridization of spread metaphases with LSI IGH Dual Color, Break Apart Rearrangement probe (Vysis) shows one chromosome 14 with a normal hybridization pattern (juxtaposed centromeric orange and telomeric green probes) and one chromosome 14 hybridizing only with the centromeric probe, indicating an IGH break (arrow); loss of the telomeric part of the probe precluded identification of the partner chromosome (G). A break apart FISH assay for CCND2 locus rearrangement (telomeric BAC clone: RP11-578L13; centromeric BAC clone: RP11-388F6) shows 2 chromosomes 12 with a normal hybridization pattern and hybridization of the telomeric green probe to 14q, indicating a cryptic t(12;14)(p13:q32) (H). The CCND2 rearrangement is confirmed in the lymph node sample by interphase FISH as the nuclei show 2 yellow (normal chromosomes 12) and one or 2 green (derivative 14) signals with the 2 BAC clones (I). H&E and immunostains images were visualized under a Nikon Eclipse 80i microscope (Nikon, Tokyo, Japan) equipped with Nikon Plan Fluor 10×, 20×/0.50 NA, 40×/ objective lenses and a CFW-1310C camera (Scion, Frederick, MD); images were acquired using Histolab 5.131.1 (Alphelys, Plaisir, France) and processed using Adobe Photoshop v7.0. FISH images were acquired with a 100× immersion objective with an Olympus BX51 fluorescence microscope equipped with the appropriate filter sets, and were documented and processed using the FISH cytovision software.
This is the first description of a cyclin D1-negative MCL with a t(12;14)(p13;q32) and cyclin D2 overexpression. Of the 4 cyclin D1-negative/cyclin D2-positive MCL previously reported, 2 were found to harbor a genetic alteration of the CCND2 gene, due to a translocation to the IGK locus.4,5 Our findings in the current case confirm that CCND2 is recurrently targeted by chromosomal rearrangements in cyclin D1-negative MCL, and identify IGH as a previously undescribed translocation partner. By analogy to other translocations involved in B-cell lymphomas, one would have expected to find this translocation more often. Interestingly, the t(12;14)(p13;q32) translocation has, to date, not been described. This is likely due to the rarity of true cyclin D1-negative MCL with CCND2 alterations but also to the cryptic nature of this rearrangement. Systematic FISH investigation of suspected cyclin D1-negative MCL overexpressing cyclin D2 without obvious 12p13 and/or 14q32 rearrangements might lead to the identification of additional cases harboring this hitherto unrecognized translocation.
Authorship
This work was supported by the Belgian National Fund for Scientific Research (FNRS). Laurence de Leval and Bettina Bisig are senior research associate and scientific research worker of the FNRS, respectively. Leticia Quintanilla-Martinez is supported by the Mantle Cell Consortium of the Leukemia Research Foundation.
Contribution: C.H. and L.dL. contributed and analyzed data and wrote the paper. F.L., L.Q.-M., B.B., and C.D. contributed and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laurence de Leval, Department of Pathology, CHU Sart-Tilman, Tour de Pathologie, +1, 4000 Liège, Belgium; e-mail: l.deleval@ulg.ac.be.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal