Abstract
Centrosome amplification is common in myeloma and may be involved in disease pathogenesis. We have previously derived a gene expression–based centrosome index (CI) that correlated with centrosome amplification and was an independent prognostic factor in a small cohort of heterogeneously treated patients. In this study, we validated the prognostic significance of the CI in 2 large cohorts of patients entered into clinical trials and showed that a high CI is a powerful independent prognostic factor in both newly diagnosed and relapsed patients, whether treated by intensive therapy (total therapy II) or novel agents (bortezomib). Tumors with high CI overexpressed genes coding for proteins involved in cell cycle, proliferation, DNA damage, and G2-M checkpoints, and associated with the centrosome and kinetochore/ microtubules. In particular, aurora kinases are significantly overexpressed in patients with high CI, with concordant increase in protein expression. Human myeloma cell lines with higher CI are more responsive to treatment with a novel aurora kinase inhibitor. Aurora kinase may represent novel therapeutic targets in these patients with very poor prognosis.
Introduction
Centrosomes are cellular microtubule-organizing centers whose normal function is crucial for chromosome segregation and cytokinesis during mitosis.1 Centrosome amplification has been detected in a broad range of solid tumors2-7 and more recently in leukemia8,9 and lymphoma.10-12 Previously, we and others have shown that centrosome amplification is common in multiple myeloma (MM) and is already present in monoclonal gammopathy of undetermined significance (MGUS), suggesting an early role in myelomagenesis.13,14 In addition, we derived a gene expression–based centrosome index (CI) that correlated closely with centrosome amplification detected by immunofluorescence. We also showed in a heterogeneously treated and relatively small cohort of patients that the CI is an independent prognostic factor.13
In the current study, we wanted to validate the prognostic importance of the CI in large cohorts of homogenously treated patients entered into clinical trials and study the differences in the gene expression profiles (GEP) of patients with high and low CI in order to understand the underlying biological difference and possibly identify therapeutic targets.
Methods
Patients
Group 1: University of Arkansas (UAMS) patients treated with total therapy II.
These are newly diagnosed patients entered into total therapy II studies (n = 351). These patients have GEP performed on CD138+ selected plasma cells (PCs) from pretreatment bone marrows (BMs) using the Affymetrix U133plus 2.0 chip (Affymetrix, Santa Clara, CA). The GEP data of these patients have been published.15 (These data are available from the National Institutes of Health Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/, under accession #GSE2658.) These patients were assigned Translocations and Cyclin (TC) class, a gene expression–based proliferation index (PI), and a recently published gene expression–based molecular classification using published methods.16,17
Group 2: Patients entered into bortezomib trials.
These are patients entered into a phase III study comparing bortezomib with dexamethasone in relapse myeloma patients who had informative gene expression data procured as part of a pharmacogenomics study (kindly provided by Millennium Pharmaceuticals18 ; n = 188). GEP was performed on PCs purified using a negative selection cocktail from BMs at trial entry using the Affymetrix U133A and U133B chip (Affymetrix).
Group 3: Mayo Clinic cohort and human myeloma cell lines (HMCLs).
The expression of STK6 and AURKB during different stages of disease from normal plasma cells (NPCs) to HMCL was assessed in 15 healthy donors, 22 with MGUS, 24 with smoldering multiple myeloma (SMM), and 101 MM (Mayo Clinic cohort) and 42 with HMCL. GEP of the Mayo Clinic cohort was performed on CD138+ selected bone marrow PCs using the Affymetrix U133A chip (GEP data are available from GEO, accession #GSE6477). The relevant biological and clinical information related to the newly diagnosed MM patients in this dataset is appended in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Samples were collected after informed consent was obtained in accordance with the Declaration of Helsinki, and the study was approved by the Mayo Clinic Institutional Review Board. GEP for the HMCL patient was performed on the U133plus 2.0 chips.
Derivation of CI and gene-expression analysis
Gene-expression intensity values were generated using the Affymetrix MAS 5.0 software, log transformed, normalized to the median, and analyzed using GeneSpring GX 7.3.1 (Agilent Technologies, Palo Alto, CA). The derivation of the CI has been reported previously.13 Briefly, it is calculated by adding the normalized expression of the following genes: CETN2, TUBG1, and the median expression of PCNT1 and PCNT2, which encodes the major components of the centrosome: centrin, gamma-tubulin, and pericentrin. We use a cut-off CI of 4.5 to differentiate patients with high and low CI. This cut-off has been previously established and corresponds to mean + 2 SD of CI of normal PCs and identify patients with centrosome amplification by immunofluorescence and poor prognosis.13
Differentially expressed genes between patients with high and low CI were identified by a 2-step procedure. First, Student t test with Benjamini and Hochberg multiple testing corrections was applied. Genes passing false discovery rate of 1% were then further filtered by fold change such that only genes with 2-fold or greater difference in expression between tumors with high and low CI were selected.
Gene set enrichment analysis (GSEA)
Gene set enrichment analysis (GSEA) has been described elsewhere.19 Briefly, the method requires 2 inputs: (1) a list of genes (L) ranked based on the correlation between their expression and the class distinction by using a suitable metric and (2) a priori defined gene sets (S) (eg, pathways or promoter motif sequences extracted from published experimental data or curated databases). The goal of GSEA is to determine whether the members of S are randomly distributed throughout L or primarily found at the top or bottom, in which case the gene set is correlated with the phenotypic class distinction (in our case, high or low CI). The ranking metric used was Signal2Noise, and the phenotype was permutated with 1000 permutations to estimate the statistical significance and false discovery rate (FDR) of enrichment.
Immunohistochemistry for aurora kinase-A and -B
Immunohistochemistry for aurora-A and -B was performed on paraffin sections of BM biopsy specimens. Mouse monoclonal antibodies were used for both aurora-A (clone 4, cat. #610939, BD Transduction Labs, San Jose, CA) and aurora-B (clone 6, cat. #611083, BD Transduction Labs). Briefly, the paraffin sections were pretreated at 100°C for 30 minutes with 1 mM Tris-EDTA pH 8.0 buffer. An automated immunostaining platform was used (Autostainer; Dako, Carpinteria, CA). Endogenous peroxidase was blocked for 5 minutes using Dako peroxidase block (Dako). Primary antibody (aurora-A or -B) was applied for 30 minutes in background reducing diluent (Dako). Dako ADVANCE detection chemistry (Dako) was used according to the manufacturer's instructions to visualize the primary antibody. The sections were counterstained with hematoxylin for 1 minute, cover slipped, and examined using a bright-field microscope.
For each tumor assessed, a section stained with CD138 antibody (Dako Cytomation) was first inspected, and areas of high plasma cell involvement were chosen for scoring. Two high-powered field containing more than 100 cells were scored by 2 hematopathologists. Plasma cells with cytoplasmic staining for aurora-A (consistent with its association with the centrosome, which has a cytoplasmic location) or nuclear staining for aurora-B (consistent with its known cellular localization and association with the kinetochore)20 were scored as positive.
Aurora kinase inhibition in HMCLs
Growth inhibition (IC50) values from 12 HMCLs (H929, JJN3, KMS11, KMS12, KMS18, MM1S, OCIMY5, OPM2, SKMM2, UTMC2, U226, and 8226) were determined by methyl-thiazol tetrazolium (MTT) assay. A total of 2 × 104 cells/well in RPMI 1640 media (supplemented with fetal bovine serum 1%, l-glutamine, and antibiotics) were grown in 96-well plates. MLN8054, a novel aurora kinase inhibitor,21 diluted in RPMI 1640 media, was added in concentrations ranging from 0.125 μM to 100 μM. Additionally, untreated cells of each HMCL were used as controls. All the experiments were performed in triplicate. After 72 hours' growing at 37°C, 10 μL of MTT reagent (3-(4, 5- dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide) was added. The plates were incubated for 2 to 4 hours until purple precipitate was visible. Hybridization buffer was added and incubated overnight in the dark. Finally, the absorbance was recorded at 570 nm and 690 nm. The measurements were determined from the average of the triplicate readings, and a relative value was calculated, considering the control readings as 1.
Statistics
Student t test or ANOVA was used for comparison of continuous variables. The distribution for overall survival (OS) and progression-free survival (PFS) was estimated using the method of Kaplan and Meier. The log-rank test was used to test for differences in survival between groups. OS was calculated from trial entry to death for both groups of patients. PFS for group 2 patients was calculated from trial entry to disease progression. Cox proportional hazards models were used to assess the association of several prognostic factors with survival. A P value of less than .05 is considered significant.
Results
The CI is a powerful prognostic factor in MM
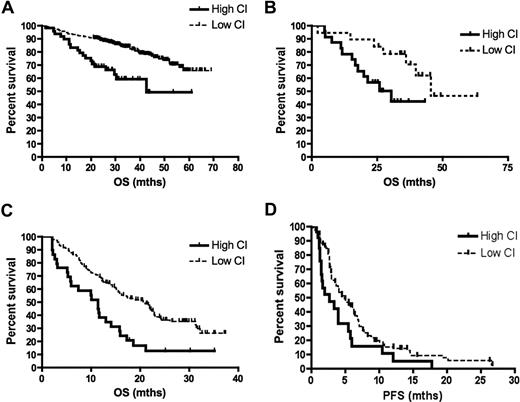
Forty-nine (14%) of 351 patients in the UAMS dataset and 29 (15%) of 188 patients in the bortezomib dataset had high CI. Patients with high CI have significantly shorter OS in both datasets (Figure 1), validating our previous observations. In the UAMS dataset, a PI greater than 2, t(4;14), and CSK1B expression above median were all also strong prognostic factors (Supplementary Figure 1). In a Cox proportional hazard analysis including these factors, a high CI (hazard ratio 1.95 [95% CI 1.07 to 3.46], P value .03) and t(4;14) (hazard ratio 1.92 [95% CI 1.13 to 3.12], P value .02) independently predicted poor survival, whereas the PI and CKS1B gene expression level did not (Table 1). Furthermore, among the patients with high PI in the UAMS dataset, those with high CI had significantly shorter OS (30.6 vs 45.6 months, log-rank P = .04), whereas survival of patients with high CI was similar regardless of PI (log-rank P = .06; Figure 1B).
Survival according to CI. (A) Patients treated with total therapy II who had a high CI had significantly shorter OS than those with low CI (median survival, 42.7 months vs not yet reached, log-rank P < .0001). (B) The OS of patients with PI greater than 2 from the UAMS dataset is further dichotomized by CI levels (median OS of high CI vs low CI is 30.6 months vs 45.6 months, log-rank P = .04). Patients entered into bortezomib trials with a high CI had significantly shorter (C) OS (median survival, 11.5 months vs 20.9 months, log-rank P = .0002) and (D) PFS (median, PFS 2.8 months vs 4.9 months, log-rank P = .02) than those with low CI.
Survival according to CI. (A) Patients treated with total therapy II who had a high CI had significantly shorter OS than those with low CI (median survival, 42.7 months vs not yet reached, log-rank P < .0001). (B) The OS of patients with PI greater than 2 from the UAMS dataset is further dichotomized by CI levels (median OS of high CI vs low CI is 30.6 months vs 45.6 months, log-rank P = .04). Patients entered into bortezomib trials with a high CI had significantly shorter (C) OS (median survival, 11.5 months vs 20.9 months, log-rank P = .0002) and (D) PFS (median, PFS 2.8 months vs 4.9 months, log-rank P = .02) than those with low CI.
Cox proportional hazard analysis of available prognostic in the UAMS dataset (group 1, N = 351)
| Parameter . | Risk ratio . | 95% CI . | Chi-square . | P . |
|---|---|---|---|---|
| t(4;14) | 1.92 | 1.13-3.12 | 5.7 | .017 |
| CI>4 | 1.95 | 1.07-3.46 | 4.66 | .039 |
| CKS1B>1 | 1.19 | 0.70-1.97 | 0.41 | .52 |
| PI<2 | 0.73 | 0.3-1.00 | 3.97 | .05 |
| Parameter . | Risk ratio . | 95% CI . | Chi-square . | P . |
|---|---|---|---|---|
| t(4;14) | 1.92 | 1.13-3.12 | 5.7 | .017 |
| CI>4 | 1.95 | 1.07-3.46 | 4.66 | .039 |
| CKS1B>1 | 1.19 | 0.70-1.97 | 0.41 | .52 |
| PI<2 | 0.73 | 0.3-1.00 | 3.97 | .05 |
In the bortezomib dataset, patients with high CI had significantly shorter OS independent of response to therapy, which was a significant determinant of survival (Figure 1C, Figure S2).22,23 When considering only bortezomib-treated patients, there was no difference in response rate between patients with high or low CI (58% vs 53%, Fisher exact P = .76), yet patients with high CI had a significantly shorter PFS (Figure 1D), suggesting a short duration of response and aggressive disease course upon progression.
Relation between the CI, other high-risk features, and genomic instability
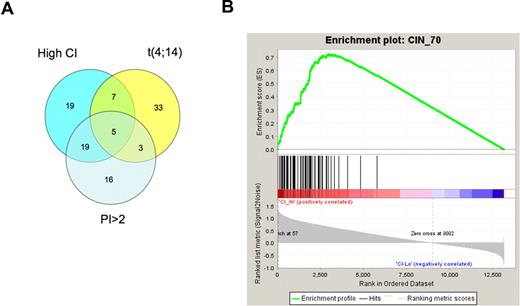
In our previous paper, we found a high CI to be associated with poor risk factors such as higher PCLI and t(4;14) genetic subtypes. Using the much larger UAMS dataset, we examined the relation of high CI, high PI (using gene expression-based PI), and t(4;14) (using TC classification) in more detail. Although there is an association between these factors, the overlap is only partial. Among the 102 patients with any of these features, only 5 had all these features. Twenty-nine patients had any 2 of the features, while 68 had only 1 of these features (Figure 2A). Similarly, when analyzed according to the recently proposed molecular classification from UAMS, among patients with high CI, 37% belonged to the PR (proliferation) group, followed by 16% belonging to the MS (MMSET, which are patients with t(4;14)), whereas it is relatively rare in HY (hyperdiploid) patients (Table S2).
Association of a high CI with other high-risk factors and genomic instability. (A) A Venn diagram showing overlap between tumors with high CI, high PI, and t(4;14). The overlap is less than 50% for 2 of these features, and only 5 (4%) patients have all 3 features. (B) An enrichment plot showing clustering of genes constituting a chromosome instability signature among the top-ranking genes overexpressed in tumors with high CI.
Association of a high CI with other high-risk factors and genomic instability. (A) A Venn diagram showing overlap between tumors with high CI, high PI, and t(4;14). The overlap is less than 50% for 2 of these features, and only 5 (4%) patients have all 3 features. (B) An enrichment plot showing clustering of genes constituting a chromosome instability signature among the top-ranking genes overexpressed in tumors with high CI.
Recently, the UAMS groups have identified a powerful GEP-based multigene prognostic classifier.24 High-risk patients identified by high CI or this GEP-based classifier overlap partially. Twenty-eight patients had high CI and high-risk GEP index, 21 patients had high CI only, 33 patients had high-risk GEP index only, and 269 patients had neither. When comparing the survival of these patients classified according to the CI and the GEP-based classifier, there was no statistical difference in survival between patients with one or both prognostic index elevated, but these patients had significantly shorter survival than those with neither index elevated (Figure S3).
While there is no direct means of measuring aneuploidy and genomic instability in these datasets, we found using GSEA that genes overexpressed in tumors with high CI are enriched (normalized enrichment score 1.67, q-value 0.03; Figure 2B) for genes forming a recently published chromosome instability signature,25 which was found to be strongly correlated with aneuploidy across a range of malignancies.
Molecular consequences of a high CI
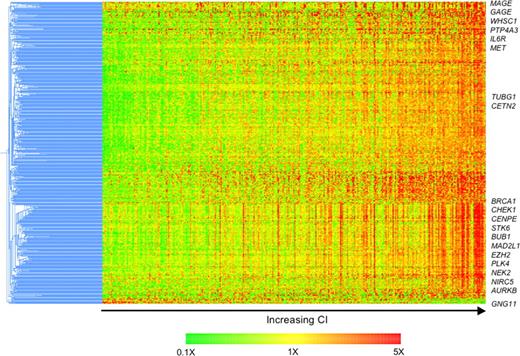
Next, we delineated the molecular phenotype of these patients with a view to identifying potential novel therapeutic targets. Six hundred sixty-two probe sets representing 434 known genes were differentially expressed (FDR 1% and greater than 2-fold difference in expression). All but 3 of these genes were overexpressed in tumors with high CI, compared with those with low CI (Table S3). The expression of these genes increases with increasing CI (Figure 3).
Differences in molecular profiles of tumors with high and low CI identify possible therapeutic targets. Four hundred thirty-four differentially expressed genes were clustered using the hierarchical agglomerative algorithm. Pearson correlation coefficient and centroid linkage were used as similarity and linkage methods, respectively. The samples are ordered from left to right by increasing CI. The expression of almost all these differentially expressed genes increases with increasing CI. Genes of interest are highlighted.
Differences in molecular profiles of tumors with high and low CI identify possible therapeutic targets. Four hundred thirty-four differentially expressed genes were clustered using the hierarchical agglomerative algorithm. Pearson correlation coefficient and centroid linkage were used as similarity and linkage methods, respectively. The samples are ordered from left to right by increasing CI. The expression of almost all these differentially expressed genes increases with increasing CI. Genes of interest are highlighted.
Overexpressed are genes coding for proteins associated with the centrosome (TUBG1, CETN2, TACC3, NEK2, PRKRA, STK6, AURKB, and PLK4), cell cycle (CCNB1, CCNB2, CCND2, E2F2, CDC gene family, CDK5, CDK6, CDKN2C), proliferation (RAN, CKS1B, TOP2A, TTK, TYMS, MCM gene family, ASPM), involvement in DNA repair/G2 cell cycle checkpoints (BRCA1, CHEK1, CHEK2, MAD2L1, BUB1, BUB1B, FANCD2, REV1L), kinetochore and microtubule attachment (AURKB, BIRC5, CENPA, CENPE, CENPH, ZWINT), cancer testis antigens (GAGE and MAGE family), and implicated in other cancer (PTP4A3, EZH2). Consistent with this, the most significantly associated gene ontology biological processes are cell cycle, mitosis, proliferation, DNA repair, and microtubule related (Table S4). These differentially expressed genes overlap to a large degree with genes found to be associated with centrosome aberrations in acute leukemia by GEP.9
Aurora kinases are potential therapeutic targets in patients with high CI
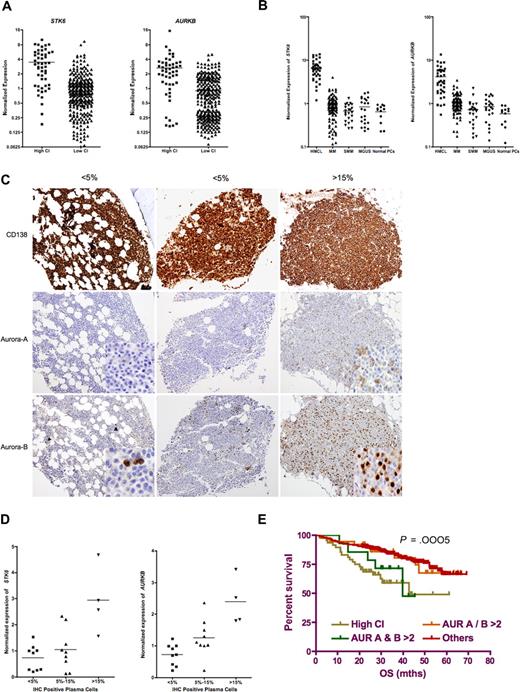
Genes coding for several proteins previously implicated in mediating centrosome amplification (STK6, AURKB, PLK4, NEK2, CDK2, and HMMR coding for aurora-A and -B, polo-like kinase, NEK2, CDK2, and RHAMM, respectively) are overexpressed.26,27 In particular, STK6 (aurora-A; from here on the gene will be referred as STK6 and the protein aurora-A) and AURKB (aurora-B; from here on the gene will be referred as AURKB and the protein aurora-B) are significantly overexpressed in high CI tumors (Figure 4A).
Aurora-A and -B are overexpressed in patients with high CI and are suitable therapeutic targets for these high-risk patients. (A) Among the differentially expressed genes are some candidate genes previously implicated in mediating centrosome amplification. Of these, the expression of both STK6 and AURKB, which code for aurora-A and aurora-B, respectively, was significantly higher in patients with high CI (3.46 ± 2.49 vs 1.07 ± 1.05, t test P < .0001, and 2.60 ± 2.43 vs 0.9 ± 0.85, t test P < .001, respectively) and may represent potentially novel therapeutic targets in these poor-prognosis patients. (B) The expression of aurora-A and -B was significantly higher in HMCLs, and a significant number of tumors (MGUS, SMM, and MM) have expression above normal plasma cells (normalized gene expression greater than 1). (C) Expression of aurora-A and -B was detected by IHC. Staining for aurora-A and -B is cytoplasmic and nuclear, respectively. The staining is generally stronger for aurora-B than -A. For aurora-B staining in the left panel, the positive cells are indicated by close arrows. For both aurora-A and -B staining in the left and right panels, image inserts show area of staining at higher magnification. Images are at 10 × magnification except for image inserts that are at 40×. The images are captured on the Olympus BX51 microscope using the Olympus DP71 CCD camera and MicroSuite 5 software (Olympus Imaging America). Adobe Photoshop CS version 8 is then used to resize the images. (D) Protein expression of aurora-A and -B correlated with gene expression. When the cases with different percentage of positive-staining cells are grouped together, there is a corresponding increase in gene expression in the patients with higher percentage of positive cells (P = .006 for aurora-A and P = .002 for aurora-B), suggesting correlation between gene and protein expression. (E) Survival of patients with high CI (regardless of STK6 or AURB expression, although 80% of patients with high CI have high STK6 and/or AURKB expression) and those overexpressing both STK6 and AURKB is significantly shorter (n = 49 and n = 14, median OS, 42.7 and 39.8 months, respectively, log-rank P = .0005) than patients overexpressing just one of these genes (n = 33, median OS not reached) or those not overexpressing either gene (n = 255, median OS not reached).
Aurora-A and -B are overexpressed in patients with high CI and are suitable therapeutic targets for these high-risk patients. (A) Among the differentially expressed genes are some candidate genes previously implicated in mediating centrosome amplification. Of these, the expression of both STK6 and AURKB, which code for aurora-A and aurora-B, respectively, was significantly higher in patients with high CI (3.46 ± 2.49 vs 1.07 ± 1.05, t test P < .0001, and 2.60 ± 2.43 vs 0.9 ± 0.85, t test P < .001, respectively) and may represent potentially novel therapeutic targets in these poor-prognosis patients. (B) The expression of aurora-A and -B was significantly higher in HMCLs, and a significant number of tumors (MGUS, SMM, and MM) have expression above normal plasma cells (normalized gene expression greater than 1). (C) Expression of aurora-A and -B was detected by IHC. Staining for aurora-A and -B is cytoplasmic and nuclear, respectively. The staining is generally stronger for aurora-B than -A. For aurora-B staining in the left panel, the positive cells are indicated by close arrows. For both aurora-A and -B staining in the left and right panels, image inserts show area of staining at higher magnification. Images are at 10 × magnification except for image inserts that are at 40×. The images are captured on the Olympus BX51 microscope using the Olympus DP71 CCD camera and MicroSuite 5 software (Olympus Imaging America). Adobe Photoshop CS version 8 is then used to resize the images. (D) Protein expression of aurora-A and -B correlated with gene expression. When the cases with different percentage of positive-staining cells are grouped together, there is a corresponding increase in gene expression in the patients with higher percentage of positive cells (P = .006 for aurora-A and P = .002 for aurora-B), suggesting correlation between gene and protein expression. (E) Survival of patients with high CI (regardless of STK6 or AURB expression, although 80% of patients with high CI have high STK6 and/or AURKB expression) and those overexpressing both STK6 and AURKB is significantly shorter (n = 49 and n = 14, median OS, 42.7 and 39.8 months, respectively, log-rank P = .0005) than patients overexpressing just one of these genes (n = 33, median OS not reached) or those not overexpressing either gene (n = 255, median OS not reached).
We further compared the gene expression of STK6 and AURKB across disease stages (using the Mayo dataset) and HMCLs. The HMCLs have very high expression of both genes, whereas the malignant samples (MM, SMM, MGUS) have a larger percentage of patients with normalized expression greater than 1 than normal PCs (Figure 4B). We validated the significance of these expression values using immunohistochemistry (IHC) in 22 MM cases with varying STK6 and AURKB expression. The staining is generally stronger for aurora-B than aurora-A (Figure 4C). We found that the percentage of malignant plasma cells staining positive for both aurora-A and -B expression increases with increasing expression of these gene (Figure 4C,D). Importantly, protein staining, particularly for aurora-B, was present in more than 5% of malignant plasma cells when the normalized gene expression was greater than 1, a gene expression level that was never seen in normal plasma cells, and staining was present in more than 15% of malignant plasma cells when the normalized gene expression exceeds 2 for both STK6 and AURKB, which is a level associated with poor prognosis. IHC assessment of tumor samples therefore offers a useful clinical means of identifying cases with abnormal level of expression as well as level of expression associated with poor prognosis.
Next we examined the relation between STK6 and AURKB expression and the CI. STK6 and AURKB expression are correlated (Pearson coefficient r = 0.5, P < .001). Of the 86 patients who overexpressed (normalized expression > 2) either gene, 38 overexpressed both, 26 overexpressed only STK6, and 22 overexpressed only AURKB. Among the 49 patients with high CI, 24 overexpressed both STK6 and AURB, 15 overexpressed one of these genes, and 10 overexpressed neither. In contrast, only 14 and 33 patients overexpressed both and one of these genes, respectively, in the 302 patients with a low CI. Therefore, expression of one or both of STK6 and AURKB is highly enriched in patients with high CI (80% vs 15%; 2-tailed chi-square P < .001). In terms of impact on survival, patients with high CI have significantly poorer prognosis regardless of whether they express one or both or none of the aurora genes. Interestingly, those who overexpressed both STK6 and AURKB without a high CI also have a similarly short survival (Figure 4E).
As the poor-prognostic patients with high CI are enriched for high expression of STK6 and AURKB, which also corresponds to detectable proteins by IHC, we hypothesized that tumors with high CI may respond to aurora kinase inhibition. Indeed, when we treated a panel of 12 HMCLs with a novel aurora kinase inhibitor, the 3 most resistant HMCLs (with the highest IC50 for the drug) had significantly lower CI, with 2 of them having CI less than 4 (Figure S4).
Discussion
In this study, we validated the CI as an important prognostic factor in MM. A high CI identifies patients with poor prognosis regardless of disease phase (newly diagnosed or relapsed) or treatment (chemotherapy, high-dose therapy with stem cell transplantation, total therapy II, and even novel agents such as bortezomib). Since the CI is essentially a 4-gene index, and we have previously shown that a high CI is a surrogate for centrosome amplification by immunofluorescence (IF), it can be a clinically applicable test using multiplex-PCR- or IF-based methods.
Although gene expression profiling reveals overexpression of a number of cell cycle and proliferation-related genes in tumors with high CI, we showed in a number of ways that the CI is probably not simply a surrogate for proliferation, and hence its impact on prognosis is due to other intrinsic tumor properties associated with centrosome amplification and not just proliferation. First, in the current analysis, using a large homogenously treated dataset and including prognostic factors such as CKS1B expression, PI, and t(4;14), we confirm our previous observation that a high CI is an independent prognostic factor. Second, a high CI is able to further dichotomize the survival of patients with high PI, whereas the inverse was not seen. Third, there is little overlap between tumors with high CI and high PI. Lastly, there is significant enrichment of genes contained within a recently defined chromosomal instability (CIN) signature among the overexpressed genes in tumors with high CI. This signature was strongly correlated with a validated “functional aneuploidy” score, which allows one to infer chromosome imbalance at sub-band resolution. This signature also was strongly associated with poor survival across a number of tumor types. Importantly, these authors further dissected away the potential effect of overlapping proliferation genes within this signature and showed that the main prognostic effect of the CIN signature is mediated by nonproliferation-related genes.25
Although we believe that part of the prognostic impact of a high CI is due to genomics instability, we were unable to demonstrate a difference in CI between hyperdiploid and nonhyperdiploid MM. The reason for this is becoming clearer as our array comparative genomics hybridization (aCGH) analysis of a cohort of myeloma patients shows that both hyperdiploid and nonhyperdiploid MM are equally unstable genetically, with similar numbers of genetic abnormalities per tumor (manuscript in preparation).
Our results also suggest that novel therapies are urgently needed for patients with high CI. Our gene expression analysis reveals STK6 and AURKB as significantly overexpressed in tumors with high CI. This was further validated at the protein level by IHC. Furthermore, elevation of STK6 and AURKB at levels corresponding to IHC detection is significantly enriched in tumors with high CI, accounting for 80% of these tumors. Therefore, the recent clinical development of aurora kinase inhibitors28 may provide a means of targeting these highly aggressive tumors that seem to do poorly with current therapy. Indeed, our in vitro HMCL study, using a novel aurora kinase inhibitor, showed that most of the cell lines tested are sensitive to the aurora kinase inhibition (all these have CI greater than 4), and the 3 resistant lines have the lowest CI, with 2 having CI less than 4. These promising in vitro data will need to be confirmed in human studies.
There are several other lines of evidence that suggest targeting aurora kinases may be beneficial in myeloma. Recent papers and meeting reports have demonstrated the efficacy of a number of aurora kinase inhibitors in HMCLs, including those resistant to various anti-MM agents such as dexamethasone, alkylating agents, anthracyclines, bortezomib, and immunomodulatory thalidomide derivatives, primary patient samples, and murine xenograft model.29-31 These agents achieve significantly lower IC50 values in malignant cells compared with normal hematopoietic cells. They are able to remain effective in the presence of protective bone marrow–derived cytokines such as interleukin 6 (IL-6) or activating RAS mutation.29,31 Furthermore, the sensitivity to these agents seems to be higher in cells overexpressing RHAMM that has been correlated with centrosome amplification in MM.14,31 It is conceivable that aurora kinase inhibition has more general efficacy in MM and is not limited to tumors with high CI, which makes them even more attractive for future development in MM.
Aurora-A is a centrosome-associated protein that regulates both centrosome function and mitotic control. Overexpression of aurora-A causes centrosome amplification, aneuploidy, and tumorigenesis.32 In addition, aurora-A overexpression has been shown to abrogate taxol and nocadozole-induced spindle checkpoint33,34 and also functionally inhibit p53,35 and seems to mediate genomic instability via multiple pathways. Aurora-B is important for cytokinesis and the stable activation of the spindle checkpoint in response to loss of tension. Chinese hamster ovary (CHO) cells overexpressing aurora-B produce progeny that are aneuploid and develop into aggressive tumors in mice.32 These aurora kinases are overexpressed in various epithelial tumors including breast, colon, bladder, ovarian, and pancreatic cancers,32 although data in hematological malignancies are lacking. Therefore, our data provide the first evidence for the possible role for the overexpression of aurora kinases in mediating centrosome amplification and tumor aggression in MM.
The mechanisms leading to STK6 and AURKB overexpression are unclear. Analysis of our aCGH data suggests no amplification of these genomic loci. It is conceivable that STK6 and AURKB overexpression is due to other upstream signaling pathways similar to that observed in breast cancer, where the overexpression of aurora kinase may be mediated by estrogen receptor signaling.36
In conclusion, a high CI identifies a group of patients with abysmal prognosis regardless of current therapy. GEP identifies overexpression of aurora kinases in this cohort of patients who may therefore benefit from treatment with aurora kinase inhibitors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We would like to thank Millennium Pharmaceuticals for providing MLN8054 for the study.
R.F. is a Clinical Investigator of the Damon Runyon Cancer Research Fund.
Authorship
Author contribution: W.J.C. designed the study, analyzed data, and wrote the manuscript; E.B. performed the drug study in HMCLs and approved the manuscript; B.B. and G.M. provided data and approved the manuscript; E.R., R.V., and A.D. performed immunohistochemistry and approved the manuscript; and R.F. designed the study and wrote the manuscript.
Conflict-of-interest statement: G.M. and B.B. are employees of Millennium Pharmaceuticals, whose drug bortezomib was discussed in the paper. R.F. is currently testing a novel aurora-A–specific inhibitor from Millenium Pharmaceuticals in myeloma.
Correspondence: Wee J. Chng, Department of Haematology-Oncology, National University Hospital, 5 Lower Kent Ridge Rd, Singapore 119074, Singapore; e-mail: mdccwj@nus.edu.sg.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal