Abstract
Pre-B lymphocytes consist of 2 distinct cell populations: large pre-B and small pre-B. The large pre-B cells are newly generated pre-B cells that express pre–B-cell receptor (pre-BCR) on the surface and are highly proliferative; small pre-B cells are derived from large pre-B cells that have down-regulated pre-BCR and withdrawn from cell cycle. The molecular events that mediate the transition from cycling pre-B to small, resting pre-B have not been fully elucidated. Here, we show that interferon regulatory factors 4 and 8 (IRF4,8) suppress surrogate light chain expression and down-regulate pre-BCR in pre-B cells. Our studies further reveal that IRF4,8 induce the expression of Ikaros and Aiolos in pre-B cells, and reconstitution of expression of either one is sufficient to suppress surrogate light chain expression and down-regulate pre-BCR in pre-B cells lacking IRF4,8. Interestingly, our results also indicate that pre-B cells undergo growth inhibition and cell-cycle arrest in the presence of IRF4,8. Moreover, we provide evidence that Ikaros and Aiolos are indispensable for the down-regulation of pre-BCR and the cell-cycle withdrawal mediated by IRF4,8. Thus, IRF4,8 orchestrate the transition from large pre-B to small pre-B cells by inducing the expression of Ikaros and Aiolos.
Introduction
B lymphocyte development in the bone marrow features a sequential rearrangement of the heavy and light chain loci and a transient expression of pre–B-cell receptor (pre-BCR). After a productive immunoglobulin heavy chain rearrangement at the pro-B stage, heavy chain protein mu pairs with the surrogate light chain (SLC) λ5 and Vpre-B. Together with the signaling molecules Igα and Igβ, they form the pre-BCR on the cell surface.1 The activation of the pre-BCR is cell autonomous and independent of ligand binding.2 Signal emanated from the pre-BCR stimulates pre–B-cell proliferation and the formation of so-called large, cycling pre-B cells. After a limited number of cell divisions, cycling pre-B cells exit the cell cycle and become small, resting pre-B cells. Light chain rearrangement and transcription takes place primarily in those quiescent pre-B cells. Pre-BCR–induced B-cell self-propagation is an important event in B-cell development through which pre-B cells expressing successfully rearranged heavy chains are clonally expanded prior to light chain rearrangement.3 In addition, pre-BCR signaling is also important for inhibiting the expression of Rag1 and Rag2, thus facilitating the maintenance of allelic exclusion of the heavy chain locus.4 Moreover, pre-BCR signaling increases the accessibility of the light chain loci, thereby promoting light chain rearrangement and transcription.5
The initial burst of cell proliferation at the large pre–B-cell stage and the subsequent passage into the quiescent, small pre–B-cell stage are critical events in pre–B-cell development. Disruption of the transition from large, cycling pre-B cells to small, resting pre-B cells often leads to a block in pre–B-cell development.6-8 However, the molecular mechanisms that control pre–B-cell expansion, and therefore, the transition from cycling pre-B to resting pre-B cells, are still not clear. It has been shown that the pre-BCR is only expressed on cycling pre-B cells but not on small, resting pre-B cells.9 Thus, down-regulation of pre-BCR has been linked to cessation of cell proliferation and cell-cycle withdrawal.3,10
Ikaros and Aiolos are members of the Ikaros family of transcription factors.11 The Ikaros family transcription factors interact with each other and other members of the Ikaros family. The N-terminal domain of Ikaros family proteins is responsible for DNA binding, whereas the C-terminal domain is involved in dimerization. The formation of Ikaros homo- and heterodimers through the C-terminal dimerization domain increases their affinity for DNA.12,13 It has been demonstrated that expression of Ikaros and Aiolos are increased in pre-B cells relative to pro-B cells, suggesting that Ikaros and Aiolos may play an important role in pre–B-cell development.14 Indeed, Aiolos has been shown to be directly involved in the silencing of the λ5 gene in pre-B cells.15 It has been reported that pre-BCR signaling induces the expression of Aiolos, which in turn, competes with EBF, an essential transcriptional activator of the λ5 gene, for binding to an overlapping region on the λ5 promoter.15 Ikaros family transcription factors silence the expression of their target genes via recruitment of transcriptional repressor complexes such as the NuRD histone deacetylase complex.16
Interferon regulator factors 4 and 8 (IRF4,8) are closely related members of the IRF family of transcription factors that have been shown to play a critical role in both innate and adaptive immunity.17 IRF4,8 are predominantly expressed in the immune system, where they display a largely overlapping expression pattern. Previous studies show that IRF4,8 can function redundantly to control an overlapping set of target genes.18-20 For example, it has been demonstrated that IRF4 and IRF8 can form complexes with the Ets family of transcription factors PU.1 and Spi-B to regulate activity of kappa 3′ enhancer and Lambda enhancers.18,20 In addition, IRF4 and IRF8 can interact with transcription factor E2A to regulate activity of kappa 3′ enhancer.19,21
Previous studies have shown that B-cell development is blocked at the pre–B-cell stage in the IRF4,8 double-mutant mice (IRF4,8−/−).6 IRF4,8−/− pre-B cells resemble cycling pre-B cells and fail to rearrange the light chain gene. Further molecular analysis revealed that compared with wild type pre-B cells, IRF4,8−/− pre-B cells express higher levels of SLC and fail to down-regulate pre-BCR.6 These results collectively suggest that IRF4,8 may negatively regulate pre–B-cell expansion. There is growing evidence indicating that pre-BCR signaling induces the expression of IRF4.15,22 However, the exact role of IRF4,8 in the transition from large, cycling pre-B to small, resting pre-B remains unclear. Here, we show that IRF4,8 suppress SLC expression, down-regulate pre-BCR, and inhibit pre–B-cell proliferation. We further provide evidence that Ikaros and Aiolos are indispensable for IRF4,8-mediated pre-BCR down-regulation and cell-cycle withdrawal.
Methods
Mice
IRF4 and IRF8 compound mutant mice (IRF4,8−/−) have been previously described.6 The mice were bred and maintained under specific pathogen–free conditions. Experiments were performed according to guidelines from the National Institutes of Health and with an approved Institutional Animal Care and Use Committee (IACUC) protocol from the University of Nebraska Medical Center. Mice of 6 to 10 weeks of age were used for this study.
FACS analysis and cell sorting
Cells were preincubated with either 2% rat serum or Fc-Block (2.4G2), and stained with optimal amounts of specific antibodies, either biotinylated or directly fluorophore-conjugated. Antibodies against B220 (RA3-6B2), CD19 (ID3), pre-BCR (SL156), and λ5 (LM34) were purchased from BD PharMingen (San Diego, CA); biotinylated anti-kappa antibody (H139-52.1) was obtained from Southern Biotechnology (Birmingham, AL). Fluorescence-activated cell sorter (FACS) analysis was performed with a FACS Calibur or a BD LSR II flow cytometer. The cells were sorted with a BD FACSAria (BD Biosciences, San Jose, CA).
Pre–B-cell culture
Pre–B-cell cultivation was carried out as described before.23 Briefly, B220+ cells were isolated from bone marrow of wild-type or IRF4,8−/− mice using a magnetic-activated cell sorter (MACS) separation column (Miltenyi Biotec, Auburn, CA). Purified cells were overlaid on top of an irradiated S17 stromal cell layer. The cells were cultivated in Opti-MEM (Invitrogen, Carlsbad, CA) medium containing 5% fetal bovine serum (FBS), 50 μM β-mercaptoethanol, 2 mM l-glutamine, 100 U penicillin-streptomycin, and 5 ng/mL IL-7 (R&D Systems, Minneapolis, MN). The pre-B cells were passaged every 3 days onto a new S17 stromal layer. Cells with fewer than 5 passages were used for the experiments.
Retroviral constructs and retroviral infection
IRF4 and IRF8 retroviral-expressing vectors have been described before.23 The inducible IRF4 and IRF8 fusion proteins were generated by fusing estrogen receptor (ER) ligand-binding domain with the N-terminal of IRF4 or IRF8. The cDNAs encoding ER and IRF4 or IRF8 fusion (IRF4ER, IRF8ER) were inserted into a MigR1 retroviral vector. Human cDNAs encoding Aiolos and Ikaros were inserted into a MigR1 vector to generate the Ikaros and Aiolos expression vector used in this study. A dominant-negative Ikaros mutant (IkarosDN) was constructed by inserting the last 468 nucleotides of the coding region of human Ikaros cDNA into a MigR1 vector. MigR1 bicistronic retroviral vector was engineered to coexpress either green fluorescent protein (GFP) or yellow fluorescent protein (YFP).
To infect primary pre-B cells, the retroviral vectors containing the genes of interest were transfected into the ecotropic retroviral packaging cell line PLAT-E using FuGene 6 (Roche, Indianapolis, IN). The cell-free supernatants are collected at 24 and 48 hours after transfection. The virus was concentrated by centrifugation at 20 000g for 1 hour and was typically used the same day to infect target cells via spin infection. The infection was carried out in a 24-well plate at 640 g for 1 hour in the presence of 10 μg/mL polybrene. The infected cells were analyzed by FACS at different time points afterward.
Cell-cycle analysis with Hoechst dye
Cell-cycle analysis with live cells was conducted using Hoechst 33342, trihydrochloride, and trihydrate (H3570; Invitrogen). The cells were washed twice with phosphate-buffered saline (PBS) and resuspended in buffer containing PBS plus 0.1% bovine serum albumin (BSA) at 106 cells/mL. Hoechst dye was added to the cells at a concentration of 10 μg/mL. The cells were incubated at 37°C water bath for 15 minutes and then analyzed in a BD LSR II flow cytometer.
Western blot analysis
The sorted GFP+ cells and cultivated wild-type pre-B cells were lysed and used for Western blot analysis. The signals were visualized using the SuperSignal West Dura HRP Detection kit (Pierce, Rockford, IL). Antibodies directed against IRF4 (M-17), IRF8 (C-19), Ikaros (H-100), and β-actin (AC-15) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Real-time PCR analysis
The cells were lysed using Trizol. Total RNA was extracted and reverse-transcribed with a single-strand cDNA synthesis kit (GE Healthcare, Little Chalfont, United Kingdom). Quantitative real-time polymerase chain reaction (PCR) analysis was carried out in a 7500 real-time PCR system (Applied Biosystems [ABI], Foster City, CA) using SYBR Green PCR Core Reagents (ABI). All samples were tested in triplicate, and average CT values were calculated and normalized to the housekeeping gene GAPDH. Each primer set was independently repeated 3 times, and average values and standard deviations were calculated. The sequences of primers used in this study are described in detail in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Results
IRF4,8 suppress SLC expression and down-regulate pre-BCR in pre-B cells
Previous studies have shown that IRF4,8−/− pre-B cells express high levels of SLC gene λ5 and Vpre-B and fail to down-regulate pre-BCR.6 To determine the effect of IRF4,8 on pre-BCR expression, we reconstituted IRF4, 8 expression individually in IRF4,8−/− pre-B cells. In order to study the kinetics of IRF4,8 effects on pre-BCR expression, we generated inducible forms of IRF4,8 by fusing the ligand-binding domain of the ER with IRF4 (IRF4ER) and IRF8 (IRF8ER). The cDNAs were inserted into a retroviral vector. The ER ligand-binding domain only was also inserted into the vector and used as a control. Our previous studies have shown that IRF4,8 stimulate CD25 expression and promote light chain rearrangement and transcription in pre-B cells.23 Our results show that IRF4ER and IRF8ER behave like wild-type IRF4,8 and can rescue light chain rearrangement and transcription as well as CD25 expression in IRF4,8−/− pre-B cells in the presence but not absence of tamoxifen (1 μM; Figure S1).
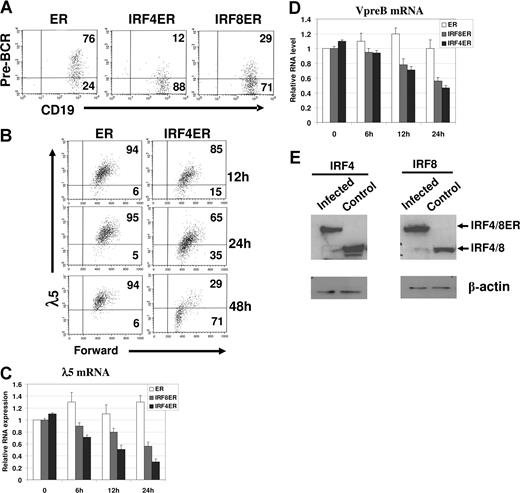
To determine the effect of IRF4,8 on pre-BCR expression, IRF4,8−/− pre-B cells were infected with retrovirus expressing either ER, IRF4ER, or IRF8ER and were analyzed by FACS at different time points afterward. Effects of IRF4,8 were analyzed in infected cells expressing a high level of GFP (GFPhi). As shown in Figure 1A, expression of IRF4 and IRF8 leads to down-regulated pre-BCR expression within 48 hours of tamoxifen treatment in IRF4,8−/− pre-B cells. The kinetics of pre-BCR down-regulation by IRF4 was further analyzed by measuring λ5 expression. The down-regulation of λ5 can be detected as early as 12 hours; by 48 hours, 71% cells have down-regulated λ5 expression (Figure 1B). The GFPhi cells were also isolated via sorting at different time points for RNA extraction and real-time PCR analysis. The result show that SLC transcripts are suppressed in the presence of IRF4,8; the effect can be detected at as early as 6 hours after tamoxifen treatment, and after 24 hours of treatment, only 30% of λ5 and 45% Vpre-B transcripts remain in IRF4-infected cells (Figure 1C,D). Western blot analysis was performed to assess the relative expression levels of IRF4ER and IRF8ER in infected cells (GFPhi). As shown in Figure 1E, the fusion proteins are expressed at levels that are comparable with their wild-type counterparts in wild-type pre-B cells, indicating that IRF4,8 are expressed at physiologic levels in the infected cells. In addition, our results also indicate that IRF4 appears to be more potent than IRF8 at suppressing SLC expression. Collectively, our results indicate that reconstitution of IRF4,8 suppress SLC expression and down-regulate pre-BCR in IRF4,8−/− pre-B cells.
IRF4,8 suppress SLC expression and down-regulate pre-BCR in pre-B cells. IRF4,8−/− pre-B cells were isolated from the bone marrow of IRF4,8−/− mice and were cultivated in Opti-MEM medium containing 5 ng/mL IL-7. The cells were infected with virus expressing ER, IRF4ER, or IRF8ER. After 24 hours, tamoxifen (1 μm) was added, and the cells were analyzed at different time points as indicated. (A) The cells were analyzed by FACS 48 hours after tamoxifen addition. The expression of pre-BCR was analyzed in cells expressing a high level of GPF (GFPhi). The numbers in the quadrants represent percentages of gated cells. (B) Time-course study was conducted after tamoxifen treatment to analyze λ5 expression in ER- and IRF4ER-infected cells. The λ5 expression in gated (GFPhi) cells was analyzed by FACS. The numbers in the quadrants represent percentages of gated cells. (C,D) Real-time PCR analysis on expression of λ5 and Vpre-B mRNA. The infected cells (GFPhi) were isolated via sorting at different time points after tamoxifen treatment, and total RNA was isolated for PCR analysis. The value of ER-infected cells at 0 hours was arbitrarily set as 1. Each primer set was independently repeated 3 times, and average values and standard deviations were calculated. (E) IRF4ER- and IRF8ER-infected cells (GFPhi) were isolated by sorting for protein extraction. Western blot analysis was performed to measure the expression of IRF4ER and IRF8ER in the sorted cells and IRF4 and IRF8 in the cultivated wild-type pre-B cells.
IRF4,8 suppress SLC expression and down-regulate pre-BCR in pre-B cells. IRF4,8−/− pre-B cells were isolated from the bone marrow of IRF4,8−/− mice and were cultivated in Opti-MEM medium containing 5 ng/mL IL-7. The cells were infected with virus expressing ER, IRF4ER, or IRF8ER. After 24 hours, tamoxifen (1 μm) was added, and the cells were analyzed at different time points as indicated. (A) The cells were analyzed by FACS 48 hours after tamoxifen addition. The expression of pre-BCR was analyzed in cells expressing a high level of GPF (GFPhi). The numbers in the quadrants represent percentages of gated cells. (B) Time-course study was conducted after tamoxifen treatment to analyze λ5 expression in ER- and IRF4ER-infected cells. The λ5 expression in gated (GFPhi) cells was analyzed by FACS. The numbers in the quadrants represent percentages of gated cells. (C,D) Real-time PCR analysis on expression of λ5 and Vpre-B mRNA. The infected cells (GFPhi) were isolated via sorting at different time points after tamoxifen treatment, and total RNA was isolated for PCR analysis. The value of ER-infected cells at 0 hours was arbitrarily set as 1. Each primer set was independently repeated 3 times, and average values and standard deviations were calculated. (E) IRF4ER- and IRF8ER-infected cells (GFPhi) were isolated by sorting for protein extraction. Western blot analysis was performed to measure the expression of IRF4ER and IRF8ER in the sorted cells and IRF4 and IRF8 in the cultivated wild-type pre-B cells.
IRF4,8 induce the expression of Ikaros and Aiolos in pre-B cells
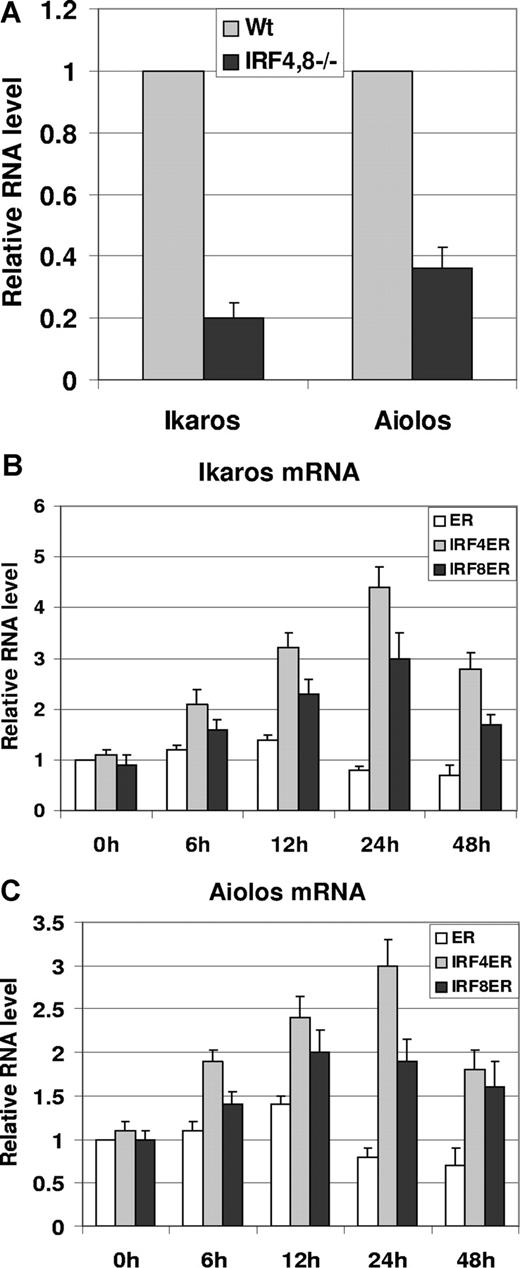
It has been shown that compared with pro-B cells, expression of Ikaros and Aiolos is elevated in pre-B cells.14 Moreover, Aiolos has been shown to be critical in the silencing of the λ5 gene.15 Therefore, we sought to determine whether there are defects in the expression of Ikaros and Aiolos in IRF4,8−/− pre-B cells. To test this possibility, pre-B cells were isolated via FACS sorting from IRF4,8−/− and wild-type control mice based on the surface markers of B220+ CD43low/− IgM−. Total RNA was extracted from sorted pre-B cells, and real-time PCR was performed. Interestingly, our results show that both Ikaros and Aiolos are expressed at lower levels in IRF4,8−/− pre-B cells, down to 20% and 34% of the levels found in wild-type pre-B cells, respectively (Figure 2A). This result suggests that IRF4,8 might regulate the expression of Ikaros and Aiolos in pre-B cells. To further explore this possibility, we measured the expression of Ikaros and Aiolos in IRF4ER- and IRF8ER-infected cells described in Figure 1C,D. As shown in Figure 2B and C, both IRF4 and IRF8 are capable of inducing Ikaros and Aiolos expression in IRF4,8−/− pre-B cells. The induction could be detected as early as 6 hours after tamoxifen treatment and peaked after 24 hours. Ikaros appears to be more highly induced than Aiolos by either IRF4 or IRF8. Interestingly, IRF4 appears to be a stronger inducer of Ikaros and Aiolos expression than does IRF8; Ikaros and Aiolos expression can be induced up to 4.5-fold and 3-fold, respectively, in IRF4-infected cells compared with 3-fold and 2-fold, respectively, in IRF8-infected cells. We were able to repeat this result using at least 2 different primer sets each targeting different region of Ikaros and Aiolos (data not shown). In addition, the primer sets we used amplified either Ikaros or Aiolos, but not both. We did not detect significant changes in the expression levels of other Ikaros family members such as Helios, Eos, and Pegasus (data not shown). Furthermore, we were also able to detect the induction of Ikaros and Aiolos expression by wild-type IRF4,8 in pre-B cells, indicating that our result is not an artifact of ER fusion proteins (data not shown). Taken together, our results indicate that IRF4,8 selectively induce Ikaros family members Ikaros and Aiolos in pre–B-cell development.
IRF4,8 induce the expression of Ikaros and Aiolos in pre-B cells. (A) Expression of Ikaros and Aiolos is defective in IRF4,8−/− pre-B cells. Bone marrow cells were isolated from IRF4,8−/− and wild-type control mice. Cells were stained with antibodies against B220, CD43, IgM, and the pre-B cells (B220+, CD43low/−, and IgM−) were isolated by sorting. The relative expression of Ikaros and Aiolos was measured in wild-type and IRF4,8−/− pre-B cells by real-time PCR. The values in the control pre-B cells were arbitrarily set as 1. (B,C) Total RNA isolated from ER-, IRF4ER-, and IRF8ER-infected cells described in Figure 1C and D were subjected to real-time analysis to measure the expression of Ikaros (Figure 1C) and Aiolos (Figure 1D). The values are average and SD of 3 independent experiments.
IRF4,8 induce the expression of Ikaros and Aiolos in pre-B cells. (A) Expression of Ikaros and Aiolos is defective in IRF4,8−/− pre-B cells. Bone marrow cells were isolated from IRF4,8−/− and wild-type control mice. Cells were stained with antibodies against B220, CD43, IgM, and the pre-B cells (B220+, CD43low/−, and IgM−) were isolated by sorting. The relative expression of Ikaros and Aiolos was measured in wild-type and IRF4,8−/− pre-B cells by real-time PCR. The values in the control pre-B cells were arbitrarily set as 1. (B,C) Total RNA isolated from ER-, IRF4ER-, and IRF8ER-infected cells described in Figure 1C and D were subjected to real-time analysis to measure the expression of Ikaros (Figure 1C) and Aiolos (Figure 1D). The values are average and SD of 3 independent experiments.
Reconstituting expression of either Ikaros or Aiolos is sufficient to down-regulate pre-BCR in IRF4,8−/− pre-B cells
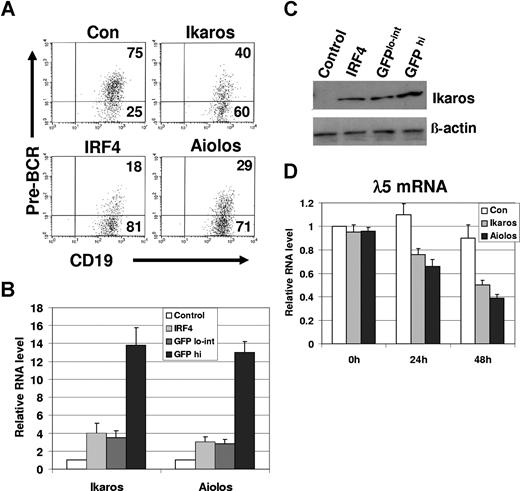
It has been shown that Aiolos suppresses λ5 expression in wild-type pre-B cells that express IRF4,8.15 To determine whether Aiolos and Ikaros are sufficient to suppress SLC expression in the absence of IRF4,8, IRF4,8−/− pre-B cells were infected with retrovirus expressing either control, IRF4, Aiolos, or Ikaros. The infected cells expressing a high level of GFP (GFPhi) and a low to intermediate level of GFP (GFPlo-int) were analyzed separately. Our result shows that in GFPhi cells, infected Aiolos or Ikaros is sufficient to down-regulate pre-BCR in IRF4,8−/− pre-B cells (Figure 3A). It has been reported that Aiolos is a more potent transcriptional regulator than Ikaros.14 Consistent with this report, our results show that Aiolos is found to be more potent than Ikaros at down-regulation of pre-BCR (Figure 3A). However, IRF4 is found to be the strongest suppressor of pre-BCR expression (Figure 3A). In contrast, in GFPlo-int cells, expression of pre-BCR is not significantly affected by Ikaros or Aiolos (data not shown).
Reconstituting the expression of either Ikaros or Aiolos is sufficient to down-regulate pre-BCR in IRF4,8−/− pre-B cells. (A) IRF4,8−/− pre-B cells were infected with virus expressing control, IRF4, Ikaros, and Aiolos. At 48 hours after infection, the cells were stained with antibodies against pre-BCR and CD19 and analyzed by FACS. The GFPhi cells were gated and further analyzed. The numbers represent percentages of the gated cells. (B) Relative expression levels of Ikaros and Aiolos were assessed by real-time PCR. The GFPhi and GFPlo-int cells were isolated via sorting from control-vector, Ikaros-, and Aiolos-infected cells (48 hours after infection). In addition, GFPhi cells from IRF4ER-infected cells were isolated via sorting 24 hours after tamoxifen treatment. Ikaros-infected cells were used to measure Ikaros expression, whereas Aiolos-infected cells were used solely to measure Aiolos expression. However, the expression levels of both endogenous Ikaros and Aiolos were measured in the control-vector and IRF4ER-infected cells (GFPhi). The value in the control-vector infected cells was set as 1. (C) Western blot analysis of Ikaros protein from GFPhi and GFPlo-int cells isolated from Ikaros-infected cells, and the sorted control-vector and IRF4ER-infected cells (GFPhi). (D) The infected cells (GFPhi) were sorted at different time points, and total RNA was isolated for real-time PCR analysis. λ5 expression was measured, and the value of control-vector infected cells at 0 hours was arbitrarily set as 1. The values are average and SD of 3 independent experiments.
Reconstituting the expression of either Ikaros or Aiolos is sufficient to down-regulate pre-BCR in IRF4,8−/− pre-B cells. (A) IRF4,8−/− pre-B cells were infected with virus expressing control, IRF4, Ikaros, and Aiolos. At 48 hours after infection, the cells were stained with antibodies against pre-BCR and CD19 and analyzed by FACS. The GFPhi cells were gated and further analyzed. The numbers represent percentages of the gated cells. (B) Relative expression levels of Ikaros and Aiolos were assessed by real-time PCR. The GFPhi and GFPlo-int cells were isolated via sorting from control-vector, Ikaros-, and Aiolos-infected cells (48 hours after infection). In addition, GFPhi cells from IRF4ER-infected cells were isolated via sorting 24 hours after tamoxifen treatment. Ikaros-infected cells were used to measure Ikaros expression, whereas Aiolos-infected cells were used solely to measure Aiolos expression. However, the expression levels of both endogenous Ikaros and Aiolos were measured in the control-vector and IRF4ER-infected cells (GFPhi). The value in the control-vector infected cells was set as 1. (C) Western blot analysis of Ikaros protein from GFPhi and GFPlo-int cells isolated from Ikaros-infected cells, and the sorted control-vector and IRF4ER-infected cells (GFPhi). (D) The infected cells (GFPhi) were sorted at different time points, and total RNA was isolated for real-time PCR analysis. λ5 expression was measured, and the value of control-vector infected cells at 0 hours was arbitrarily set as 1. The values are average and SD of 3 independent experiments.
Our result suggests that the expression level of Ikaros and Aiolos may be critical for the down-regulation of pre-BCR. To confirm this conclusion, we analyzed the relative expression levels of infected Ikaros or Aiolos cDNA in the GFPlo-int and the GFPhi cells. Ikaros or Aiolos transcripts in GFPlo-int and GFPhi cells isolated from either Ikaros- or Aiolos-infected cells were assessed by real-time PCR and compared with the endogenous Ikaros or Aiolos in the control-vector or IRF4ER-infected cells (GFPhi). Our results show that Ikaros or Aiolos is expressed at a level that is 2 to 3-fold above that of the control in GFPlo-int cells. However, Ikaros or Aiolos is expressed at a level that is 12- to 13-fold higher than that of the control when GFPhi cells were used (Figure 3B). When compared with their levels induced by IRF4, GFPhi cells isolated from Ikaros- or Aiolos-infected cells expresses Ikaros or Aiolos at a level that is about 4-fold higher (Figure 3B). Western blot analysis is consistent with real-time PCR analysis and shows that Ikaros protein isolated from Ikaros infected GFPhi cells is expressed at a level that is about 3-fold higher than that of GFPlo-int(Figure 3C). Interestingly, Ikaros protein induced by IRF4 is expressed at a level that is similar to that in the GFPlo-int cells isolated from the Ikaros-infected cells.
To determine the effects of Ikaros and Aiolos on the expression of SLC transcript, infected cells (GFPhi) were isolated by sorting at different time points for RNA isolation and real-time PCR analysis. Consistent with the FACS analysis, reconstitution of either Ikaros or Aiolos expression is sufficient to inhibit λ5 gene expression in IRF4,8−/− pre-B cells (Figure 3D). Indeed, λ5 expression was down to about 38% and 48% of the control in Aiolos- and Ikaros-infected cells, respectively. In summary, our results indicate that Ikaros and Aiolos suppress SLC expression and down-regulate pre-BCR in IRF4,8−/− pre-B cells, and that the effectiveness of Ikaros and Aiolos is dependent on their expression levels.
Reconstituting expression of IRF4, Ikaros, or Aiolos is sufficient to inhibit the proliferation of IRF4,8−/− pre-B cells
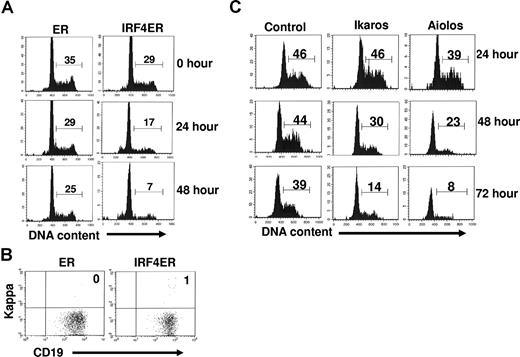
The transition of large pre-B to small pre-B features the down-regulation of pre-BCR and the cell-cycle withdrawal. Our results show that IRF4,8 down-regulate pre-BCR in pre-B cells. To determine if reconstitution of IRF4,8 expression is sufficient to cause growth arrest in pre-B cells, IRF4,8−/− pre-B cells were infected with virus expressing control (ER only) or IRF4ER fusion protein. The effect of IRF4 on cell-cycle progression was visualized directly with a Hoechst dye at different time points after tamoxifen treatment. Our results show that IRF4 inhibits cell-cycle progression; percentage of cycling cells is decreased from 29% at the start of the treatment to only 7% 48 hours later (Figure 4A). In contrast, only a slight decrease in the percentage of cycling cells in ER-infected cells was observed during the same time period. It is worth noting that cell-cycle arrest induced by IRF4 is not an indirect result of pre–B-cell differentiation, as very few kappa-positive B cells were detected in culture at the end of the experiment (Figure 4B). To further determine if reconstitution of expression of Ikaros or Aiolos is sufficient to inhibit pre–B-cell proliferation in the absence of IRF4,8, IRF4,8−/− pre-B cells were infected with virus expression either control, or Ikaros, or Aiolos, and their effects on cell cycle progression were examined daily for 3 days. Our results show that reconstituting expression of either Ikaros or Aiolos (in GFPhi cells) is sufficient to inhibit pre–B-cell proliferation. Aiolos appears to be a stronger growth inhibitor than Ikaros; only 8% of cells were still cycling in Aiolos-infected cells compared with 14% in Ikaros-infected cells at day 3 (Figure 4C). Similar to the effect on pre-BCR expression, it is apparent that high levels of Ikaros and Aiolos are required to block cell-cycle progression as evidenced by the finding that very little effect on cell-cycle progression was observed when GFPlo-int cells were analyzed (data not shown). Taken together, our results indicate that IRF4, Ikaros, or Aiolos each is sufficient to inhibit the proliferation of IRF4,8−/− pre-B cells.
Reconstituting expression of IRF4, Ikaros, or Aiolos is sufficient to inhibit the proliferation of IRF4,8−/− pre-B cells. (A) IRF4,8−/− pre-B cells were infected with virus expressing ER and IRF4ER. At 24 hours after infection, tamoxifen (1μM) was added to activate IRF4. The cell-cycle status was examined at different time points by staining the cells with Hoechst dye (10 μg/mL). (B) The effect of IRF4 on pre–B-cell proliferation is uncoupled from pre–B-cell differentiation. After 48 hours of treatment with Tamoxifen, the infected cells were stained with anti-kappa and anti-CD19 antibodies and analyzed by FACS. (C) IRF4,8−/− pre-B cells were infected with control, Ikaros, and Aiolos. The cell-cycle status of the infected cells was analyzed daily for 3 days. Cell-cycle status of the GFPhi cells was shown, and the numbers represent percentages of the cycling cells. The result is a representative of 3 independent experiments.
Reconstituting expression of IRF4, Ikaros, or Aiolos is sufficient to inhibit the proliferation of IRF4,8−/− pre-B cells. (A) IRF4,8−/− pre-B cells were infected with virus expressing ER and IRF4ER. At 24 hours after infection, tamoxifen (1μM) was added to activate IRF4. The cell-cycle status was examined at different time points by staining the cells with Hoechst dye (10 μg/mL). (B) The effect of IRF4 on pre–B-cell proliferation is uncoupled from pre–B-cell differentiation. After 48 hours of treatment with Tamoxifen, the infected cells were stained with anti-kappa and anti-CD19 antibodies and analyzed by FACS. (C) IRF4,8−/− pre-B cells were infected with control, Ikaros, and Aiolos. The cell-cycle status of the infected cells was analyzed daily for 3 days. Cell-cycle status of the GFPhi cells was shown, and the numbers represent percentages of the cycling cells. The result is a representative of 3 independent experiments.
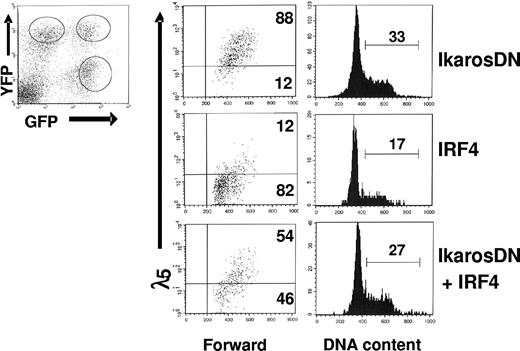
IkarosDN antagonizes the down-regulation of pre-BCR and blocks the cell-cycle withdrawal mediated by IRF4 in pre-B cells
Our results show that Ikaros and Aiolos can substitute for IRF4,8 to down-regulate pre-BCR and promote cell-cycle withdrawal, suggesting that the effect of IRF4,8 may be mediated by Ikaros and Aiolos. To evaluate this possibility, we examined the impact of IkarosDN on pre-BCR down-regulation and cell-cycle withdrawal mediated by IRF4,8. The formation of Ikaros homo- and heterodimers through the C-terminal dimerization domain increases their affinity for DNA. However, the heterodimers between wild-type Ikaros and the mutated form are unable to bind DNA. Thus, the Ikaros protein containing only the C-terminal dimerization domain can negatively interfere with the activity of wild-type Ikaros family members.13,14 The last 156 amino acids at the C-terminal region of human Ikaros protein, which contains the dimerization domain but lacks the DNA-binding domain, was amplified for use as a dominant-negative mutant. The dominant-negative effect of IkarosDN was tested and confirmed (Figure S2). To determine the effect of IkarosDN on IRF4-mediated pre-BCR down-regulation and cell-cycle arrest, IRF4,8−/− pre-B cells were coinfected with virus expression IRF4 (YFP) and IkarosDN (GFP). At 3 days after infection, infected cells were analyzed by FACS. As shown in Figure 5, infected cells consist of 3 distinct populations: GFP+YFP−, GFP−YFP+, and GFP+YFP+. While expression of IkarosDN by itself (GFP+YFP−) has little effect on λ5 expression, the expression of λ5 is dramatically down-regulated in the GFP−YFP+ population, which expresses IRF4. Interestingly, the down-regulation of λ5 is attenuated in cells coexpressing both IRF4 and IkarosDN (GFP+YFP+), suggesting that down-regulation of pre-BCR by IRF4 is dependent on Ikaros and Aiolos. Cell-cycle analysis further shows that IkarosDN almost completely blocks the cell-cycle inhibition mediated by IRF4. Similar results were obtained when cells were coinfected with IRF8 and IkarosDN (data not shown). Thus, Ikaros and Aiolos are critical for the down-regulation of pre-BCR and the cell-cycle withdrawal mediated by IRF4,8.
Effect of IRF4 on pre-BCR down-regulation and cell-cycle withdrawal is attenuated in the presence of IkarosDN. IRF4,8−/− pre-B cells were coinfected with IRF4 (YFP) and IkarosDN (GFP). The infected cells consist of 3 distinct populations: YFP+GFP−, YFP+GFP+, and YFP−GFP+. At 3 days after infection, the cells were stained with anti-λ5 antibody and Hoechst dye and analyzed by FACS. The λ5 expression and cell-cycle status in the 3 distinct populations of cells were analyzed separately. The result is a representative of 3 independent experiments, and the numbers represent percentages of the cycling cells.
Effect of IRF4 on pre-BCR down-regulation and cell-cycle withdrawal is attenuated in the presence of IkarosDN. IRF4,8−/− pre-B cells were coinfected with IRF4 (YFP) and IkarosDN (GFP). The infected cells consist of 3 distinct populations: YFP+GFP−, YFP+GFP+, and YFP−GFP+. At 3 days after infection, the cells were stained with anti-λ5 antibody and Hoechst dye and analyzed by FACS. The λ5 expression and cell-cycle status in the 3 distinct populations of cells were analyzed separately. The result is a representative of 3 independent experiments, and the numbers represent percentages of the cycling cells.
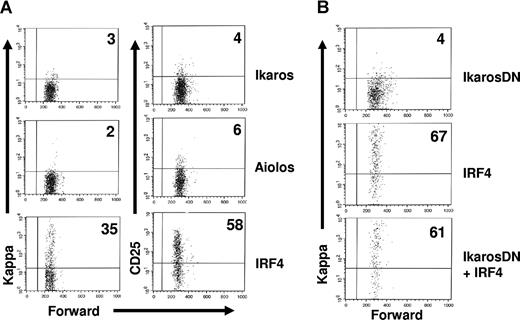
IkarosDN fails to antagonize the effect of IRF4 on light chain rearrangement and transcription in pre-B cells
Our previous studies have shown that IRF4,8 promote light chain rearrangement and transcription in pre-B cells.23 Since Ikaros and Aiolos can mediate the effects of IRF4,8 on the down-regulation of pre-BCR and the cell-cycle withdrawal, we asked if reconstitution of Ikaros or Aiolos expression is also able to rescue light chain rearrangement and transcription in IRF4,8−/− pre-B cells. To test this, IRF4,8−/− pre-B cells were infected with virus expressing Ikaros, Aiolos, and IRF4. As reported previously, IRF4 rescues kappa rearrangement and transcription as well as the expression of the pre–B-cell maturation marker CD25.23 However, neither Aiolos nor Ikaros is able to rescue the expression of kappa light chain and CD25 in IRF4,8−/− pre-B cells (Figure 6A). This result suggests that IRF4,8-mediated light chain rearrangement and transcription is independent of Ikaros and Aiolos. To further confirm this finding, we coinfected IRF4,8−/− pre-B cells with virus expressing IRF4 (YFP) and IkarosDN (GFP). The effect of IRF4 on kappa light chain expression was analyzed in the presence or absence of IkarosDN. Our results show that IRF4 is capable of promoting light chain rearrangement and transcription in the presence of IkarosDN (Figure 6B). In fact, IkarosDN had little effect on IRF4-mediated kappa rearrangement and transcription. In summary, our results indicate that Ikaros and Aiolos are dispensable for IRF4,8-mediated light chain rearrangement and transcription.
IkarosDN fails to antagonize the effect of IRF4 on light chain rearrangement and transcription in pre-B cells. (A) IRF4,8−/− pre-B cells were cultivated in Opti-MEM medium containing 5 ng/mL IL-7 and infected with virus expressing IRF4, Ikaros, and Aiolos. At 24 hours after infection, the infected cells were washed and replated on top of an irradiated S17 stromal layer in the absence of IL7 for another 36 hours. The cells were then stained with antibodies against kappa and CD25 and analyzed by FACS. The expression of kappa and CD25 in the GFPhi cells are shown. (B) IRF4,8−/− pre-B cells were coinfected with IRF4 (YFP) and IkarosDN (GFP) as described in Figure 5. At 24 hours after infection, IL-7 was removed from the culture media. After another 72 hours, the cells were stained with anti-kappa antibody and Hoechst dye and analyzed by FACS. The infected cells were analyzed using strategies described in Figure 5. The result is a representative of 3 independent experiments. Numbers in quadrants represent percentages of gated cells.
IkarosDN fails to antagonize the effect of IRF4 on light chain rearrangement and transcription in pre-B cells. (A) IRF4,8−/− pre-B cells were cultivated in Opti-MEM medium containing 5 ng/mL IL-7 and infected with virus expressing IRF4, Ikaros, and Aiolos. At 24 hours after infection, the infected cells were washed and replated on top of an irradiated S17 stromal layer in the absence of IL7 for another 36 hours. The cells were then stained with antibodies against kappa and CD25 and analyzed by FACS. The expression of kappa and CD25 in the GFPhi cells are shown. (B) IRF4,8−/− pre-B cells were coinfected with IRF4 (YFP) and IkarosDN (GFP) as described in Figure 5. At 24 hours after infection, IL-7 was removed from the culture media. After another 72 hours, the cells were stained with anti-kappa antibody and Hoechst dye and analyzed by FACS. The infected cells were analyzed using strategies described in Figure 5. The result is a representative of 3 independent experiments. Numbers in quadrants represent percentages of gated cells.
Discussion
Previous studies have shown that B-cell development is blocked at the large pre–B-cell stage in IRF4,8−/− mice, suggesting that IRF4,8 are critical regulators of pre–B-cell development.6 Here, we provide insights into the molecular mechanisms through which IRF4,8 orchestrate pre–B-cell development. Collectively, our results indicate that IRF4,8 induce the expression of Ikaros and Aiolos to suppress SLC expression and down-regulate pre-BCR. The activity of SLC loci is developmentally regulated.24 Silencing of SLC loci at the pre-B stage is accompanied by relocation of SLC loci to pericentromeric heterochromatin, an event that is likely dependent on the Ikaros family of transcription factors.15,25,26 Silencing of SLC also has been linked to pre-BCR signaling.25 A recent study has identified Aiolos as the mediator of pre-BCR–induced suppression of λ5 expression.15 Since IRF4 expression can be rapidly induced by pre-BCR signaling, our results establish IRF4 and possibly IRF8 as the nuclear effectors of the pre-BCR signaling pathway that control the expression of Aiolos and Ikaros in pre-B cells. Interestingly, we observed that IRF4 is more potent than IRF8 at inducing the expression of Ikaros and Aiolos, which may explain why IRF4 is more effective than IRF8 at down-regulation of pre-BCR.
It has been shown that expression levels of Ikaros and Aiolos are elevated from pro-B to pre-B cells.14 Our results show that Ikaros and Aiolos expression are rapidly induced in the presence of IRF4,8, suggesting that Ikaros and Aiolos might be direct targets of IRF4,8. A recent study has shown that SLP65/Blnk activation leads to rapid induction of IRF4 and Aiolos in SLP65/Blnk−/− pre-B cells.15 Interestingly, induction of IRF4 is more rapid and can be detected several hours earlier than that of Aiolos. However, in contrast to our results with IRF4,8, Blnk activation did not affect Ikaros expression. One possible explanation of these disparate results is that the basal level of IRF4,8 in Blnk−/− pre-B cells may be high enough to maintain relatively normal Ikaros expression. Since Blnk is just one of the signaling molecules downstream of pre-BCR, it is possible that other signaling molecules, such as Btk, may be able to mediate pre-BCR signaling and to induce IRF4 expression. Consistent with this idea, B-cell development is blocked at the cycling pre–B-cell stage in mice lacking both Blnk and Btk.7 Given our finding that expression of Ikaros and Aiolos are reduced in IRF4,8−/− pre-B cells and the observation that reconstitution of physiologic levels of IRF4,8 in pre-B cells stimulates Ikaros and Aiolos expression, we conclude that IRF4,8 regulate the expression of both these genes during pre–B-cell development.
Our results show that reconstitution of IRF4,8 expression leads to growth inhibition and cell-cycle arrest in IRF4,8−/− pre-B cells, suggesting that IRF4,8 are critical for terminating pre–B-cell expansion. Pre-BCR is thought to be critical for pre–B-cell expansion because mutations of components of SLC lead to a reduction in pre–B-cell numbers.27-29 IL-7 signaling is also important for pre–B-cell expansion and survival. It has been demonstrated that pre-BCR signaling enhances the response of pre-B cells to IL-7.30 Indeed, IRF4,8−/− pre-B cells are hypersensitive to IL-7 stimulation (Figure S3A). Large pre-B cells lose responsiveness to IL-7 during the transition to small, resting pre-B cells. It has been suggested that expression of IL-7 receptor alpha chain (IL-7R) can be regulated by Ikaros.31 However, reconstitution of the expression of either IRF4,8, Ikaros, or Aiolos in IRF4,8−/− pre-B cells has no effect on IL-7R expression (Figure S3B,C).
Ikaros has been shown to directly inhibit G1-S cell-cycle transition, and the extent of the inhibition is dependent on Ikaros protein level.32 In fact, Ikaros and family members can function as tumor suppressor genes; a reduction in their expression level enhances lymphocyte activation and facilitates lymphomagenesis.33,34 The molecular mechanism by which Ikaros inhibits cell-cycle progression remains to be determined. It has been reported that reconstitution of Ikaros expression in Ikaros-null T leukemia cells blocks cell-cycle progression and induces the expression of cell cycle–dependent kinase inhibitor p27kip1.35 Our results show that an IkarosDN mutant can block the cell-cycle withdrawal mediated by IRF4,8, indicating that Ikaros and Aiolos mediate the growth inhibitory effect of IRF4,8 in pre-B cells. Therefore, in pre–B-cell development, IRF4,8, through inducing Ikaros and Aiolos, may target 2 critical events to inhibit pre-B proliferation: down-regulation of pre-BCR and the inhibition of the G1-S transition. It is conceivable that the ability to simultaneously targeting pre-BCR expression and G1-S transition would render IRF4,8 more effective and efficient at shutting down pre–B-cell expansion.
Our findings also indicate that high levels of Ikaros and Aiolos are required to down-regulate pre-BCR and repress pre–B-cell proliferation. Our results show that when expressed at a high level, as we observed in the GFPhi cells, either Ikaros or Aiolos is sufficient to down-regulate pre-BCR and inhibit pre–B-cell proliferation. However, when expressed at low to medium levels as observed in the GFPlo-int cells, neither Ikaros nor Aiolos alone is sufficient to inhibit pre–B-cell proliferation. Interestingly, Western blot analysis indicates that Ikaros in GFPlo-int cells is expressed at a level comparable to that induced by IRF4. This finding would suggest that the IRF4-directed Ikaros expression alone might not be sufficient to suppress SLC expression and cause cell-cycle withdrawal. Therefore, the simultaneous induction of Ikaros and Aiolos by IRF4 may be necessary to repress pre–B-cell proliferation. Indeed, the numbers of pre-B cells are dramatically increased in Aiolos−/− mice, suggesting that there is a defect in pre–B-cell proliferation and that Ikaros alone is not sufficient to terminate pre–B-cell expansion.36 Similarly, our results predict that Aiolos by itself also would be insufficient to rein in pre–B-cell expansion. In addition, our results also show that IRF4 appears to be more potent than either Aiolos or Ikaros alone at down-regulating pre-BCR. This finding would suggest that either down-regulation of pre-BCR is more efficient in the presence of both Ikaros and Aiolos or, that IRF4 also uses Ikaros- and Aiolos-independent pathways to facilitate pre-BCR down-regulation.
Our previous studies have demonstrated that IRF4,8 promote light chain rearrangement and transcription.23 In this study, we show that unlike the effect on pre–B-cell expansion, this aspect of IRF4,8 function cannot be substituted by Ikaros and Aiolos. Importantly, IRF4 is able to promote light chain rearrangement and transcription in the presence of the IkarosDN mutant, indicating that Ikaros and Aiolos are dispensable for IRF4,8-mediated pre–B-cell differentiation. In summary, our studies reveal 2 critical roles of IRF4,8 in pre–B-cell development: to limit pre–B-cell expansion via Ikaros- and Aiolos-dependent pathways and to promote pre–B-cell differentiation via Ikaros- and Aiolos-independent processes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs Karen Gould and Neena Haider for critical reading of the manuscript. We thank Dr Rob Lewis for providing the YFP expression plasmid and Dr Tim McKeithan for help with construction of estrogen receptor fusion proteins.
This work was supported by National Institutes of Health (NIH) grant no. AI-67891(R.L.).
National Institutes of Health
Authorship
Contribution: S.M. designed and performed experiments. S.P. performed experiments and data analysis. L.T. performed experiments. R.L. designed and performed experiments, analyzed the data, and prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Runqing Lu, Department of Genetics, Cell Biology and Anatomy, University of Nebraska Medical Center, 985805 Nebraska Medical Center, Omaha, NE 68198-5805; e-mail: rlu@unmc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal