Kawamata and colleagues combine single nucleotide polymorphism microarrays (SNP-Chips) with biostatistical algorithms to explore gene copy number in 399 cases of ALL from the ALL–BFM (Berlin-Frankfurt-Münster) 2000 trial. They coin the term molecular allelokaryotyping to describe this technology.
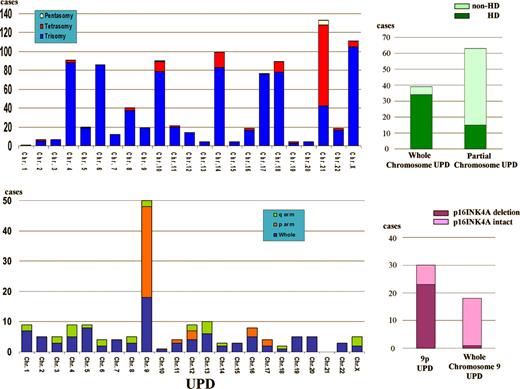
Molecular allelokaryotyping reveals more deletions and duplications than previously appreciated (see figure). It confirms common nonrandom chromosomal abnormalities: greater than 50 chromosome hyperdiploidy, deletion of 9p (site of p16INK4A), and deletion of 12 p (site of EVT6/AML1), and their established associations with age, lineage, tumor burden, ploidy, and prognosis. Among hyperdiploid samples, absence of gain of chromosome 17 or gain of chromosome 18 or both abrogate the usual favorable outcomes of hyperdiploid ALL, but in this study, gains of chromosomes 17 and 18 do not associate with gains of chromosomes 4 and 10, which confer the favorable outcomes documented in the Children's Oncology Group classification.1 The authors also detect new common abnormalities, such as 1q deletion (11%), frequently beginning at the PBX1 gene, and 6q deletion (11.5%), as well as 24 other nonrandom duplications or deletions that occur in more than 1.5% of patients. However, in contrast to the recent data from Mullighan et al,2 deletion of PAX5 (9p13) and other transcriptional factors associated with lymphocyte development are relatively rare.
While these findings are interesting, by far the most fascinating discovery is that UPD occurs in one-quarter of the cases of ALL studied. Whole-chromosome UPD, probably a result of faulty chromosome segregation, arises almost exclusively in hyperdiploid ALL, whereas partial UPD, a recombination event, occurs in nonhyperdiploid ALL. UPD is especially common in patients with T-cell ALL with 9p deletion, where partial UPD associates with p16INK4A deletions, whereas whole-chromosome UPD generally excludes p16INK4 deletion.
UPD was first noted in the leukemic clones of patients with Down syndrome (DS) more than 12 years ago3 and, more recently, in the leukemic cells of patients with neurofibromatosis type (NF1)4 and myeloproliferative disorders.4 In these children constitutionally predisposed to develop leukemia, UPD is clearly the second hit that allows the full expression of a leukemic phenotype. By finding UPD in a large number of ALL patients, molecular allelokaryotyping has probably revealed a new group of tumor suppressor genes in ALL. The associations with hyperdiploid ALL may provide the missing link in the pathogenesis of hyperdiploid ALL.
This study whets the appetite, is easy to digest, and leaves us hungry for more.
Numerical chromosomal changes and uniparental disomy in pediatric ALL.
Numerical chromosomal changes and uniparental disomy in pediatric ALL.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal