Hundreds of articles have been published that describe the induction of leukemia or lymphoma in rodents by chronic infection with murine leukemia viruses (MuLV). MuLV induce malignancy by acting as insertional mutagens, activating proto-oncogenes or inactivating tumor suppressor genes.1 Non-random clusters of proviral insertion sites, called common sites of proviral insertion (CIS), represent selection for the rare insertions that can alter leukemia genes, causing disease. Many important leukemia/lymphoma genes, including many involved in human cancer, have been identified in this way.1 Nevertheless, the field has never formally agreed on what criteria should be adopted to identify the cancer gene(s) at a given CIS, so a variety of circumstantial data are collected to find the most promising candidates to pursue functionally. A more systematic approach is warranted.
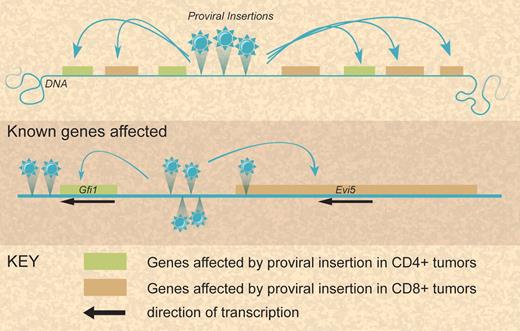
In the report from Sauvageau et al, the authors used low-density Taqman arrays to perform qRT-PCR on the CIS-associated genes within 50 kilobase pairs (kb) of the median integration site for 20 CIS to determine how many and which genes are affected by proviral insertions at CIS. This is the first time large-scale expression analysis around CIS has been undertaken in such a way. Sauvageau et al suggest that several nearby and some more distant genes are usually affected at each CIS. Furthermore, for the same CIS a different set of genes seems to be up-regulated depending on the lineage of the transformed cell (see figure).
This paper indicates layers of unexpected complexity in retroviral mutagenesis experiments. Ideally, one would have complete saturation of all proviral insertion sites and gene expression microarray data so that one could cluster like cases and deduce altered transcriptional networks resulting from changes in the expression of specific target genes at CIS. However, complicating these analyses are the choice of controls for comparisons of gene expression levels. What is the appropriate noncancer cell type to use? Should phenotypically similar cancers from the same screen, but lacking a proviral insertion at this CIS, be used? Both approaches have faults. Appropriate normal cells for comparison are hard to identify. Expression may be high for a gene in a control leukemia that lacks a proviral insertion at the relevant CIS, because that gene is altered as a result of another insertion mutation in the same pathway. Indeed, Sauvageau et al find this may often be the case. Finally, the field has never adopted standard criteria to identify statistically significant CIS, making results from different screens hard to compare.2 These complexities indicate that care must be taken in interpreting the results of retrovirus-based insertional mutagenesis screens.
Sauvageau et al performed a sensitized cancer screen in which some mice infected with MuLV were carrying hypomorphic Eed genes.3 Their work verifies that Eed has tumor suppressor activity, emphasizing the role of polycomb complexes in controlling self renewal and cancer. Interestingly, no specific MuLV-induced mutations were uncovered that correlated with Eed genotype, suggesting that its loss generally predisposes to leukemogenesis, without altering the subsequent genetic pathways chosen. It remains to be seen whether most such sensitized screens will result in the identification of specific cooperating genes or whether most predisposing mutations will only influence the latency/susceptibility to MuLV induced cancer but not the genetic pathway along which the leukemia will evolve.
Modes of leukemia gene alteration at complex loci after proviral insertion. Illustration by Debra Tyler.
Modes of leukemia gene alteration at complex loci after proviral insertion. Illustration by Debra Tyler.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal