Proviral insertional mutagenesis is a powerful tool for the discovery of cancer-associated genes. The ability of integrated proviruses to affect gene expression over long distances combined with the lack of methods to determine the expression levels of large numbers of genes in a systematic and truly quantitative manner have limited the identification of cancer genes by proviral insertional mutagenesis. Here, we have characterized a new model of proviral insertional mutagenesis-induced lymphoid tumors derived from Eed Polycomb group gene mutant mice and quantitatively determined the expression levels of all genes within 100 kb of 20 different retroviral common insertion sites (CISs) identified in these tumors. Using high-throughput quantitative reverse transcription–polymerase chain reaction (Q-RT-PCR), we document an average of 13 CIS-associated genes deregulated per tumor, half of which are leukemia subtype–specific, while the others are coordinately deregulated in the majority of tumors analyzed. Interestingly, we find that genes located distantly from common proviral integration sites are as frequently deregulated as proximal genes, with multiple genes affected per integration. Our studies reveal an unsuspected conservation in the group of genes deregulated among phenotypically similar subtypes of lymphoid leukemias, and suggest that identification of common molecular determinants of this disease is within reach.
Introduction
Proviral insertion mutagenesis screens have proven useful tools for identification of oncogenes and tumor suppressors involved in cancer.1,–3 With the advent of high-throughput retroviral insertion site cloning, identification of large numbers of common insertion sites (CISs) is readily accessible and holds great promise for elucidation of cancer-promoting pathways.4,,,,–9 A CIS is a genomic region where retroviral insertions are found in close proximity in multiple tumors. The assumption is made that such events cannot be accounted for by a random process, but are likely due to a selective advantage conferred to cells by aberrant expression of cancer genes surrounding these viral integrations. Large-scale studies have used statistical parameters to define a CIS.6,9 Suzuki et al6 established genomic interval cutoffs of 30 kb for 2 integrations, 50 kb for 3 insertions, and 100 kb for 4 or more insertions for a panel of 1200 integrations to label a locus as a CIS
To date, more than 500 CISs have been identified, and numerous others are expected as new screens are conducted in different mouse models (Retroviral Tagged Cancer Gene Database [RTCGD]).10 This constitutes an important list of candidate cancer genes and a rich source of information to improve our understanding of the molecular bases of cancer. The full potential of proviral insertional mutagenesis in the identification of oncogenic determinants remains limited by several factors. First, the retrovirus tropism limits the range of tumor types that arise after infection. Proviral insertional mutagenesis screens have mainly used Moloney murine leukemia virus (MMLV) or mammary tumor virus (MMTV), which induce hematopoietic and mammary tumors, respectively. Solid tumors, which are more prevalent in human, are more difficult to study by proviral insertion mutagenesis. Recently, the use of a modified MMLV virus encoding the platelet-derived growth factor (PDGF) β-chain allowed for identification of several CISs in brain tumors.11 Transposons have also recently been adapted to insertional mutagenesis screens, which included hematopoietic and solid tumors.12,–14 Second, the enhancer elements in the proviral long terminal repeats (LTRs) can influence gene expression over large distances,4 and may possibly affect several genes at once. The magnitude of this effect has not been extensively studied. Third, retroviral integration is not a random process,8,15 and therefore some cancer-associated genes may not be targeted. Inversely, this selectivity in viral integrations could result in incorrect labeling of some genes as “true CISs” contributing to tumor formation. To date, the number of well-documented cases of CIS-associated genes that are aberrantly expressed or involved in tumor development remains relatively small.16,–18 An extensive and quantitative analysis of gene expression near common integration sites would thus be of great value to identify the relevant genes affected. Affymetrix-type expression arrays are of interest, but sensitivity and specificity issues may complicate the analysis. Moreover, the apparently aberrant gene expression detected by this approach has to be validated using quantitative reverse transcription–polymerase chain reaction (Q-RT-PCR). Recent development in microwell high-throughput Q-RT-PCR approaches and systematic Taqman probe set design for all mouse genes provides a new possibility of quantitatively assessing gene expression over large distances around CISs.
As a first test to this approach, we conducted proviral insertion mutagenesis screens for leukemia formation in wild-type and in a Eed Polycomb group (PcG) gene sensitized background where chromatin is expected to be altered. The Eed PcG mutant mouse was chosen because heterozygosity for a null allele of Eed (Eed3354/+) or homozygosity for a hypomorph allele (Eed1989/1989) causes the development of hyperproliferative primitive progenitor cells evolving into myeloid and lymphoid diseases, indicating that Eed possesses antiproliferative roles in the hematopoietic system.19 Eed mutant mice also show accelerated T-cell tumor development when exposed to N-methyl-N-nitrosourea.20 The studies presented here constitute the most comprehensive analysis of changes in expression levels of genes surrounding retroviral insertions. They reveal the unpredictable effect of proviral insertional mutagenesis in altering the expression levels of the genes surrounding CISs and document the potential of this approach for elucidating cancer-associated genes activated in hematopoietic tumors.
Methods
Mice and genotyping
Eed1989SB hypomorphic (Eedhypo; 101/R1 × C3Hf/R1 hybrid background) and Eed3354SB null (Eednull) mutant mice have been described before21,22 and were maintained as described.23 Eed3354SB (Eednull) mutant mice were backcrossed 13 to 15 times in a C57Bl/6J background. Animals were housed and handled in accordance with the guidelines of the Animal Care and Use Committee at the Institut de Recherche Clinique de Montréal (IRCM) and IRIC.
Retroviral insertional mutagenesis
Eedhypo/hypo, Eedhypo/+ newborn mice and wild-type littermates were injected intraperitoneally with 105 infectious units of MMLV suspended in 50 μL of Dulbecco modified Eagle medium (DMEM).3 Cells from bone marrow, spleen, thymus, and lymph nodes of moribund mice were isolated for DNA and RNA extraction and for histologic analysis.
Histologic staining and morphologic analysis of lymphomas
Morphologic features of leukemic cell populations were examined under 100-fold magnification of Wright stain–dyed cytospins or touch preparations of bone marrow, spleen, liver, lung, and kidney of diseased mice. Images were acquired using a Leica DMIRB microscope with an HCXPL FluotarL 40×/0.6 numeric aperture objective (Leica, Wetzlar, Germany) and a Retiga EKI camera (Q-Imaging, Burnaby, BC). Images were transformed directly into TIFF files using Adobe Photoshop version 6.0 software (Adobe, San Jose, CA). Percentage of infiltration of blasts in hematopoietic and nonhematopoietic organs was determined for every diseased mouse. For subsequent analyses, tissue samples containing more than 75% blasts were selected (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Inverse PCR
Amplification of viral insertion sites was carried out using inverse PCR as described by Suzuki et al.6 Inverse PCR products were separated on a 1% agarose gel, purified using the QIAEX II Gel Extraction Kit (Invitrogen, Carlsbad, CA), and directly sequenced using nested primers in the MMLV LTRs at the Montreal Genome Center. Retroviral insertion sites were mapped on the mouse genome (February 2006 assembly) using the BLAT alignment tool from the University of California, Santa Cruz (UCSC) Genome Browser. The primers used for the primary and secondary Inverse PCR and sequencing primers are available online in Document S1.
Statistical significance of CISs
The number of retroviral integrations found for each CIS in Eed mutant and wild-type tumors was used to determine the statistical significance of the difference in the number of integrations between both genotypes. The Fisher exact test was applied. The adjusted P value was estimated by applying the Benjamini-Hochberg correction to correct for multiple testing.24 A P value of .05 or less was considered significant.
Q-RT-PCR
Total RNA was isolated using Trizol according to the manufacturer's protocol (Invitrogen) and treated with DnaseI (Invitrogen) before cDNA synthesis. Reverse transcription of total RNA was performed using the MMLV-RT and random hexamer primers according to the manufacturer's protocol (Invitrogen). The relative quantification of genes was performed by quantitative real-time PCR using 384-well Taqman low-density arrays (Applied Biosystems, Foster City, CA) and custom Taqman assays (designed with the Primer Xpress program from Applied Biosystems) for genes not available with low-density arrays (Lenep, Cks1b, BC061194, and Prm3). Amplification efficiency of custom assays was measured over a 5-log dilution range. Primer and probe sequences are available on request. Q-RT-PCR reactions were done on a high-throughput ABI 7900HT Fast Real-Time PCR System (Applied Biosystems) using the method described in the User Bulletin No. 2 (P/N 4303859B).43 Briefly, the Ct (threshold cycle) values of each gene were normalized to the endogenous control gene 18S rRNA (Applied Biosystems; ρCT = Cttarget − Ctendogenous) and compared with the corresponding wild-type nontransformed tissue (calibrator sample) using the “ΔΔCt” method (ΔΔCT = ΔCtSample − ΔCtCalibrator). Relative fold difference values for all samples are provided in Table S3.
PCR reactions were carried out using 25 to 50 ng of cDNA and Taqman Universal Master Mix according to the manufacturer's recommendations (Applied Biosystems). Q-RT-PCR cycling conditions were 2 minutes at 50°C and 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 59°C. All reactions were done in triplicate. Average values were used for quantification.
To select genes for Q-RT-PCR analyses, the median chromosomal location of each CIS found in 3 or more tumors was determined. The UCSC Genome browser Tables tool was used to identify every gene found within a window of 50 kb upstream and downstream of the median CIS position, according to the RefSeq genes track on the mouse genome February 2006 assembly.
URLs
Further information about the retroviral insertion sites and tumors is available at the Retroviral Tagged Cancer Gene Database (http://RTCGD.ncifcrf.gov).10
GenBank accession numbers
Results
Increased incidence of irradiation-induced leukemias in Eed mutant mice
To further document the tumor repressive function of Eed,20 large cohorts of Eedhypo/hypo and Eednull/+ mice were given a single dose of ionizing radiation (600 cGy) at 5 weeks of age and were monitored for leukemia development (Figure S1). Heterozygous Eednull/+ and homozygous Eedhypo/hypo mutant mice developed T- and B-cell leukemia with higher penetrance and shorter latency than control littermates (Table 1; Figure S1). Interestingly, the median survival time of Eednull/+ mutants was almost twice as long as homozygous Eedhypo/hypo mice. Together, this suggests that loss of function of Eed increases susceptibility of cells to oncogenic insult. A detailed characterization of the leukemias observed in these mice is provided in Figure S1 and in Document S1. In agreement with previous genotoxic studies performed in the Eed mutant background,20 our studies demonstrate that Eed exerts tumor suppressive activity in lymphoid cells.
Survival of Eed mutant and wild-type mice and phenotypic characterization of their lymphomas
| Mouse line . | Treatment . | n . | Median survival, d . | Penetrance, % . | Lymphoma, % . | Tumor-free, %* . | ||
|---|---|---|---|---|---|---|---|---|
| B cell . | T cell . | Not determined . | ||||||
| Wild-type B6 | Irradiated | 13 | 504 | 62 | — | 23 | 38 | 38 |
| Eednull/+ | Irradiated | 13 | 338 | 92 | 31 | 38 | 23 | 8 |
| Wild-type 101 × C3Hf | Irradiated | 16 | > 600 | 19 | — | 13 | 6 | 81 |
| Eedhypo/hypo | Irradiated | 11 | 184 | 73 | 9 | 46 | 18 | 27 |
| Wild-type 101 × C3Hf | MMLV | 27 | 190 | 100 | 19 | 70 | 11 | — |
| Eedhypo/+ | MMLV | 47 | 151 | 100 | 13 | 64 | 23 | — |
| Eedhypo/hypo | MMLV | 30 | 128 | 100 | 9 | 69 | 22 | — |
| Mouse line . | Treatment . | n . | Median survival, d . | Penetrance, % . | Lymphoma, % . | Tumor-free, %* . | ||
|---|---|---|---|---|---|---|---|---|
| B cell . | T cell . | Not determined . | ||||||
| Wild-type B6 | Irradiated | 13 | 504 | 62 | — | 23 | 38 | 38 |
| Eednull/+ | Irradiated | 13 | 338 | 92 | 31 | 38 | 23 | 8 |
| Wild-type 101 × C3Hf | Irradiated | 16 | > 600 | 19 | — | 13 | 6 | 81 |
| Eedhypo/hypo | Irradiated | 11 | 184 | 73 | 9 | 46 | 18 | 27 |
| Wild-type 101 × C3Hf | MMLV | 27 | 190 | 100 | 19 | 70 | 11 | — |
| Eedhypo/+ | MMLV | 47 | 151 | 100 | 13 | 64 | 23 | — |
| Eedhypo/hypo | MMLV | 30 | 128 | 100 | 9 | 69 | 22 | — |
Penetrance is equal to 100% minus the percentage of mice that died of nonhematologic etiology. B cell indicates B-ALL; T cell, T-ALL; Not determined, lymphoma cell type not determined; and —, not applicable.
*Tumor-free mice were killed at 600 days.
Retroviral insertional mutagenesis in Eed mutant mice
Insertional mutagenesis screens in sensitized backgrounds have been conducted using mice mutant for cell-cycle regulators.5,25 However, the impact of a sensitized background for a chromatin modifier with tumor suppressive activity, such as Eed, has not yet been evaluated in these screens. Cohorts of heterozygous and homozygous Eed hypomorph mice and wild-type littermates (controls) were therefore injected with MMLV and subsequently monitored for disease occurrence (Figure 1A). In accordance with the irradiation studies (Figure S1), MMLV-infected Eedhypo/+ mice developed tumors more rapidly than wild-type mice, and loss of the second Eed allele further accelerated the rate of tumor onset (Figure 1B; blue survival curve). As shown in Table 1, Eedhypo/hypo mice displayed a median survival time of 128 days compared with 151 and 190 days for Eedhypo/+ and wild-type mice, respectively (P < .001). Interestingly, we could not detect loss of heterozygosity in 10 of 10 Eedhypo/+ tumors analyzed (Figure 1C).
Increased incidence of MMLV-induced lymphomas in Eed mutant mice. (A) Schematic representation of the experimental design. (B) Survival curves showing increased incidence of MMLV-induced tumors in Eedhypo/+ and Eedhypo/hypo mutant mice compared with control littermates. (C) Presence of both Eedhypo mutant and wild-type alleles in MMLV-induced Eedhypo/+ tumors. Top shows the hypomorph T > A point mutation at position 1031 of Eed. Bar graphs show the number of clones sequenced, containing the Eedhypo or wild-type allele, per Eedhypo/+ tumor. Clones were obtained by amplifying the Eed gene from DNA (middle) or RNA (cDNA; bottom) extracted from heterozygous tumors. (D) Wright-Giemsa stainings of representative cytologic preparations of T and B lymphomas and their healthy counterparts from wild-type and Eed mutant mice. *No healthy lymph node preparations could be obtained.
Increased incidence of MMLV-induced lymphomas in Eed mutant mice. (A) Schematic representation of the experimental design. (B) Survival curves showing increased incidence of MMLV-induced tumors in Eedhypo/+ and Eedhypo/hypo mutant mice compared with control littermates. (C) Presence of both Eedhypo mutant and wild-type alleles in MMLV-induced Eedhypo/+ tumors. Top shows the hypomorph T > A point mutation at position 1031 of Eed. Bar graphs show the number of clones sequenced, containing the Eedhypo or wild-type allele, per Eedhypo/+ tumor. Clones were obtained by amplifying the Eed gene from DNA (middle) or RNA (cDNA; bottom) extracted from heterozygous tumors. (D) Wright-Giemsa stainings of representative cytologic preparations of T and B lymphomas and their healthy counterparts from wild-type and Eed mutant mice. *No healthy lymph node preparations could be obtained.
Mice from all 3 genotypes (Eedhypo/hypo, Eedhypo/+ and wild-type) developed T-cell leukemias at approximately the same frequency, varying between 64% and 73%. B-cell leukemias were also observed, but at much lower frequencies (10% Eedhypo/hypo, 13% Eedhypo/+, and 19% wild-type). Flow cytometric analysis confirmed the T and B lymphoid origin of the tumors (data not shown). Histologic analysis of the hematopoietic tissues of these mice showed infiltration by blastlike lymphoid cells reminiscent of acute lymphoid leukemia (Figure 1D). In most cases, tumor cells had infiltrated the bone marrow, spleen and thymus, liver, lung, and kidney of the animals (Figure 1D; Table S1). Analysis of the T-cell receptor β chain and the immunoglobulin KC light chain rearrangement patterns by Southern blot indicated that the majority of MMLV-induced lymphomas were monoclonal (data not shown). The phenotype of 5 Eedhypo/hypo, 11 Eedhypo/+, and 3 wild-type hematopoietic tumors could not be precisely characterized, although histologic staining confirmed death from lymphoid leukemia (Table S1; data not shown).
Identification of CIS in Eed leukemia/lymphoma
Inverse PCR was used to locate MMLV integrations in a panel of 25 Eedhypo/hypo and 26 wild-type lymphoid tumors (Figures 2A, S2). Mapping by UCSC BLAT searches on the mouse genome revealed 321 retroviral insertion sites (RISs) (Figure S2). To determine which insertions constitute a CIS, we used criteria proposed by Suzuki et al.6 When considering a total of 321 insertions, the probability of 2 or more insertions to occur in a window of 41 kb is less than .005. Based on this analysis, 33 CISs were identified (Table 2; Figure 2A), of which 14 were novel, including the Fgfr3, Sh2d3c, and Zbtb7b loci (Table 2). The Bmi1, Zbtb7b, and Zfpn1a1 loci were specifically targeted in the Eedhypo/hypo-sensitized background (12%-16% of the tumors analyzed), whereas the Nnp1 and Sh2d3c loci were targeted only in wild-type tumors. However, expression of these genes did not differ significantly between Eed mutant and wild-type tumors (Figure 2; see next paragraph). The CIS near the Pim1 gene showed a bias, although not statistically significant, for the Eedhypo/hypo genotype (44% Eedhypo/hypo tumors vs 12% wild-type; Table 2; see adjusted P value). However, Pim1 overexpression does not cooperate with loss of function of Eed in tumor development, because no acceleration of the tumor onset was observed in compound Pim1 Tg/Eedhypo mutant mice compared with Pim1 transgenic (Tg) mice after more than 400 days (Pim1 Tg, n = 6 mice; Pim1 Tg/Eedhypo/+, n = 26 mice; Pim1 Tg/Eedhypo/hypo, n = 9 mice; data not shown). For all the other CISs analyzed, the difference between the number of integrations found in Eed mutant and wild-type tumors was not statistically significant either (Table 2). It remains possible that some of the identified CISs, like Zbtb7b or Fgfr3, effectively cooperates with the loss of function of Eed. Further functional studies will be required to clarify this possibility. Therefore, we conclude that the frequency and specificity of viral integrations in the genome is not affected by the functional status of Eed.
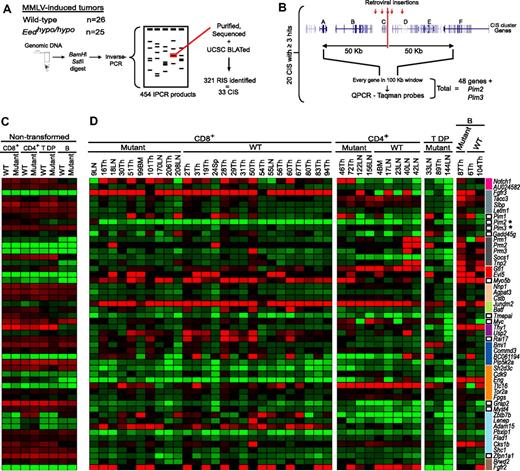
Quantitative analysis of gene-expression levels surrounding CIS clusters in Eedhypo/hypo and wild-type tumors. (A) Schematic representation of the retroviral insertional mutagenesis screen. Briefly, DNA was extracted from Eed mutant or wild-type MMLV-induced lymphomas, and inverse PCR was used to amplify genomic DNA flanking the retroviral integrations. Amplicons were purified, sequenced, and BLATed against the mouse genome. RIS indicates retroviral insertion site; and CIS, common insertion site. (B) Schematic representation of the strategy used to assay expression of genes surrounding a CIS. The median position for every CIS that was found in at least 3 independent MMLV-induced lymphomas was first determined. Genes encompassed in a region spanning 50 kb upstream and downstream of the median CIS (100-kb window) were then assayed for their expression levels by quantitative RT-PCR using Taqman assays. A total of 48 different genes, plus 2 other CISs unrelated to this study (Pim2, Pim3) were analyzed. (C) Color gradient representation of the ΔCt values of CIS-associated genes in nontransformed CD4+, CD8+, CD4+CD8+ (T DP), or B200+IgM+ (B) lymphoid cells from healthy mice. Phenotype and genotypes of the analyzed cells are shown on top. Genes analyzed are shown to the right of panel D. * indicates known CISs unrelated to this study. Reactions were done in triplicate, and average values were calculated. Average ΔCt values were determined using the 18S rRNA as the internal endogenous control gene to normalize the levels of target gene expression. Low ΔCt values denote high expression levels. Average Ct values for the 18S rRNA endogenous control was approximately 9. Mutant indicates Eedhypo/hypo; WT, wild-type. (D) Relative fold differences in gene expression levels of Eedhypo/hypo mutant and wild-type MMLV-induced lymphomas compared with nontransformed wild-type lymphoid cells. The phenotype, genotype, and ID of the analyzed tumor cells are shown on top. Genes analyzed are shown to the right. Genes are grouped by CIS cluster, indicated by a colored bar beside the gene name. Reactions were done in triplicate, and average values were calculated. Relative fold differences were determined using the ΔΔCt method. The corresponding wild-type nontransformed cells were used as the reference calibrators to assess relative fold differences in expression levels in tumors. Differences of less than −5-fold or more than 5-fold were considered significant. Tumor tissues taken for quantitative RT-PCR contained 75% to 100% lymphoid blasts. T DP indicates CD4+CD8+; B, B220+IgM+; Mutant, Eedhypo/hypo; and WT, wild-type.
Quantitative analysis of gene-expression levels surrounding CIS clusters in Eedhypo/hypo and wild-type tumors. (A) Schematic representation of the retroviral insertional mutagenesis screen. Briefly, DNA was extracted from Eed mutant or wild-type MMLV-induced lymphomas, and inverse PCR was used to amplify genomic DNA flanking the retroviral integrations. Amplicons were purified, sequenced, and BLATed against the mouse genome. RIS indicates retroviral insertion site; and CIS, common insertion site. (B) Schematic representation of the strategy used to assay expression of genes surrounding a CIS. The median position for every CIS that was found in at least 3 independent MMLV-induced lymphomas was first determined. Genes encompassed in a region spanning 50 kb upstream and downstream of the median CIS (100-kb window) were then assayed for their expression levels by quantitative RT-PCR using Taqman assays. A total of 48 different genes, plus 2 other CISs unrelated to this study (Pim2, Pim3) were analyzed. (C) Color gradient representation of the ΔCt values of CIS-associated genes in nontransformed CD4+, CD8+, CD4+CD8+ (T DP), or B200+IgM+ (B) lymphoid cells from healthy mice. Phenotype and genotypes of the analyzed cells are shown on top. Genes analyzed are shown to the right of panel D. * indicates known CISs unrelated to this study. Reactions were done in triplicate, and average values were calculated. Average ΔCt values were determined using the 18S rRNA as the internal endogenous control gene to normalize the levels of target gene expression. Low ΔCt values denote high expression levels. Average Ct values for the 18S rRNA endogenous control was approximately 9. Mutant indicates Eedhypo/hypo; WT, wild-type. (D) Relative fold differences in gene expression levels of Eedhypo/hypo mutant and wild-type MMLV-induced lymphomas compared with nontransformed wild-type lymphoid cells. The phenotype, genotype, and ID of the analyzed tumor cells are shown on top. Genes analyzed are shown to the right. Genes are grouped by CIS cluster, indicated by a colored bar beside the gene name. Reactions were done in triplicate, and average values were calculated. Relative fold differences were determined using the ΔΔCt method. The corresponding wild-type nontransformed cells were used as the reference calibrators to assess relative fold differences in expression levels in tumors. Differences of less than −5-fold or more than 5-fold were considered significant. Tumor tissues taken for quantitative RT-PCR contained 75% to 100% lymphoid blasts. T DP indicates CD4+CD8+; B, B220+IgM+; Mutant, Eedhypo/hypo; and WT, wild-type.
MMLV CISs identified in Eedhypo/hypo and wild-type lymphomas
| CIS candidate gene . | Protein family . | Mouse chromosome . | Nb insertions . | Total insertions . | P . | Adjusted P . | Human chromosome . | Human diseases* . | |
|---|---|---|---|---|---|---|---|---|---|
| Eedhypo/hypo . | Wild-type . | ||||||||
| Notch1† | Transmembrane receptor | 2qA3 | 18 | 12 | 30 | .999 | .999 | 9q34.3 | T-ALL |
| Fgfr3‡ | NA/Tyr kinase receptor | 5qB1 | 6 | 9 | 15 | .541 | .965 | 4p16.3 | Multiple myeloma, T lymphoma |
| Pim1 | Ser/Thr kinase | 17qA3.3 | 11 | 3 | 14 | .027 | .545 | 6p21.2 | NHL, DLCL |
| Gadd45g | DNA-damage inducible gene | 13qA5 | 6 | 6 | 12 | .743 | .999 | 9q22.2 | — |
| Prm2‡ | Histone protein | 16qA1 | 5 | 4 | 9 | .999 | .999 | 16p13.13 | — |
| Gfi1 | Zinc finger protein | 5qE5 | 2 | 5 | 7 | .419 | .914 | 1p22.1 | — |
| Myo5b‡ | Myosin protein | 18qE2 | 4 | 3 | 7 | .703 | .999 | 18q21.1 | — |
| Nnp1‡ | Nuclear protein | 10qC1 | 0 | 6 | 6 | .023 | .545 | 21q22.3 | — |
| Jundm2 | Transcription factor | 12qD1 | 2 | 4 | 6 | .668 | .999 | 14q24.3 | — |
| Tmepai | Signal transduction | 2qH4 | 3 | 3 | 6 | .999 | .999 | 20q13.32 | — |
| Myc | Transcription factor | 15qD1 | 2 | 3 | 5 | .999 | .999 | 8q24.21 | Burkitt lymphoma, B-CLL |
| Thy1‡ | Membrane glycoprotein | 9qA5.2 | 1 | 4 | 5 | .350 | .914 | 11q23.3 | — |
| Rai17 | SP-RING Zinc finger protein | 14qA3 | 1 | 4 | 5 | .350 | .914 | 10q22.3 | — |
| Bmi1 | Polycomb protein | 2qA3 | 4 | 0 | 4 | .110 | .755 | 10p12.2 | — |
| Sh2d3c‡ | Signal transduction | 2qB | 0 | 4 | 4 | .110 | .755 | 9q34.11 | — |
| Grap2‡ | Signal transduction | 15qE1 | 1 | 3 | 4 | .999 | .999 | 22q13.1 | — |
| Myst4‡ | Histone acetyltransferase | 14qA2 | 2 | 2 | 4 | .999 | .999 | 10q22.2 | AML |
| Zbtb7b‡ | POZ/Krueppel factor/NA | 3qF1 | 3 | 0 | 3 | .110 | .755 | 1q22 | — |
| Zfpn1a1 | Zinc finger protein | 11qA1 | 3 | 0 | 3 | .110 | .755 | 7p12.2 | ALL, DLBL |
| Brwd2 | WD-repeat protein | 7qF3 | 1 | 2 | 3 | .999 | .999 | 6q14.1 | — |
| Fli1/Ets1 | Transcription factor/transcription factor | 9qA4 | 2 | 0 | 2 | .235 | .914 | 11q24.3 | — |
| Runx3 | Transcription factor | 4qD3 | 2 | 0 | 2 | .235 | .914 | 1p36.11 | — |
| Hirip3‡ | NA | 7qF3 | 0 | 2 | 2 | .490 | .914 | 16p11.2 | — |
| Hhex | Homeobox protein | 19qC2 | 0 | 2 | 2 | .490 | .914 | 10q23.33 | — |
| Idb3/E2F2 | Transcription factor/HLH factor | 4qD3 | 0 | 2 | 2 | .490 | .914 | 1p36.12 | — |
| Lfng | Glycosyltransferase | 5qG2 | 0 | 2 | 2 | .490 | .914 | 7p22.3 | — |
| Pex5 | Peroxisomal targeting signal | 6qF2 | 0 | 2 | 2 | .490 | .914 | 12p13.31 | — |
| Supt4h2/Bzrap1 | Transcription initiation factor/NA | 11qC | 0 | 2 | 2 | .490 | .914 | 17q23.2 | — |
| Eml4 | WD-repeat protein | 17qE4 | 1 | 1 | 2 | .999 | .999 | 2p21 | — |
| Ris2 | DNA replication factor | 8qE1 | 1 | 1 | 2 | .999 | .999 | 16q24.3 | — |
| Supt6h‡ | Transcription initiation factor | 11qB5 | 1 | 1 | 2 | .999 | .999 | 17q11.2 | — |
| Zfp217 | Zinc finger protein | 2qH3 | 1 | 1 | 2 | .999 | .999 | 20q13.2 | — |
| Zfp644‡ | Zinc finger protein | 5qE5 | 1 | 1 | 2 | .999 | .999 | 1qp22.2 | — |
| CIS candidate gene . | Protein family . | Mouse chromosome . | Nb insertions . | Total insertions . | P . | Adjusted P . | Human chromosome . | Human diseases* . | |
|---|---|---|---|---|---|---|---|---|---|
| Eedhypo/hypo . | Wild-type . | ||||||||
| Notch1† | Transmembrane receptor | 2qA3 | 18 | 12 | 30 | .999 | .999 | 9q34.3 | T-ALL |
| Fgfr3‡ | NA/Tyr kinase receptor | 5qB1 | 6 | 9 | 15 | .541 | .965 | 4p16.3 | Multiple myeloma, T lymphoma |
| Pim1 | Ser/Thr kinase | 17qA3.3 | 11 | 3 | 14 | .027 | .545 | 6p21.2 | NHL, DLCL |
| Gadd45g | DNA-damage inducible gene | 13qA5 | 6 | 6 | 12 | .743 | .999 | 9q22.2 | — |
| Prm2‡ | Histone protein | 16qA1 | 5 | 4 | 9 | .999 | .999 | 16p13.13 | — |
| Gfi1 | Zinc finger protein | 5qE5 | 2 | 5 | 7 | .419 | .914 | 1p22.1 | — |
| Myo5b‡ | Myosin protein | 18qE2 | 4 | 3 | 7 | .703 | .999 | 18q21.1 | — |
| Nnp1‡ | Nuclear protein | 10qC1 | 0 | 6 | 6 | .023 | .545 | 21q22.3 | — |
| Jundm2 | Transcription factor | 12qD1 | 2 | 4 | 6 | .668 | .999 | 14q24.3 | — |
| Tmepai | Signal transduction | 2qH4 | 3 | 3 | 6 | .999 | .999 | 20q13.32 | — |
| Myc | Transcription factor | 15qD1 | 2 | 3 | 5 | .999 | .999 | 8q24.21 | Burkitt lymphoma, B-CLL |
| Thy1‡ | Membrane glycoprotein | 9qA5.2 | 1 | 4 | 5 | .350 | .914 | 11q23.3 | — |
| Rai17 | SP-RING Zinc finger protein | 14qA3 | 1 | 4 | 5 | .350 | .914 | 10q22.3 | — |
| Bmi1 | Polycomb protein | 2qA3 | 4 | 0 | 4 | .110 | .755 | 10p12.2 | — |
| Sh2d3c‡ | Signal transduction | 2qB | 0 | 4 | 4 | .110 | .755 | 9q34.11 | — |
| Grap2‡ | Signal transduction | 15qE1 | 1 | 3 | 4 | .999 | .999 | 22q13.1 | — |
| Myst4‡ | Histone acetyltransferase | 14qA2 | 2 | 2 | 4 | .999 | .999 | 10q22.2 | AML |
| Zbtb7b‡ | POZ/Krueppel factor/NA | 3qF1 | 3 | 0 | 3 | .110 | .755 | 1q22 | — |
| Zfpn1a1 | Zinc finger protein | 11qA1 | 3 | 0 | 3 | .110 | .755 | 7p12.2 | ALL, DLBL |
| Brwd2 | WD-repeat protein | 7qF3 | 1 | 2 | 3 | .999 | .999 | 6q14.1 | — |
| Fli1/Ets1 | Transcription factor/transcription factor | 9qA4 | 2 | 0 | 2 | .235 | .914 | 11q24.3 | — |
| Runx3 | Transcription factor | 4qD3 | 2 | 0 | 2 | .235 | .914 | 1p36.11 | — |
| Hirip3‡ | NA | 7qF3 | 0 | 2 | 2 | .490 | .914 | 16p11.2 | — |
| Hhex | Homeobox protein | 19qC2 | 0 | 2 | 2 | .490 | .914 | 10q23.33 | — |
| Idb3/E2F2 | Transcription factor/HLH factor | 4qD3 | 0 | 2 | 2 | .490 | .914 | 1p36.12 | — |
| Lfng | Glycosyltransferase | 5qG2 | 0 | 2 | 2 | .490 | .914 | 7p22.3 | — |
| Pex5 | Peroxisomal targeting signal | 6qF2 | 0 | 2 | 2 | .490 | .914 | 12p13.31 | — |
| Supt4h2/Bzrap1 | Transcription initiation factor/NA | 11qC | 0 | 2 | 2 | .490 | .914 | 17q23.2 | — |
| Eml4 | WD-repeat protein | 17qE4 | 1 | 1 | 2 | .999 | .999 | 2p21 | — |
| Ris2 | DNA replication factor | 8qE1 | 1 | 1 | 2 | .999 | .999 | 16q24.3 | — |
| Supt6h‡ | Transcription initiation factor | 11qB5 | 1 | 1 | 2 | .999 | .999 | 17q11.2 | — |
| Zfp217 | Zinc finger protein | 2qH3 | 1 | 1 | 2 | .999 | .999 | 20q13.2 | — |
| Zfp644‡ | Zinc finger protein | 5qE5 | 1 | 1 | 2 | .999 | .999 | 1qp22.2 | — |
P value is based on the Fisher exact test. Adjusted P value applies the Benjamini-Hochberg correction.
T-ALL indicates T-cell acute lymphoblastic leukemia; NA, not available; NHL, non-Hodgkin leukemia; DLCL, diffuse large-cell leukemia; B-CLL, B-cell chronic lymphocytic leukemia; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; DLBL, diffuse large B-cell lymphoma; and —, not identified in Human blood diseases.
Genes implicated in human blood diseases according to the Cancer Gene Census (www.sanger.ac.uk/genetics/CGP/Census).
Some tumors contained more than 1 MMLV integration in Notch1. Frequency of tumors with Notch1 MMLV integration: Wild-type, 10 of 26; Eedhypo/hypo, 9 of 25. See Table S2 for details.
New CIS identified in this screen.
Common and cell type–specific expression of CIS-associated genes in lymphoid tumors
Because viral enhancers can affect gene expression over long distances,4 we determined the median genomic location of each CIS identified in 3 or more Eedhypo/hypo and wild-type tumors and designed Q-RT-PCR assays for each gene (hereafter called CIS-associated genes; n = 48) located within 50 kb of distance from the median integration site (Figure 2B). The basal expression level of 48 CIS-associated genes was first determined in sorted nontransformed T cells (CD4+, CD8+, and CD4+CD8+) and B cells isolated from healthy Eedhypo/hypo mice (before MMLV infection), and compared with that of wild-type littermates. Results from these studies indicated that the loss of 2 functional alleles of Eed does not alter the expression level of any of these CIS-associated genes (Figure 2C; compare wild-type [WT] vs Mutant lanes).
Tissues that were massively (75%-100%) infiltrated with neoplastic cells (Table S1) were also evaluated for expression of CIS-associated genes relative to untransformed (control) tissues. Analyses of these Q-RT-PCR studies performed in triplicate are presented in Figure 2D (levels are expressed as fold difference compared with untransformed cells; light green and bright red indicates ≤ or ≥ 10-fold above values determined in control cells shown in Figure 2C, respectively). Strikingly, most tumors contained a large number of aberrantly expressed CIS-associated genes (average = 13 with ≥ 5-fold difference). Overall, and in agreement with the results recently obtained by Theodorou et al,26 approximately half (ie, 54%) of the recurrent and proven retroviral integrations lead to detectable deregulation of at least 1 CIS-associated gene (≥ 5-fold difference). In contrast, several CIS-associated genes were deregulated in tumors for which no proviral integrations in the corresponding CISs could be identified. For example, proviral integrations in the Myo5b gene were found in 7 different tumors (Table 2), of which only 2 (tumors 6Th and 72Th; Figure 2D) showed deregulated expression of this gene. However, significant overexpression of this gene was detected in 10 other tumors, for which no proviral insertions in this region could be detected (for details on RIS and expression levels of genes in each individual tumors, see Tables S2–S3). Similar findings were also reported by Theodorou et al26 for 4 different CIS-associated genes. Overexpression of these genes in absence of nearby retroviral integrations could be caused by the activation of upstream regulators in the same pathway.
As previously mentioned, it is difficult to predict aberrant gene expression based on integration data alone (Figure 3A-C; see MMLV plus signs and arrows in left and right panels and compare impact of expression of the genes for each tumor). Nevertheless, the Q-RT-PCR studies showed that genes located near a CIS are more frequently overexpressed in tumors than genes found in nontargeted regions of the genome (Figures 2D, S3A). The data also show that multiple genes are often deregulated for a given CIS. For the studies presented in Figure 3A,B, 58% and 54% of the tumors aberrantly express 2 or more genes within approximately 100 kb of the Zbtb7b (Figure 3A) and Sh2d3c (Figure 3B) loci, respectively. For all the other groups of CIS-associated genes tested (Figure 2D; see color code in last row), less than 16% of the tumors showed deregulated expression of 2 or more CIS-associated genes.
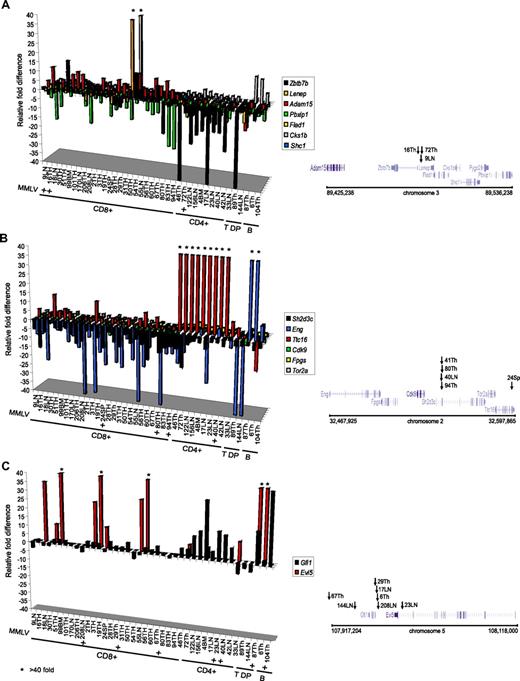
Cell type–specific expression patterns of CIS-associated genes in lymphomas. Relative fold differences in expression levels of genes surrounding a CIS at the Zbtb7b/Adam15/Pbxip1 (A), Sh2d3c/Ttc16/Eng (B), and Gfi1/Evi5 loci (C) in MMLV-induced lymphoid tumors from Eed mutant and wild-type mice. Expression data corresponds to data in Figure 2D. Tumor IDs and phenotypes are shown at the bottom of each graph. * indicates samples with expression level differences higher than 40-fold; and + indicates MMLV retroviral insertions identified by inverse PCR. Positioning of genes and retroviral insertions (arrows with tumor ID) identified at each CIS cluster are depicted on the right panels. Chromosome number and location (in bp) for each CIS cluster are indicated at the bottom of the right panel. B indicates B220+IgM+; T DP, CD4+CD8+.
Cell type–specific expression patterns of CIS-associated genes in lymphomas. Relative fold differences in expression levels of genes surrounding a CIS at the Zbtb7b/Adam15/Pbxip1 (A), Sh2d3c/Ttc16/Eng (B), and Gfi1/Evi5 loci (C) in MMLV-induced lymphoid tumors from Eed mutant and wild-type mice. Expression data corresponds to data in Figure 2D. Tumor IDs and phenotypes are shown at the bottom of each graph. * indicates samples with expression level differences higher than 40-fold; and + indicates MMLV retroviral insertions identified by inverse PCR. Positioning of genes and retroviral insertions (arrows with tumor ID) identified at each CIS cluster are depicted on the right panels. Chromosome number and location (in bp) for each CIS cluster are indicated at the bottom of the right panel. B indicates B220+IgM+; T DP, CD4+CD8+.
Another important finding was that the aberrant expression of the CIS-associated genes is independent of Eed's activity in tumor cells. As shown in Figure 2D, no obvious differences in gene expression patterns were observed between Eed mutant and wild-type tumors. However, we observed a remarkable degree of similarity in gene expression patterns within tumor subgroups (eg, CD4+ vs CD8+; Figure 2D). For example, the Fgfr3 gene, which is expressed at very low levels in nontransformed T or B cells (ΔCT value > 26; Figure 2C), is highly expressed (up to 1000-fold) in 92% of CD8+ tumors, while only 33% of CD4+ and no double-positive (T DP) tumors show expression of this gene (Figure 2D). Eng, Myo5b, and Thy1 show a similar pattern in nontransformed and transformed pre-B cells (Figure 2D). When arbitrarily considering a more than 5-fold difference threshold as significant, the desintegrin metalloproteinase Adam15 is overexpressed in 54% CD8+ tumors, but not in other lymphoid cell types (Figure 2D; red bars in Figure 3A). Interestingly, the Adam15 gene is located approximately 45 kb upstream of the median CIS chromosomal location (Figure 3A right). The CIS-associated genes Zbtb7b and Pbxip1 (Figure 3A right) are located at 43 and 86 kb away from Adam15 and show very different expression patterns. While Zbtb7b is down-regulated in most CD4+ tumors analyzed (8 of 9; see CD4+ in Figure 3A left), its expression is not significantly affected in the majority of CD8+ samples (Figure 3A). In contrast, Pbxip1 is down-regulated (11-fold average) in CD8+ (77%) tumors, and to a lesser extent in CD4+ tumors (6-fold average in 56% of tumors).
A similar analysis is shown in Figure 3B,C for 2 additional sets of CIS-associated genes, Sh2d3c/Eng/Ttc16 and Gfi1/Evi5 (Figure 3B,C, respectively). Sh2d3c/Eng/Ttc16 distribution around a CIS located between exons 1 and 2 of Sh2d3c is shown in Figure 3B (right). All CD4+ tumors exhibit an average of 309-fold increase in the tetratricopeptide repeat gene Ttc16 compared with nontransformed controls, whereas only a fraction (less than 20%) of CD8+ tumors show overexpression (less than 9-fold average; red bars in Figure 3B indicate a more than 40-fold difference in expression levels). Sh2d3c (targeted in 4 tumors; Figure 3B right) is down-regulated on average 9-fold in 69% of CD8+ and 44% of CD4+ tumors, whereas Eng (Figure 3B white bars) is strongly down-regulated in double positives and in several CD8+ tumors. The expression analysis of the Gfi1/Evi5 group of CIS-associated genes provides further evidence for cell type–specific regulation of gene expression (Figure 3C). For example, Gfi1 is overexpressed in 88% of CD4+ and in less than 4% CD8+ tumors, while Evi5 expression shows the opposite pattern. These data suggest that the cellular context has an important impact on CIS-associated gene expression in the tumors analyzed.
Our studies also led to the identification of genes for which expression is affected in a large spectrum of lymphoid tumors. Jundm2 is overexpressed in almost every lymphoid tumor analyzed (88%), independently of the subtype (Figure 2C). Although the Fgfr2 gene is expressed at relatively low levels in tumors (Table S3), it is overexpressed in 54% of CD8+ and 67% of CD4+ tumors compared with nontransformed cells (Figure 2C). The Pim2 gene is down-regulated in 92% of CD8+, 100% of CD4+, and 2 of 3 double-positive (CD4+CD8+) leukemias. This suggests that these genes may have a broader role in T-cell tumor formation.
We next performed a series of control experiments to determine whether the expression levels of genes not associated with CISs differ between tumor and control cells. For this, we randomly selected 23 genes across the genome, located distantly from known common integration sites, as well as 8 Polycomb (and PcG-related) genes and assessed their expression levels in both nontransformed cells and MMLV-infected lymphoid tumors. Results from these studies showed that 16% (5 of 31) of these randomly selected genes were down-regulated in tumorigenic cells, whereas none of these genes were consistently overexpressed (except for Ece2; Figure S3B). This is in contrast with most of the CIS-associated genes tested, which were consistently up-regulated in tumor cells (8 of 50 CIS-associated genes vs 1 of 31 randomly selected genes). According to the Kolmogorov-Smirnov test, this increase in the proportion of CIS-associated genes that are up-regulated in tumorigenic cells is statistically significant (P < .001; Document S1; Figure S3C). Importantly, using an alternative threshold-based test, this result was confirmed and shown to be independent from the specific threshold used (Document S1; Figure S3D). Together, these data indicate that heterogeneity between the tumor cells and their corresponding normal counter-parts does not explain the global up-regulation in CIS-associated gene expression.
The results presented here identify Fgfr3, Fgfr2, Ttc16, Adam15, Evi5, Jundm2, and Myo5b as candidate leukemia-associated genes, some of which are novel. Q-RT-PCR on CIS-associated genes thus reveals a series of candidate leukemia-associated genes and suggests an unsuspected complexity in the regulation of defined gene clusters with oncogenic relevance. Data presented in this report also suggests conservation in the genetic programs leading to lymphoid tumor formation.
Discussion
In this study, we have developed a combinatorial approach which exploits proviral insertional mutagenesis, bioinformatics, and high-throughput Q-RT-PCR for the efficient identification of 7 candidate leukemia-associated genes (Fgfr3, Fgfr2, Ttc16, Adam15, Evi5, Jundm2, and Myo5b) from a list of 50 prospects (located within 50 kb in average of common integration sites). The power of this new approach is validated by studies which confirmed implication of at least 4 of these 7 genes in human cancers (Fgfr3 in multiple myeloma, Fgfr2 in breast cancer, and Adam15 in breast, prostate, and stomach cancers)27,,,,–32 and in the transformation of chicken embryonic fibroblasts (Jundm2).33 Results from our high-throughput expression analysis also revealed an unsuspected complexity in the deregulation of genes at candidate cancer loci, and underline the unpredictable effect of proviral insertional mutagenesis on the expression levels of CIS-associated genes. Most interestingly, we identified genes which deregulation is common to all subtypes of lymphoid leukemias analyzed (Jundm2, Fgfr2), thus suggesting the existence of shared genetic alterations in these tumors. Finally, we successfully identified 14 new CISs in this study (Table 2). The distribution of insertions found in our wild-type tumors differs substantially from most tumor sets in the RTCGD database. The use of a hybrid (101/C3Hf) background mouse strain, different from other reported proviral insertional mutagenesis screens, likely explains this high number of new CISs identified and the difference in the distribution of the insertions in this study.
The power of insertional mutagenesis in predicting a region containing cancer genes appears relatively high because 95% (19 of 20) of CIS clusters analyzed in this study (Figure 2D right, color-coded groups) contained at least 1 gene that was aberrantly expressed in a large proportion of our leukemias (summarized in Table 3). Moreover, results presented in Figure 3 indicate that several cancer genes are often clustered in a given CIS, and that aberrant expression frequently correlates with tumor type. Thus, a CIS identified in a CD8+ leukemia sometimes implies deregulation of a different gene than if identified in a CD4+ tumor. Aberrant expression of Ttc16 in CD4+ leukemia and of Eng in CD8+ leukemia best illustrates this point for the Sh2d3c CIS (Figure 3B). These conclusions are obviously dependent on the difference in gene-expression levels between normal and neoplastic cells, and explain our initial focus on lymphoid tumors for which normal (untransformed) counterparts are easily purified. Indeed, the expression levels of the CIS-associated genes did not vary much between the 3 different types of normal T cells (Figure 2C; CD8+ vs CD4+ and T DP) and pre-B cells, except for the T-cell marker Thy1. This kind of analysis might be difficult to perform with myeloid tumors, although recent purification strategies for stem and myeloid progenitors may provide new possibilities for extending these studies.34 When interpreting the gene-expression results, one must also keep in mind that retroviral insertions can create truncated dominant gain-of-function mutations that might contribute to leukemogenesis without obvious alterations in transcript levels. Point mutations acquired during tumorigenic process can also affect normal gene function. A parallel screen assessing transcript integrity of CIS-associated genes will likely uncover new oncogenes and will be complementary to expression studies to further understand the events leading to leukemia.
Percentage of tumors aberrantly expressing 1 or more gene per CIS cluster
| CIS cluster ID* . | CD8+, % . | CD4+, % . | T DP, % . | B, % . | Total, % . |
|---|---|---|---|---|---|
| 1 (Bmi1) | 57.7 | 100.0 | 100.0 | 33.3 | 68.3 |
| 2 (Fgfr3) | 92.3 | 88.9 | 66.7 | 66.7 | 87.8 |
| 3 (Pim1) | 19.2 | 55.6 | 66.7 | 66.7 | 34.1 |
| 4 (Pim2) | 92.3 | 100.0 | 66.7 | 33.3 | 87.8 |
| 5 (Pim3) | 3.8 | 0.0 | 0.0 | 0.0 | 2.4 |
| 6 (Sh2d3c) | 96.2 | 100.0 | 100.0 | 100.0 | 97.6 |
| 7 (Zbtb7b) | 96.2 | 100.0 | 66.7 | 100.0 | 95.1 |
| 8 (Jundm2) | 88.5 | 88.9 | 66.7 | 66.7 | 85.4 |
| 9 (Gfi1) | 34.6 | 77.8 | 33.3 | 100.0 | 48.8 |
| 10 (Nnp1) | 7.7 | 0.0 | 0.0 | 0.0 | 4.9 |
| 11 (Brwd2) | 61.5 | 66.7 | 100.0 | 33.3 | 63.4 |
| 12 (Thy1) | 34.6 | 11.1 | 33.3 | 0.0 | 26.8 |
| 13 (Prm2) | 19.2 | 77.8 | 100.0 | 100.0 | 43.9 |
| 14 (Notch1) | 50.0 | 55.6 | 66.7 | 33.3 | 51.2 |
| 15 (Grap2) | 26.9 | 66.7 | 66.7 | 33.3 | 39.0 |
| 16 (Myc) | 0.0 | 55.6 | 33.3 | 33.3 | 17.1 |
| 17 (Zfpn1a1) | 23.1 | 11.1 | 33.3 | 0.0 | 19.5 |
| 18 (Tmepai) | 46.2 | 44.4 | 66.7 | 0.0 | 43.9 |
| 19 (Myst4) | 7.7 | 33.3 | 33.3 | 0.0 | 14.6 |
| 20 (Rai17) | 23.1 | 0.0 | 33.3 | 0.0 | 17.1 |
| 21 (Gadd45g) | 15.4 | 44.4 | 66.7 | 0.0 | 24.4 |
| 22 (Myo5b) | 65.4 | 33.3 | 33.3 | 100.0 | 58.5 |
| CIS cluster ID* . | CD8+, % . | CD4+, % . | T DP, % . | B, % . | Total, % . |
|---|---|---|---|---|---|
| 1 (Bmi1) | 57.7 | 100.0 | 100.0 | 33.3 | 68.3 |
| 2 (Fgfr3) | 92.3 | 88.9 | 66.7 | 66.7 | 87.8 |
| 3 (Pim1) | 19.2 | 55.6 | 66.7 | 66.7 | 34.1 |
| 4 (Pim2) | 92.3 | 100.0 | 66.7 | 33.3 | 87.8 |
| 5 (Pim3) | 3.8 | 0.0 | 0.0 | 0.0 | 2.4 |
| 6 (Sh2d3c) | 96.2 | 100.0 | 100.0 | 100.0 | 97.6 |
| 7 (Zbtb7b) | 96.2 | 100.0 | 66.7 | 100.0 | 95.1 |
| 8 (Jundm2) | 88.5 | 88.9 | 66.7 | 66.7 | 85.4 |
| 9 (Gfi1) | 34.6 | 77.8 | 33.3 | 100.0 | 48.8 |
| 10 (Nnp1) | 7.7 | 0.0 | 0.0 | 0.0 | 4.9 |
| 11 (Brwd2) | 61.5 | 66.7 | 100.0 | 33.3 | 63.4 |
| 12 (Thy1) | 34.6 | 11.1 | 33.3 | 0.0 | 26.8 |
| 13 (Prm2) | 19.2 | 77.8 | 100.0 | 100.0 | 43.9 |
| 14 (Notch1) | 50.0 | 55.6 | 66.7 | 33.3 | 51.2 |
| 15 (Grap2) | 26.9 | 66.7 | 66.7 | 33.3 | 39.0 |
| 16 (Myc) | 0.0 | 55.6 | 33.3 | 33.3 | 17.1 |
| 17 (Zfpn1a1) | 23.1 | 11.1 | 33.3 | 0.0 | 19.5 |
| 18 (Tmepai) | 46.2 | 44.4 | 66.7 | 0.0 | 43.9 |
| 19 (Myst4) | 7.7 | 33.3 | 33.3 | 0.0 | 14.6 |
| 20 (Rai17) | 23.1 | 0.0 | 33.3 | 0.0 | 17.1 |
| 21 (Gadd45g) | 15.4 | 44.4 | 66.7 | 0.0 | 24.4 |
| 22 (Myo5b) | 65.4 | 33.3 | 33.3 | 100.0 | 58.5 |
CD8+, n = 26; CD4+, n = 9; T DP (CD4+CD8+), n = 3; and B (B220+IgM+), n = 3.
Closest gene to median CIS location.
The systematic exploitation of quantitative expression arrays as shown herein may set new standards in retroviral insertional mutagenesis screens. Importantly, special care in selecting only tumor samples that are massively infiltrated (75%-100%) with neoplastic cells also helps to prevent contamination from untransformed cells in the expression analyses. In this study, the expression of the CIS-associated genes found in only 2 independent tumors (Table 2) was not analyzed by Q-RT-PCR, even if we knew of the potential importance of some of the genes involved. For example, the histone acetyltransferase Myst4 gene (also known as Morf) is fused to Cbp in acute myeloid leukemia,35 and transgenic mice overexpressing the Ris2 gene (or Cdt1) in the absence of p53 develop thymic lymphoblastic lymphoma.36 The role of these “2 hits” CISs is therefore likely to be also important in tumorigenesis. Moreover, extending gene-expression analysis over larger chromosomal distances (> 50 kb in both directions surrounding the CIS) might be informative, because a significant proportion of the retroviral integrations in CISs analyzed in this study was not accompanied by deregulation in gene expression. Extending our protocol to the 510 CISs already identified in the literature should also be informative in revealing additional cancer genes. According to the RTCGD retroviral database, a complete study of all genes encoded within 100 kb of distance of the 35 different CISs containing at least 10 hits would include approximately 120 candidate genes. Replicate analysis in a set of 100 tumors from different phenotypes would thus require 24 000 Q-RT-PCR reactions, approximately 3 times the size of this study. Considering that we identified on average 13 deregulated CIS-associated genes per tumor analyzed in our study, 2 to 3 times as many candidate genes would easily be accessible with such an extended approach.
In our gene-expression analysis, we found that the proportion of genes down-regulated in tumorigenic cells was similar between the CIS-associated genes and the randomly selected genes (5 of 50 of the CIS-associated genes compared with 4 of 31 of the randomly genes, a greater than 5-fold difference). On the contrary, a significantly higher proportion of the CIS-associated genes was up-regulated compared with the randomly selected genes (> 5-fold difference; P < .001). The exact nature of this discrepancy remains unclear, but underlies the bias of retroviral insertional mutagenesis screens and of our approach for the identification of oncogenes over tumor suppressor genes.
Using radiation and retrovirus-induced tumor strategies, we confirmed in 2 different sensitized mouse models that Eed exerts tumor-suppressive activity in lymphoid tissues. Mutation of only 1 Eed allele is sufficient to sensitize cells to transformation. A dominant-negative effect of the mutant protein is unlikely, because it is unstable and undetectable in Eednull/null cells.37 Moreover, homozygous mutants have a stronger phenotype and develop tumors more rapidly than heterozygotes. This suggests that gene dosage is important, and that slight changes in Eed levels may be sufficient to predispose mice to tumor formation. Similarly, targeted deletion of only 1 allele of the Suv39h1 H3K9 methyltransferase predisposes mice to develop B-cell lymphomas.38 Although FACS and histologic analysis of tumors suggests that loss of Eed function does not change the phenotype of lymphomas compared with wild-type counterparts, there could be some subtle differences in the differentiation state of the lymphomas that have not been detected in this study. The increased susceptibility to oncogenic transformation in Eed mutant cells is not well understood. No specific CISs were identified in Eed mutant tumors, suggesting that Eed might not sensitize cells toward distinct oncogenic pathways, but instead has a more general effect in cellular transformation. Recently, Eed was shown to interact with the DDB1-CUL4 complex, which is involved in cell-cycle progression, replication, and DNA damage repair.39 A link between Eed's main interacting partner, Ezh2, and the homologous recombination pathway of DNA repair was also established.40 Moreover, EZH2 and SUZ12 interact with PARP3, a protein involved in base excision repair.41 This suggests that increased tumor development in Eed mutants may be due to defects in DNA repair. Further experiments are needed to clarify the mechanism underlying the tumor-suppressive role of Eed. Moreover, in the era of chromatin modulation for therapeutic targets, it is postulated that the elimination of the Polycomb genes Eed, Suz12, or Ezh2 might be highly efficient to induce apoptosis in transformed cells.42 In contrast, the studies presented here document that loss of function of Eed sensitizes cells to transformation and therefore raises important concerns about potential therapeutic approaches targeting this gene.
In summary, high-throughput Q-RT-PCR to assess expression of CIS-associated genes is a powerful addition to insertional mutagenesis to identify novel cancer-associated genes. We conclude that extension of this approach to other CIS and tumor subtypes will be instrumental in uncovering novel tissue-specific and common cancer gene networks for the design of better-targeted molecular therapies.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Dr Neal Copeland for critical reading of the manuscript and to Jana Krosl and Jalila Chagraoui for insightful discussions and comments. Thanks to Pierre Chagnon and Jean Duchaine from the IRIC Genomics and High-throughput platforms, Amélie Faubert for help with the analysis of the irradiation-induced tumors, Melanie Frechette for her expertise and help with the maintenance and manipulation of the animals, Danièle Gagné from the IRIC flow cytometry platform, and Martine Dupuis and Éric Massicotte from the IRCM flow cytometry service for assistance in flow cytometry and cell sorting.
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to G.S. and by a grant from the Cancer Research Network of Fonds de la Recherche en Santé du Québec to J.H. M.S. is a recipient of a CIHR Canada Graduate Scholarship Doctoral award. G.S. holds a Canada Research Chair in Molecular Genetics of Stem Cells and is a scholar of the Leukemia & Lymphoma Society of America.
Authorship
Contribution: M.S. designed and performed research, analyzed data, and wrote the paper; M.M., J.L., and J.H. performed research; S.L did statistical analysis; and G.S. designed research and helped with writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guy Sauvageau, Institute for Research in Immunology and Cancer (IRIC), University of Montreal, CP 6128, Downtown Station, Montreal, QC, Canada H3C 3J7; e-mail: guy.sauvageau@umontreal.ca.