Abstract
Hepcidin is the principal iron regulatory hormone, controlling the systemic absorption and remobilization of iron from intracellular stores. Recent in vivo studies have shown that hepcidin is down-regulated by erythropoiesis, anemia, and hypoxia, which meets the need of iron input for erythrocyte production. Erythropoietin (EPO) is the primary signal that triggers erythropoiesis in anemic and hypoxic conditions. Therefore, a direct involvement of EPO in hepcidin regulation can be hypothesized. We report here the regulation of hepcidin expression by EPO, in a dose-dependent manner, in freshly isolated mouse hepatocytes and in the HepG2 human hepatocyte cell model. The effect is mediated through EPOR signaling, since hepcidin mRNA levels are restored by pretreatment with an EPOR-blocking antibody. The transcription factor C/EBPα showed a pattern of expression similar to hepcidin, at the mRNA and protein levels, following EPO and anti-EPOR treatments. Chromatin immunoprecipitation experiments showed a significant decrease of C/EBPα binding to the hepcidin promoter after EPO supplementation, suggesting the involvement of this transcription factor in the transcriptional response of hepcidin to EPO.
Introduction
Hepcidin has been described as the central regulator of iron homeostasis, and deficiencies in hepcidin are associated with several iron-related disorders.1 Hepcidin modulates iron homeostasis by inducing the internalization and degradation of ferroportin,2 the single known cellular iron exporter, expressed by duodenal enterocytes as well as by macrophages and hepatocytes.
Hypoxia and anemia are the 2 main signals that trigger the erythroid regulator of intestinal iron absorption, independently of iron stores.3 These signals also regulate the production of erythrocytes through synthesis of the hormone erythropoietin (EPO).4,5 The hypothesis that hypoxia could act both on erythropoiesis induction and on hepcidin down-regulation via EPO signaling was first advanced in 2002,6 based on the evidence that liver hepcidin gene expression is strongly decreased by EPO injection in vivo. The first evidence concerning a possible direct role of EPO on the regulation of hepcidin synthesis by hepatocytes, the main hepcidin-producing cells, was provided by Fein et al,7 who demonstrated a down-regulation of this protein in a hepatoma and in a pancreatic cell line after stimulation with EPO.
With the objective of clarifying the possible direct role of EPO on hepcidin regulation, we analyzed the dose-dependent effect of EPO on hepcidin levels on freshly isolated mouse hepatocytes and on the human hepatocyte cell line HepG2, which express endogenous hepcidin, EPO, and EPOR.8-10 The involvement of EPOR signaling and of the transcription factor C/EBPα was also investigated.
Methods
Animals
C57BL/6 mice 10 to 14 weeks of age were used as the source of hepatocytes. Animals were acclimatized in polyethylene cages lined with wood shavings, under a 12-hour light/12-hour dark cycle. Mice had free access to standard rat chow and drinking water. An acclimatizing period of at least 1 week was performed, before starting the experiments. Animals were anesthetized with diethyl ether before the start of the surgical procedures.
Isolation and incubation of hepatocytes
Hepatocyte isolation was performed by collagenase perfusion, as described by Moldéus et al,11 with the modifications described in Carvalho et al.12
Immediately after isolation, cell viability was determined with the trypan blue exclusion test. Viability was always more than 83%.
Since previous reports have shown that recombinant human EPO (rEPO) mimics the effect of murine EPO on mouse cells,13,14 mouse hepatocytes were incubated, immediately following isolation, with 0.01 to 2 U/mL rEPO (Sigma-Aldrich, St Louis, MO), and/or 1 or 5 μg/mL goat anti-EPO receptor (EPOR) polyclonal antibody (Sigma-Aldrich) for 3 hours, which corresponds to the incubation period where hepcidin response to rEPO was maximum (data not shown). To test for responsiveness of hepcidin transcription to an exogenous stimulus, incubation with 20 ng/mL human IL-6 (Sigma-Aldrich), for 3 hours, was performed.
Cell viability was determined after each experiment by the lactate dehydrogenase (LDH) leakage method, which was randomly confirmed by the trypan blue exclusion test. No statistical differences in cell viability were observed between any of the treatments and the nontreated control (data not shown). Viability values of 74% plus or minus 7% were obtained.
HepG2 culture and treatments
HepG2 cells were maintained in complete DMEM, (DMEM supplemented with 10% FCS and 1% penicillin/streptomycin/amphotericin). One day before treatments, 3 × 105 cells were seeded in 6-well plates and incubated overnight (O/N). Cells were then treated with 0.01 to 2.5 U/mL rEPO for 3 hours, in complete DMEM. For anti-EPOR treatments, cells were incubated with 0.1 to 10 μg/mL goat anti-EPOR for 30 minutes and then treated with 1 or 2 U/mL rEPO, when appropriate, for 3 hours. Negative control was performed by replacing the primary antibody with an irrelevant antibody (polyclonal anti-CD3, A0452; DAKO, Glostrup, Denmark). Specificity control was performed for EPOR by absorption of the primary antibody with a specific blocking peptide (sc-697-P; Santa Cruz Biotechnology, Santa Cruz, CA). The blocking peptide was combined with the primary antibody with a 5-fold excess of the peptide in BSA/TBS buffer overnight at 4°C. After treatment, cells were harvested by trypsinization, pelleted, and frozen at −80°C until required. All experiments were performed at least 3 times.
Determination of gene expression
Total RNA was extracted from cells using RNeasy Midi Kit (Qiagen, Valencia, CA) and 0.5 μg used for reverse-transcription with SuperScript First-Strand kit (Invitrogen, Frederick, MD), following the manufacturer's instructions. Assessment of mRNA levels was performed in an iCycler iQ5 polymerase chain reaction (PCR) detection system (Bio-Rad, Hercules, CA), using specific oligonucleotides (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). As an internal control, GAPDH mRNA levels were determined simultaneously. Relative expression levels were calculated as 2(Ct human GAPDH endogenous control gene − Ct gene)*10 000. For every quantitative reverse-transcription (qRT)–PCR, a dilution series consisting of 4 serial dilutions was used during optimization of the procedure. All qRT-PCR experiments were done at least in triplicate, with 2 to 3 replicates each.
Western blot analysis
Cells were trypsinized and lysed in gel loading buffer (500 mM Tris-HCl, pH 6.8, 5% sodium dodecyl sulfate (SDS), 20% glycerol, 50 μL bromophenol blue). Total cell lysate (40 μg) was boiled, separated by SDS–polyacrylamide gel electrophoresis (PAGE) (12%), and transferred to a nitrocellulose Hybond-C membrane (GE Healthcare, Little Chalfont, United Kingdom). The blots were blocked at room temperature (R/T) with 5% dry milk/0.05% Tween 20 in PBS (TBS-T), incubated with rabbit polyclonal C/EBPα antibody (Santa Cruz Biotechnology), and washed 3 times with TBS-T, and the signal was detected with goat anti–rabbit IgG–HRP conjugate (Molecular Probes, Eugene, OR), followed by incubation with the enhanced chemiluminescence substrate Super Signal West Dura (Pierce, Rockford, IL). For loading control, the membrane was stripped and hybridized with anti–β-actin (Abcam, Cambridge, United Kingdom) antibody. Quantification of band intensity was performed with ImageJ (National Institutes of Health, Bethesda, MD).15 To be able to observe C/EBPα expression even after treatment with high EPO doses, the maximum amount of total protein (40 μg) was loaded in each well, coupled with long autoradiography exposure times and use of a highly sensitivity detection substrate (Pierce's SuperSignal West Dura Extended Duration Substrate; Figure 3A).
Chromatin immunoprecipitation
Two 90-mm dishes of 70% to 80% confluent HepG2 cells, cultured and treated as previously described, were used per condition for the chromatin immunoprecipitation (ChIP) analysis. Cells were cross-linked with 0.38% formaldehyde at 37°C for 10 minutes. The fixation was stopped by adding glycine to a final concentration of 0.125 M and incubating at R/T for 5 minutes. Total cell lysate was extracted using 600 μL ChIP lysis buffer (50 mM Tris-HCl, pH 8.1, containing 1% Triton X-100, 0.1% deoxycholate, 150 mM NaCl, and 5 mM EDTA), supplemented with 1:100 protease inhibitor cocktail (Sigma-Aldrich). The DNA was sheared to an average fragment size of 500 base pairs by sonication (10 pulses of 5 seconds with 55-second rest period between each). Cell lysates were precleared with protein A–agarose beads (Santa Cruz Biotechnology) and 50 μg chromatin was immunoprecipitated overnight, at 4°C, using the polyclonal rabbit anti-C/EBPα (14AA) antibody (sc-61; Santa Cruz Biotechnology) or incubated with normal rabbit serum for the serum control (5 μg chromatin was retained for use as 10% input). Antibody-chromatin complexes were collected using protein A–agarose beads. The agarose was pelleted and washed sequentially in lysis buffer, lysis buffer with 500 mM NaCl, lysis buffer with 0.25 M LiCl, and finally in Tris/EDTA buffer (10 mM/1 mM, pH 8.0). DNA and protein were eluted from the beads by washing in the elution buffer (0.1 M NaHCO3 with 1% SDS and 20 μg/mL herring sperm DNA). The cross-links were then reversed by incubating at 65°C for 4 hours, and 10 μg proteinase K was added to the samples and incubated for an additional 1 hour, at 45°C. DNA was isolated by phenol/chloroform extraction and ethanol precipitation, and dissolved in Tris/EDTA. DNA was amplified by real-time PCR using the oligonucleotides 5′-TCATCAAACTGCTTAAC-3′ and 5′-TCTGTCTGGCTGTCCCACTGCT-3′, as forward and reverse primers, respectively, at an annealing temperature of 64°C. The results are expressed as a ratio of the input (into the reaction) to the output (after immunoprecipitation) normalized to the no-EPO control.
Statistical analysis
Results are presented as the ratio of mRNA and protein levels to the average of controls. Control mRNA and protein levels were normalized to 1. In general, one-way ANOVA was used to test for significant differences among sample means. When comparisons included the simultaneous effect of more than one variable, 2-way ANOVA was applied. When significant differences were found, Fisher least significant difference (LSD) was used to identify significantly different means. All statistical analysis was performed with STATGRAPHICS Centurion XV (Herndon, VA).
Results and discussion
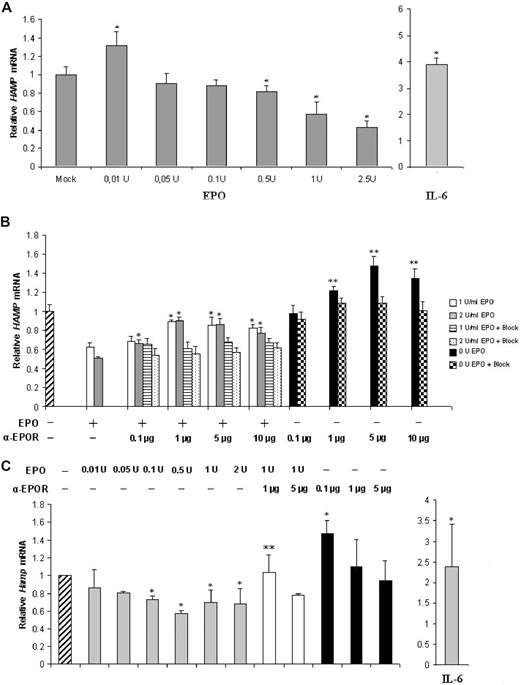
The effect of EPO on hepcidin expression was analyzed in the human liver cell line HepG2 and in freshly isolated mouse hepatocytes, which are useful cell systems for studies of hepcidin regulation in vitro.8,10 We found that hepcidin mRNA expression is modulated by EPO in HepG2 cells and in mouse hepatocytes in a dose-dependent manner (Figure 1A,C). In HepG2 cells, supplementation with 0.01 U induced a moderate but significant increase in hepcidin, while the highest EPO doses (0.5-2.5 U) caused a significant down-regulation (up to 60% decrease) of hepcidin expression (Figure 1A). Hepcidin 1 expression was down-regulated in mouse hepatocytes at all EPO concentrations examined, with the exception of the lowest 0.01 U (Figure 1C). Down-regulation was, however, less pronounced than in the HepG2 cell line, with a lowest value of approximately 40%, for 0.5 U EPO. Treatment with IL-6, a strong inducer of hepcidin transcription in both in vitro and in vivo settings,16 was performed to assess for a normal responsiveness of hepcidin transcription to exogenous stimulus in the conditions of our assays. A significant increase in hepcidin expression was observed both in HepG2 and in mouse hepatocytes (Figure 1A,C right).
EPO modulates hepcidin expression in a dose-dependent manner in HepG2 cells and in freshly isolated mouse hepatocytes. (A) qRT-PCR analysis of HAMP mRNA expression in HepG2 cells treated with rEPO or IL-6. Figure represents the average plus SD of 3 independent experiments; * denotes a statistically significant difference between treated samples and mock (P < .05; one-way ANOVA). (B) qRT-PCR analysis of HAMP mRNA expression in HepG2 cells treated with rEPO (1 or 2 U/mL) and/or with 0.1 to 10 μg/mL anti-EPOR. Bars on the right of each group correspond to HAMP mRNA levels after preabsorption of the anti-EPOR antibody with a specific peptide (+Block). * denotes a significant difference (P < .05) between EPO(+)/anti-EPOR(+)–treated samples and the EPO(+)/anti-EPOR(−) controls (one-way ANOVA) ** stands for a significant difference P < .05 level, between EPO(−)/anti-EPOR(+) or EPO(−)/anti-EPOR(+)/Block(+) and the EPO(−)/anti-EPOR(−) control (2-way ANOVA). Figure represents the average + SD of 3 independent experiments. (C) qRT-PCR analysis of Hamp1 mRNA expression in freshly isolated mouse hepatocytes treated with 0.01 to 2.0 U/mL rEPO (gray bars), and/or 0.1 to 5 μg anti-EPOR (white and black bars, respectively). Untreated HepG2 cells were used as control (hatched bar, left). Treatment with IL-6 was used to assess Hamp1 response to stimulation (gray bar, right). A total of 7 animals were used for each treatment; * denotes a significant difference (P < .05) between EPO(+)/anti-EPOR(−) or EPO(−)/anti-EPOR(+) or IL-6(+) and the nontreated control (one-way ANOVA). ** stands for a significant difference at the 95% confidence level between (EPO(+)/anti-EPOR(+) and the EPO(1 U)/anti-EPOR(−) control (one-way ANOVA).
EPO modulates hepcidin expression in a dose-dependent manner in HepG2 cells and in freshly isolated mouse hepatocytes. (A) qRT-PCR analysis of HAMP mRNA expression in HepG2 cells treated with rEPO or IL-6. Figure represents the average plus SD of 3 independent experiments; * denotes a statistically significant difference between treated samples and mock (P < .05; one-way ANOVA). (B) qRT-PCR analysis of HAMP mRNA expression in HepG2 cells treated with rEPO (1 or 2 U/mL) and/or with 0.1 to 10 μg/mL anti-EPOR. Bars on the right of each group correspond to HAMP mRNA levels after preabsorption of the anti-EPOR antibody with a specific peptide (+Block). * denotes a significant difference (P < .05) between EPO(+)/anti-EPOR(+)–treated samples and the EPO(+)/anti-EPOR(−) controls (one-way ANOVA) ** stands for a significant difference P < .05 level, between EPO(−)/anti-EPOR(+) or EPO(−)/anti-EPOR(+)/Block(+) and the EPO(−)/anti-EPOR(−) control (2-way ANOVA). Figure represents the average + SD of 3 independent experiments. (C) qRT-PCR analysis of Hamp1 mRNA expression in freshly isolated mouse hepatocytes treated with 0.01 to 2.0 U/mL rEPO (gray bars), and/or 0.1 to 5 μg anti-EPOR (white and black bars, respectively). Untreated HepG2 cells were used as control (hatched bar, left). Treatment with IL-6 was used to assess Hamp1 response to stimulation (gray bar, right). A total of 7 animals were used for each treatment; * denotes a significant difference (P < .05) between EPO(+)/anti-EPOR(−) or EPO(−)/anti-EPOR(+) or IL-6(+) and the nontreated control (one-way ANOVA). ** stands for a significant difference at the 95% confidence level between (EPO(+)/anti-EPOR(+) and the EPO(1 U)/anti-EPOR(−) control (one-way ANOVA).
To test for the specificity of the EPO effect, we blocked the EPOR with an anti-EPOR antibody that recognizes the extracellular domain of this receptor and prevents EPO-EPOR interaction. On both models, EPOR blocking abrogated the repressive effect of EPO on hepcidin expression (Figure 1B,C). The specificity of the blocking effect of anti-EPOR antibody was confirmed by the fact that preabsorption of the primary antibody with an excess of a specific blocking peptide restored hepcidin mRNA to levels not different from those induced by the 2 doses of EPO alone (Figure 1B). Similarly, coincubation of HepG2 cells with 1 and 2 U EPO and with an irrelevant antibody did not induce significant differences in hepcidin mRNA levels in comparison with cells treated with EPO alone (data not shown). Interestingly, cells treated with anti-EPOR alone showed a dose-dependent up-regulation of hepcidin mRNA, relative to the nontreated control cells (Figure 1B,C).
No significant effect of EPO supplementation or EPOR blocking on murine hepcidin 2 mRNA levels was observed (data not shown).
Analysis of the levels of CEBPA and Cebpa mRNAs showed that both CEBPA (Figure 2A) and Cebpa (Figure 2B) mRNA levels are significantly decreased after treatment with 0.5 to 2.5 U or 0.1 to 2 U EPO, respectively for the HepG2 cell line and fresh mouse hepatocytes. Blocking of EPOR and EpoR restored mRNA expression to levels similar to the control cells and significantly higher than EPO-treated (1 U) cells. The specificity of the anti-EPOR antibody was confirmed by the observation that preabsorption with a specific peptide restored CEBPA mRNA expression to levels not different from those of cells treated with 1 U EPO alone. Similarly to hepcidin mRNA expression, no rescue of CEBPA mRNA levels was observed after cotreatment with 1 U EPO and an irrelevant antibody. IL-6 treatment did not induce significant changes in CEBPA or Cebpa mRNA levels (Figure 2A,B right).
EPO modulates CEBPA expression in a dose-dependent manner in liver-derived cells. qRT-PCR assessment of CEBPA (A) and Cebpa (B) mRNA expression in HepG2 cells and in mouse hepatocytes treated with rEPO (▒, left) and/or anti-EPOR (□ and ■, respectively) and IL-6 (▒, right). Untreated cells were used as control (▫, left). Figure represents the average + SD of 3 independent experiments for HepG2 cells. A total of 7 animals were used for each mouse hepatocyte treatment; * denotes a significant difference (P < .05) between EPO(+)/anti-EPOR(−) or EPO(−)/anti-EPOR(+) or EPO(−)/anti-EPOR(+)/Block(+) or IL6(+) and the EPO(−)/anti-EPOR(−) control (one-way ANOVA). ** stands for a significant difference (P < .05) between EPO(+)/anti-EPOR(+) or EPO(1 U)/anti-EPOR(+)/Block(+) and the EPO(1 U)/anti-EPOR(−) control (2-way ANOVA).
EPO modulates CEBPA expression in a dose-dependent manner in liver-derived cells. qRT-PCR assessment of CEBPA (A) and Cebpa (B) mRNA expression in HepG2 cells and in mouse hepatocytes treated with rEPO (▒, left) and/or anti-EPOR (□ and ■, respectively) and IL-6 (▒, right). Untreated cells were used as control (▫, left). Figure represents the average + SD of 3 independent experiments for HepG2 cells. A total of 7 animals were used for each mouse hepatocyte treatment; * denotes a significant difference (P < .05) between EPO(+)/anti-EPOR(−) or EPO(−)/anti-EPOR(+) or EPO(−)/anti-EPOR(+)/Block(+) or IL6(+) and the EPO(−)/anti-EPOR(−) control (one-way ANOVA). ** stands for a significant difference (P < .05) between EPO(+)/anti-EPOR(+) or EPO(1 U)/anti-EPOR(+)/Block(+) and the EPO(1 U)/anti-EPOR(−) control (2-way ANOVA).
Since hepcidin and CEBPA response to EPO and anti-EPOR is essentially the same in HepG2 and in fresh primary mouse hepatocytes, we decided to continue the experimental work using the HepG2 cell model. The expression of C/EBPα in response to EPO and anti-EPOR treatments was analyzed. The high sensitivity detection method adopted in this study allowed for the detection of C/EBPα expression even after down-modulation by EPO treatments. Although some discrepancy exists in the literature concerning the levels of C/EBPα in HepG2 cells,17-22 the levels detected in the present study were similar to those reported by Chew et al20 and Yoon and Smart,21 using equally or less sensitive conditions.
The expression of C/EBPα protein closely mimics what was observed for CEBPA mRNA, with a mild increase in C/EBPα induced by the lowest dose of EPO and a decrease that is more pronounced from 0.5 U EPO onward, reaching approximately 50% for 2.5 U EPO (Figure 3A,B). A down-modulation of C/EBPα protein levels by EPO after only 3 hours of treatment can be explained by the short half-lives of the mRNA and protein: 2 hours for mRNA (in HepG2 cells)23 and 1 hour for the C/EBPα protein.24
C/EBPα protein levels and binding to HAMP promoter are modulated by EPO in a dose-dependent manner. (A) Western blot analysis of C/EBPα levels in HepG2 cells after treatment with physiological and supraphysiological doses of rEPO, anti-EPOR antibody, or IL-6. β-Actin was used as a loading control (bottom panel). Numbers on the left of each panel correspond to the sizes of the MW marker bands. Figure shows a representative result of 3 experiments. Vertical lines have been inserted to indicate repositioned gel lanes. (B) Scanning densitometry of C/EBPα Western blot. Data are normalized for β-actin levels and are expressed as arbitrary units (AU). (C) qRT-PCR–ChIP analysis of the effect of rEPO and anti-EPOR on the binding of C/EBPα to the HAMP promoter. Figure shows the averages and SD of 2 experiments, each with 3 replicates. * signals a significant (P < .05) difference between EPO-treated samples and the nontreated control (one-way ANOVA); ** signals a significant (P < .05) difference between anti-EPOR–treated samples and the 2.5 U EPO–treated cells (one-way ANOVA).
C/EBPα protein levels and binding to HAMP promoter are modulated by EPO in a dose-dependent manner. (A) Western blot analysis of C/EBPα levels in HepG2 cells after treatment with physiological and supraphysiological doses of rEPO, anti-EPOR antibody, or IL-6. β-Actin was used as a loading control (bottom panel). Numbers on the left of each panel correspond to the sizes of the MW marker bands. Figure shows a representative result of 3 experiments. Vertical lines have been inserted to indicate repositioned gel lanes. (B) Scanning densitometry of C/EBPα Western blot. Data are normalized for β-actin levels and are expressed as arbitrary units (AU). (C) qRT-PCR–ChIP analysis of the effect of rEPO and anti-EPOR on the binding of C/EBPα to the HAMP promoter. Figure shows the averages and SD of 2 experiments, each with 3 replicates. * signals a significant (P < .05) difference between EPO-treated samples and the nontreated control (one-way ANOVA); ** signals a significant (P < .05) difference between anti-EPOR–treated samples and the 2.5 U EPO–treated cells (one-way ANOVA).
To test if the EPO-induced down-modulation of C/EBPα would impact on the binding of this transcription factor to the HAMP promoter, ChIP analysis was performed, using lysates from HepG2 cells treated according to the protocol followed for the previous experiments (Figure 3C). Results show that treatment with the lowest (0.01 U) EPO dose induces a mild, but significant, increase in the binding of C/EBPα to HAMP promoter. In contrast, treatment with 0.01 to 2.5 U EPO causes a significant decrease of C/EBPα binding, with a maximum of 75% reduction for 2.5 U EPO.
Previous studies have invoked EPO in the regulation of hepcidin expression.6,25,26 However, they do not clarify the mechanism underlying this interaction, namely whether EPO is capable of directly signaling the hepatocyte to modulate hepcidin levels. The study from Pak et al26 suggested that during erythropoiesis, “hepcidin is not directly regulated by EPO but rather by a signal downstream from EPO” (page 3733), based on inhibition of erythropoiesis with carboplatin. Our results show that, at least in vitro, EPO, via signaling through EPOR, is able to modulate hepcidin expression in freshly isolated mouse hepatocytes and in human hepatoma cells. It is possible that the results obtained in vivo after blocking of erythropoiesis are reflecting carboplatin-induced changes in other parameters secondary to erythropoiesis inhibition. One hypothesis is that the increased hepatic nonheme iron levels observed in the study of Pak et al may be counteracting the down-regulation of hepcidin by EPO. To control for EPO-induced changes in iron levels in our in vitro models, we measured TFR1 mRNA expression in the same samples used for hepcidin and CEBPA/Cebpa assessment. No significant changes were observed after EPO or anti-EPOR treatments (data not shown), supporting the hypothesis that the mechanism underlying the modulation of hepcidin by EPO is independent of iron sensing by the hepatocyte.
We show that EPO-induced hepcidin regulation is achieved by the modulation of C/EBPα mRNA and protein levels, with significant changes on the transcriptional activation of hepcidin. Several studies have demonstrated a role for C/EBPα in the regulation of hepcidin transcription,22,27-29 but its involvement on the EPO-mediated regulation of hepcidin expression has not been suggested prior to this study. The mechanism underlying the C/EBPα regulation by EPO remains unknown. A previous study showed that the transcription factor CHOP (GADD153), which is known to impair C/EBPα binding to target promoters,30 is up-regulated in Rauscher murine erythroleukemia cells by treatment with high doses (50-100 U/mL) of rEPO.31 Such a mechanism could further amplify the role of C/EBPα in the EPO-mediated hepcidin expression. However, we failed to observe an up-regulation of CHOP mRNA, at least with the EPO doses used in our experimental protocol, some of which are already supraphysiological (results not shown). This suggests that CHOP may not be involved in the down-modulation of hepcidin by EPO in physiological conditions.
The relevance of the present results is further reinforced by previous reports showing that hepatocytes, which are the main hepcidin-producing cells, express EPO and EPOR.4,5,32,33 In addition, a variety of malignant cell lines, including HepG2, was found to synthesize both this hormone and its receptor, which suggests the importance of the EPO signaling beyond erythropoiesis.9,33
The EPO concentrations that in the present in vitro study were shown to inhibit hepcidin expression do not correspond to normal in vivo physiological concentrations but rather to concentrations that are found only in situations of severe anemia, hypoxia, and ineffective erythropoiesis or when EPO is administered therapeutically. Schmidt et al34 found that physiological (0.01 and 0.03 U) and supraphysiological (0.1-3 U) doses of EPO activate different signaling pathways, with effective MEK and Erk signaling only at physiological doses. Similarly, low and high EPO doses have opposite effects on nitric oxide production and vasodilatation.35 We propose that a similar mechanism may underlie the distinct effects in hepcidin expression of the different EPO doses used, whereby the basal EPO signal would be modulated by other signaling pathways activated by higher EPO concentrations, resulting in a repression of hepcidin expression.
In conclusion, the present results support the hypothesis of a direct role for EPO in the regulation of hepcidin, via modulation of C/EBPα expression. Although this does not argue against the existence of additional mediators of hepcidin expression in the context of erythropoiesis, as has recently been demonstrated for the TGF-β superfamily member GDF15,36 we suggest that a direct signaling of EPO to the hepatocyte could be one of the mechanisms underlying the reported in vivo hepcidin modulation by erythropoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Calouste Gulbenkian Foundation, FCT (PDCT/MGI/50407/2005), FLAD (Portugal), and Innova/APBRF (United States). J.P.P. was supported by FCT (Portugal, SFRH/BPD/19042/2004). D.T. was funded by the Wellcome Trust (073381/Z/03/Z).
Wellcome Trust
Authorship
Contribution: J.P.P. conceptualized the idea, designed and performed the research, analyzed data, and wrote the paper; S.R., H.P., and S.T. performed the research; D.T. and F.C. contributed vital analytical tools and contributed to paper writing; G.P. conceptualized the idea, analyzed data, and contributed to paper writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Graça Porto, Iron Genes and Immune System, IBMC-Instituto de Biologia Molecular e Celular, Universidade do Porto, Porto, Portugal; e-mail: gporto@ibmc.up.pt.
References
Author notes
*S.R., H.P., and S.T. contributed equally to this work.