Abstract
The most common hereditary elliptocytosis (HE) and hereditary pyropoikilocytosis (HPP) mutations are α-spectrin missense mutations in the dimer-tetramer self-association site. In this study, we systematically compared structural and functional properties of the 14 known HE/HPP mutations located in the α-spectrin tetramer binding site. All mutant α-spectrin recombinant peptides were well folded, stable structures, with only the R34W mutant exhibiting a slight structural destabilization. In contrast, binding affinities measured by isothermal titration calorimetry were greatly variable, ranging from no detectable binding observed for I24S, R28C, R28H, R28S, and R45S to approximately wild-type binding for R34W and K48R. Binding affinities for the other 7 mutants were reduced by approximately 10- to 100-fold relative to wild-type binding. Some sites, such as R28, were hot spots that were very sensitive to even relatively conservative substitutions, whereas other sites were only moderately perturbed by nonconservative substitutions. The R34W and K48R mutations were particularly intriguing mutations that apparently either destabilize tetramers through mechanisms not probed by the univalent tetramer binding assay or represent polymorphisms rather than the pathogenic mutations responsible for observed clinical symptoms. All α0 HE/HPP mutations studied here appear to exert their destabilizing effects through molecular recognition rather than structural mechanisms.
Introduction
The hereditary elliptocytosis (HE) syndromes are a common group of disorders characterized by elliptical erythrocytes on peripheral blood smear.1-4 These disorders are characterized by marked clinical, biochemical, and genetic heterogeneity. Most patients with typical HE are asymptomatic, but others have chronic hemolysis or the related disorder hereditary pyropoikilocytosis (HPP). Biochemical and genetic heterogeneity is related to qualitative and/or quantitative defects in one of several erythrocyte membrane skeleton proteins, particularly α-spectrin, β-spectrin, protein 4.1R, or glycophorin C, which leads to mechanical weakness or fragility of the erythrocyte membrane skeleton.
The majority of HE-associated defects occur in spectrin, the principal structural component of the red cell membrane skeleton. Spectrin is a flexible, rope-like molecule formed by antiparallel lateral association of 2 subunits, α- and β-spectrin, which are primarily composed of many tandem, homologous 106 amino acid motifs, or “spectrin type repeats.”5-7 Spectrin repeats are highly stable, independently folding 3 helix bundle units that are responsible for imparting much of the strength and flexibility to the erythrocyte membrane skeleton. For example, when conformational changes of membrane skeleton components are probed in intact red cells using cysteine-specific reagents, spectrin is the only membrane skeleton component showing stress-related increases in labeling indicative of tensile stress-related unfolding of specific domains.8 Assembly of spectrin heterodimers is initiated by 2 pairs of specialized repeats located near the C-terminal end of the α subunit and near the N-terminal end of the β-spectrin subunit.9-11 Two spectrin heterodimers self-associate in a head-to-head orientation to form tetramers, the predominant form of the molecule on red cell membranes. Tetramer assembly, which normally involves 2 head-to-head α-β associations per tetramer, is a moderate affinity, temperature-dependent association,12,13 involving small regions near the N-terminus of the α subunit and near the C-terminus of the β subunit that have been hypothesized to form a hybrid 3 helix bundle repeat similar to the typical spectrin type repeat.14-16 Specifically, this hybrid repeat consists of a C helix contributed by the α0 partial repeat and an A and B helix contributed by the partial β17 repeat (Figure 1). Spectrin tetramers are connected into a highly ordered 2-dimensional lattice through binding, at their tail ends, to short actin oligomers and associated proteins at a junctional complex, in an association facilitated by protein 4.1R.17-23 The moderate affinity of the spectrin tetramer interaction is apparently a critical feature of red cell deformability as local dissociation and reassociation of spectrin tetramers occurs in response to shear stress and thereby allows the cell to accommodate the distortions required for passage of the erythrocyte through the microvasculature.24
Impaired spectrin tetramer formation was one of the first abnormalities identified in the erythrocytes of patients with HE and HPP.25-27 This was followed by identification of numerous missense mutations located within the putative hybrid α-β repeat tetramer binding site that impaired tetramer formation and destabilized red cell membranes.14,28-44 Attempts to correlate spectrin mutations with clinical severity have been confounded by marked clinical, biochemical, and genetic variability. For instance, the same tetramer site missense mutation has been linked to asymptomatic HE, chronic hemolytic HE, and severe HPP phenotypes in cases where patients spanning multiple generations in larger families were studied.41,45 In many cases, inheritance of modifier alleles, such as the common αLELY polymorphism, that substantially alter the level of α-spectrin expressed by alleles with pathogenic mutations in heterozygotes can worsen or ameliorate the phenotype.46,47 Furthermore, most known HE and HPP mutations have been studied only in small numbers of patients, frequently from single families using targeted searches for potential mutations, thereby increasing the possibility that additional unidentified pathogenic mutations or polymorphisms contribute to the observed clinical phenotype.
The structural and functional effects of only a few spectrin tetramer site mutations have been studied so far. The effect on binding affinity of 3 mutations located in α0 were previously analyzed by high-performance liquid chromatography (HPLC) binding assays coupled with structural analysis using nuclear magnetic resonance.48 This study showed poor correlation between structural changes and tetramer binding affinity, leading the authors of that study to conclude that reduced spectrin tetramerization resulted from changes in molecular recognition rather than through conformational disruption. In another study, 3 β17 repeat mutants were studied using changes in circular dichroism to detect tetramer site interactions.49 However, this technique showed substoichiometric association for wild-type recombinant peptides apparently because of partial aggregation of the β-spectrin recombinant, and detectable binding was only observed for one of the 3 mutations. Furthermore, in contrast to the conclusions from analysis of α mutations using nuclear magnetic resonance, conformational perturbations were observed for both the adjacent full repeat (β16) and the repeat containing the pathogenic mutation.
The severity of the pathologic effects in the HE/HPP syndromes is heterogeneous. Severity has been associated in some cases with spectrin deficiency resulting from mRNA accumulation defects or increased degradation of mutant spectrin before its assembly on the membrane with different mutations associated with various degrees of spectrin deficiency.50 An analysis in these cases is compounded by the inheritance in cis or in trans of modifier alleles, such as αLELY, that influence the amount of mutant spectrin assembled on the membrane. Considering the wide variability in clinical severity and the variable results obtained in prior HE mutant characterization studies, it is important to structurally and functionally characterize and compare pathogenic mutations in a system that eliminates confounding effects of modifier alleles or other factors. Because the most common defects identified in HE/HPP are α-spectrin missense mutations, the present study systematically compares the 14 reported pathogenic mutations located in the α0 partial repeat. Minimum size α and β recombinant peptides retaining normal tetramer binding affinity49,51 were used to maximize detection of potential subtle changes in structure and tetramer binding affinities. The effects of these mutations on protein folding, structural stability, and thermodynamic properties of tetramer assembly provide a molecular basis for the clinical properties of these diseases and where appropriate, genotype-phenotype correlations are discussed.
Methods
Expression and purification of recombinant erythrocyte spectrin proteins
The polymerase chain reaction was used to amplify regions of interest (residues 1-158 of α-spectrin for α0-1, residues 1902-2084 of β-spectrin for β16-17) from erythrocyte α- and β-spectrin cDNA clones.6,7 DNA site-directed mutagenesis was used to introduce HE- and HPP-associated point mutations into the α0-1 construct that resulted in the following amino acid substitutions: I24S, I24T, R28C, R28H, R28L, R28S, V31A, R34W, R41W, R45S, R45T, G45V, K48R, and L49F. All constructs were expressed as glutathione S-transferase fusion protein using the pGEX-2T vector and DH5α Escherichia coli cells. The presence of expected mutations was confirmed by DNA sequencing. Wild-type β16-17, wild-type α0-1, and mutant α0-1 recombinants were expressed, purified, released from glutathione S-transferase, and repurified as previously described.11
Mass spectrometry
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) was performed by spotting 1 μL of a 1:1 mixture consisting of sinapinic acid matrix solution and purified protein onto a MALDI target plate using carbonic anhydrase as an internal standard. Purified recombinant proteins were also analyzed using electrospray ionization mass spectrometry on a ThermoFinnigan LTQ mass spectrometer (Thermo Fisher Scientific, San Jose, CA) interfaced on-line with a 75 μm × 8 cm PLRP-S protein reverse phase column used for desalting the samples. Spectra of multiple mass/charge state ions were deconvoluted to final mass values using BioWorks software. Expected masses of recombinant proteins were calculated using the GPMAW, version 4 program (Lighthouse Data, Odense, Denmark).
Circular dichroism measurements
CD spectra were performed on a Jasco J720 instrument at room temperature in a 0.2-mm path length quartz cell in 10 mM sodium phosphate, 130 mM NaCl, pH 7.3. Three consecutive scans were averaged, and final spectra were corrected for contribution of buffer. Protein concentrations were determined from optical density at 280 nm, and extinction coefficients were calculated as previously described.52
Sedimentation equilibrium analysis
The solution oligomeric state of all recombinants was determined using sedimentation equilibrium analysis in a Beckman Coulter XL-I analytical centrifuge (Fullerton, CA) as previously described.53,54 Briefly, aggregates were removed and buffer was exchanged by HPLC gel filtration (TSKgel Super SW3000, Tosoh Bioscience, Montgomeryville, PA) using a column buffer consisting of 10 mM Tris, 130 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 0.15 mM phenylmethylsulphonyl fluoride, and 1 mM tris (2-carboxyethyl) phosphine hydrochloride, pH 7.4. For each experiment, 3 initial protein concentrations at sample volumes of 110 μL were analyzed in parallel. A water blank scan was taken immediately before each experiment to correct for window distortion in the fringe displacement data.55 Each sample was analyzed at 2 speeds (typically 28 000 rpm and 40 000 rpm) at 4°C, followed by analysis at 30°C at the higher speed. Fringe displacement data were collected every 4 to 6 hours, and attainment of equilibrium was determined by comparing successive scans using WinMatch, version 0.99. Data were edited using WinReed, version 0.99, and analyzed using the Winnl, version 1.06, program56 to simultaneously fit data from all loading concentrations. The program SEDNTERP, version 1.08, was used to calculate ρ for the solvent and M and  from amino acid compositions of recombinant proteins. WinMatch, WinReed, and Winnl are available from the Analytical Ultracentrifugation Facility at the University of Connecticut via their FTP site, http://biotech.uconn.edu/auf. The program SEDNTERP was written by T. Laue, J. Hayes, and J. Philo, and is available on the website of Reversible Associations in Structural and Molecular Biology, http://www.rasmb.bbri.org.
from amino acid compositions of recombinant proteins. WinMatch, WinReed, and Winnl are available from the Analytical Ultracentrifugation Facility at the University of Connecticut via their FTP site, http://biotech.uconn.edu/auf. The program SEDNTERP was written by T. Laue, J. Hayes, and J. Philo, and is available on the website of Reversible Associations in Structural and Molecular Biology, http://www.rasmb.bbri.org.
Differential scanning calorimetry
Thermal denaturation analyses of spectrin recombinant peptides were performed using an MCS differential scanning calorimeter (MicroCal, Northampton, MA). Purified recombinants were exhaustively dialyzed against 10 mM sodium phosphate, 130 mM NaCl, 1 mM β-mercaptoethanol, pH 7.4. Samples were degassed for 5 minutes, and protein samples at 0.6 to 1.0 mg/mL in the sample cell were compared with dialysate buffer in the reference cell by scanning from 20°C to 90°C at a scan rate of 90°C/h. Data analysis was performed using the ORIGIN version 3.2 program provided with the instrument.
Isothermal titration calorimetry
Thermodynamic parameters for the binding of the α0-1 recombinants to wild-type β16-17 were determined using a VP-ITC isothermal titration calorimeter (OriginLab). Samples were dialyzed in 10 mM sodium phosphate, 130 mM NaCl, 1 mM β-mercaptoethanol, pH 7.4, shortly before analysis, and wild-type β16-17 at 23-37 μM or dialysate buffer (controls) was loaded into the reaction cell. Wild-type or mutant α0-1 spectrin was used as the titrant for experimental measurements and controls at 23°C with reaction cell stirring at 300 rpm. Typically, 25 to 29 injections of 10 μL of titrant were used. Selected experiments were performed at 30°C and 37°C. Protein concentrations were determined as described in “Sedimentation equilibrium analysis.” Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to verify the absence of degradation products before and after each experiment. Data were analyzed using OriginLab 5.0 software as previously described53,57 to calculate dissociation constants and other thermodynamic parameters.
Results
Purification and structural characterization of α0-1 spectrin mutant proteins
The 3 steps in the spectrin dimer-tetramer equilibrium and the locations of the α0-1 and β16-17 recombinants within the overall spectrin repeat structure are shown in Figure 1. These 2 peptides represent the minimum regions of the molecule that retain normal tetramer binding affinity.49,51 They are therefore appropriate experimental templates for evaluating effects of HE and HPP mutations located within the α0 region on protein folding, stability, and tetramer binding affinity. All spectrin recombinants were purified in good yields (1.2-12 mg/L of bacterial culture) and were purified to a highly homogeneous state (Figure 1B). Similarly, no atypical aggregation of purified proteins samples was observed when analyzed by analytical HPLC gel filtration, and all masses were within the expected mass error ranges of 0.05% for MALDI-MS and 0.025% for electrospray ionization mass spectrometry, which indicated that no unanticipated modifications had occurred during expression and purification (data not shown). The secondary structure of wild-type α0-1 and the 14 α0-1 mutants was compared using CD in physiologic buffer at room temperature (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). None of the HE or HPP point mutants significantly perturbed the secondary structure of α0-1 wild-type spectrin at room temperature with all α0-1 mutants exhibiting a helical content of approximately 65%.
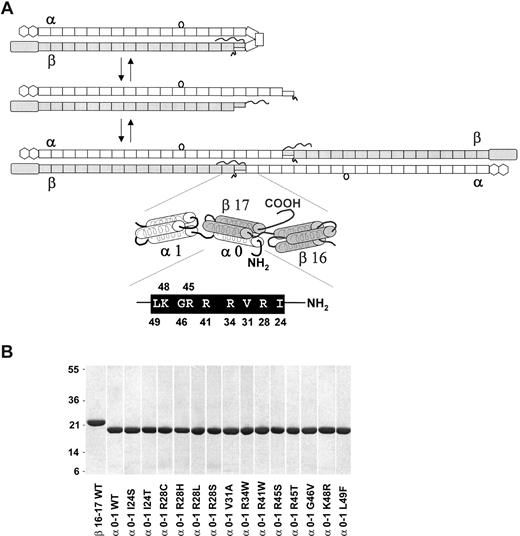
Relationship between the human red cell spectrin dimer-tetramer equilibrium and tetramer site univalent recombinant peptides. (A) A model depicting the 2 equilibria in the overall dimer-tetramer equilibrium of human red cell spectrin. The first step in tetramer formation is opening of a closed dimer (top panel), followed by head-to-head association of 2 open dimers to form a tetramer. Dimer and tetramer models schematically illustrate the repeats that comprise the α and β monomers as follows: rectangles represent the many tandem homologous “spectrin type” repeats; the loop attached to the α9 repeat depicts the SH3 domain, which is inserted in the loop between the B and C helices of repeat 9 (this SH3 domain is designated α10 for historical reasons); the hexagons at the α chain C-terminus represent EF-hand regions (calmodulin-like domains); the elongated rectangle (ABD) at the N-terminus of β-spectrin represents the actin-binding domain (calponin homology domain); the squiggly “tail” represents the nonhomologous phosphorylated C-terminal end of β-spectrin. An enlarged view of the tetramerization site schematically illustrates the α0-1 and β16-17 recombinant peptides using cylinders to represent the 3 helix bundles. In this model, the tetramer binding site is composed of a C helix from the partial α0 repeat and a B and C helix from the β17 partial repeat. The amino acid residues and residue numbers in the α0 C helix that are mutated in HE/HPP patients are shown in the black bar immediately below the tetramer site model. (B) A 1-D 12% Bis-Tris SDS gel stained with Coomassie Brilliant Blue of the purified recombinant proteins (2 μg). Molecular weights of standard proteins are indicated on the left (in kilodaltons).
Relationship between the human red cell spectrin dimer-tetramer equilibrium and tetramer site univalent recombinant peptides. (A) A model depicting the 2 equilibria in the overall dimer-tetramer equilibrium of human red cell spectrin. The first step in tetramer formation is opening of a closed dimer (top panel), followed by head-to-head association of 2 open dimers to form a tetramer. Dimer and tetramer models schematically illustrate the repeats that comprise the α and β monomers as follows: rectangles represent the many tandem homologous “spectrin type” repeats; the loop attached to the α9 repeat depicts the SH3 domain, which is inserted in the loop between the B and C helices of repeat 9 (this SH3 domain is designated α10 for historical reasons); the hexagons at the α chain C-terminus represent EF-hand regions (calmodulin-like domains); the elongated rectangle (ABD) at the N-terminus of β-spectrin represents the actin-binding domain (calponin homology domain); the squiggly “tail” represents the nonhomologous phosphorylated C-terminal end of β-spectrin. An enlarged view of the tetramerization site schematically illustrates the α0-1 and β16-17 recombinant peptides using cylinders to represent the 3 helix bundles. In this model, the tetramer binding site is composed of a C helix from the partial α0 repeat and a B and C helix from the β17 partial repeat. The amino acid residues and residue numbers in the α0 C helix that are mutated in HE/HPP patients are shown in the black bar immediately below the tetramer site model. (B) A 1-D 12% Bis-Tris SDS gel stained with Coomassie Brilliant Blue of the purified recombinant proteins (2 μg). Molecular weights of standard proteins are indicated on the left (in kilodaltons).
HPLC gel filtration analysis did not indicate abnormal irreversible aggregation, but some spectrin recombinant peptides form weak artifactual homodimers that would typically not be detected by gel filtration analysis. We therefore analyzed the solution oligomer state of each mutant using sedimentation equilibrium analysis. The wild-type and all mutant α0-1 recombinants were monomers in solution as indicated by fitting of the data to single species with molecular weights within 5% of the calculated molecular weights for α0-1 monomers (Figure S2). Interestingly, although the α R34W was monomeric at 4°C, it was prone to slow hydrolysis by low levels of contaminating proteases over time at 30°C; and as a result, the 30°C data could not be globally fit to a single species model for this mutation.
Thermal unfolding of α0-1 wild-type and mutant recombinants
DSC experiments were performed on all purified recombinant α0-1 proteins to measure thermal unfolding in phosphate-buffered saline solution at pH 7.4. Wild-type α0-1 was highly stable with a single reversible 2-state transition and a Tm of 54.6°C (Figure 2). Most single-point α0 region mutants showed thermal transitions within 2°C of the wild-type transition (Table 1). Each thermal unfolding was repeated for a total of 3 consecutive scans, and all transitions were essentially completely reversible. The only mutation that substantially destabilized the structure of the monomer was the R34W mutant, which unfolded with a Tm of 49.1°C with a broader thermal transition (Figure 2). This decreased thermal stability is consistent with the increased susceptibility of this mutant to proteolysis observed during extended incubation at 30°C during the sedimentation equilibrium experiments described in “Sedimentation equilibrium analysis.”
Thermal denaturation analysis of α0-1 recombinant peptides. Representative thermal denaturation experiments of wild-type and 2 HE mutant α0-1 peptides. DSC analysis was performed at a scan rate of 90°C/hr in 10 mM sodium phosphate, 130 mM NaCl, 1 mM β-ME, pH 7.4, at concentrations ranging from 0.6 to 1.0 mg/mL. Normalized spectra are shown for: the α0-1 wild-type (—), Tm = 54.6°C; the α0-1 R28C (···), which had the highest observed Tm of 55.6°C; and the α0-1 R34W (----), which had the lowest Tm at 49.1°C and the most atypical melting curve.
Thermal denaturation analysis of α0-1 recombinant peptides. Representative thermal denaturation experiments of wild-type and 2 HE mutant α0-1 peptides. DSC analysis was performed at a scan rate of 90°C/hr in 10 mM sodium phosphate, 130 mM NaCl, 1 mM β-ME, pH 7.4, at concentrations ranging from 0.6 to 1.0 mg/mL. Normalized spectra are shown for: the α0-1 wild-type (—), Tm = 54.6°C; the α0-1 R28C (···), which had the highest observed Tm of 55.6°C; and the α0-1 R34W (----), which had the lowest Tm at 49.1°C and the most atypical melting curve.
Structural and functional properties for wild-type and HE/HPP α0-1 mutants
| α0-1 . | Tm,* °C . | Kd,† μM . | n† . | ΔH,† kcal/mol . | −TΔS,‡ kcal/mol . | ΔG,‡ kcal/mol . | Affinity category§ . |
|---|---|---|---|---|---|---|---|
| Wild-type | 54.6 | 0.4 ± 0.1 | 0.9 ± 0.06 | −33.1 ± 3.5 | 24.5 ± 3.4 | −8.6 | wt |
| I24S | 53.2 | >100 | − | − | − | − | 3 |
| I24T | 54.4 | 41 ± 7 | 0.9 ± 0.06 | −29.0 ± 7.9 | 23.2 ± 8.0 | −5.8 | 2 |
| R28C | 55.6 | >200 | − | − | − | − | 3 |
| R28H | 53.6 | >200 | − | − | − | − | 3 |
| R28L | 53.1 | 67 ± 4 | 1.1 ± 0.03 | −21.2 ± 1.4 | 15.6 ± 1.5 | −5.6 | 2 |
| R28S | 53.3 | >200 | − | − | − | − | 3 |
| V31A | 54.5 | 23 ± 3 | 1.0 ± 0.06 | −32.1 ± 6.5 | 26.0 ± 6.6 | −6.1 | 2 |
| R34W | 49.1 | 0.1 ± 0.0 | 0.9 ± 0.02 | −40.5 ± 3.6 | 31.1 ± 3.6 | −9.4 | ∼wt |
| R41W | 53.4 | 8.7 ± 0.4 | 1.0 ± 0.05 | −29.4 ± 4.7 | 22.6 ± 4.8 | −6.8 | 1 |
| R45S | 55.1 | >200 | − | − | − | − | 3 |
| R45T | 54.6 | 14 ± 2.0 | 0.9 ± 0.01 | −29.5 ± 1.0 | 23.1 ± 1.1 | −6.4 | 1 |
| G46V | 52.8 | 3.7 ± 0.7 | 0.9 ± 0.02 | −31.3 ± 2.3 | 24.1 ± 2.4 | −7.2 | 1 |
| K48R | 54.7 | 0.5 ± 0.1 | 0.9 ± 0.05 | −33.5 ± 0.4 | 25.0 ± 3.9 | −8.5 | ∼wt |
| L49F | 55.4 | 30 ± 1 | 1.0 ± 0.03 | −14.9 ± 1.2 | 8.9 ± 1.2 | −6.0 | 2 |
| α0-1 . | Tm,* °C . | Kd,† μM . | n† . | ΔH,† kcal/mol . | −TΔS,‡ kcal/mol . | ΔG,‡ kcal/mol . | Affinity category§ . |
|---|---|---|---|---|---|---|---|
| Wild-type | 54.6 | 0.4 ± 0.1 | 0.9 ± 0.06 | −33.1 ± 3.5 | 24.5 ± 3.4 | −8.6 | wt |
| I24S | 53.2 | >100 | − | − | − | − | 3 |
| I24T | 54.4 | 41 ± 7 | 0.9 ± 0.06 | −29.0 ± 7.9 | 23.2 ± 8.0 | −5.8 | 2 |
| R28C | 55.6 | >200 | − | − | − | − | 3 |
| R28H | 53.6 | >200 | − | − | − | − | 3 |
| R28L | 53.1 | 67 ± 4 | 1.1 ± 0.03 | −21.2 ± 1.4 | 15.6 ± 1.5 | −5.6 | 2 |
| R28S | 53.3 | >200 | − | − | − | − | 3 |
| V31A | 54.5 | 23 ± 3 | 1.0 ± 0.06 | −32.1 ± 6.5 | 26.0 ± 6.6 | −6.1 | 2 |
| R34W | 49.1 | 0.1 ± 0.0 | 0.9 ± 0.02 | −40.5 ± 3.6 | 31.1 ± 3.6 | −9.4 | ∼wt |
| R41W | 53.4 | 8.7 ± 0.4 | 1.0 ± 0.05 | −29.4 ± 4.7 | 22.6 ± 4.8 | −6.8 | 1 |
| R45S | 55.1 | >200 | − | − | − | − | 3 |
| R45T | 54.6 | 14 ± 2.0 | 0.9 ± 0.01 | −29.5 ± 1.0 | 23.1 ± 1.1 | −6.4 | 1 |
| G46V | 52.8 | 3.7 ± 0.7 | 0.9 ± 0.02 | −31.3 ± 2.3 | 24.1 ± 2.4 | −7.2 | 1 |
| K48R | 54.7 | 0.5 ± 0.1 | 0.9 ± 0.05 | −33.5 ± 0.4 | 25.0 ± 3.9 | −8.5 | ∼wt |
| L49F | 55.4 | 30 ± 1 | 1.0 ± 0.03 | −14.9 ± 1.2 | 8.9 ± 1.2 | −6.0 | 2 |
Values are from single DSC experiments.
Averages and standard deviations for binding to wild-type β16-17 in replicate ITC experiments (triplicates for all mutants and four replicates for wild-type). All experiments shown here were conducted at 23° C (see text for wild-type values at other temperatures). Average protein concentrations for wild-type β16-17 (cell) and α0-1 constructs (syringe) are, respectively: 19 μM and 563 μM for WT, 24 μM and 565 μM for I24S, 30 μM and 653 μM for I24T, 35 μM and 511 μM for R28C, 33 μM and 603 μM for R28H, 28 μM and 510 μM for R28L, 38 μM and 820 μM for R28S, 24 μM and 575 μM V31A, 14 μM and 270 μM for R34W, 29 μM and 674 μM for R41W, 28 μM and 560 μM for R45S, 29 μM and 664 μM for R45T, 29 μM and 619 μM for G46V, 25 μM and 576 μM for K48R, 32 μM and 541 μM for L49F. For experiments where binding was below quantifiable limits, upper limits for Kd were estimated using Origin software for the protein concentrations used in the analysis; these experiments were performed in duplicate.
Values for −TΔS and ΔG are calculated from the experimentally determined association constant (K) and enthalpy (ΔH) using the relationships: RTlnK = ΔG = ΔH-TΔS where ΔG is the observed free energy, R is the gas constant, T is the experimental temperature in Kelvin, and ΔS is the calculated entropy.
Binding affinity groups were defined as follows: wt indicates similar to wild-type; 1, approximately 10-fold weaker than wild-type; 2, approximately 100-fold weaker than wild-type; 3, marginal or no detectable binding.
Evaluation of the effects of α0 HE and HPP mutations on spectrin tetramer assembly
The capacity of wild-type and mutant α0-1 to bind to β16-17 was initially assessed using HPLC gel filtration. HPLC gel filtration separation is a useful, rapid initial assay to check for head-to-head assembly of univalent peptides containing the spectrin tetramer binding site in a quasi-quantitative manner that does not require large amounts of protein per assay.15,48 Retention times of all α0-1 mutant proteins in the absence of β16-17 were the same as the wild-type protein, indicating that there were not any major differences in protein folding and hydrodynamic radius. HPLC binding assays of mutants exhibited a wide range of behavior, ranging from binding interactions similar to wild-type to no detectable binding (Figure S3). Six mutants exhibited no detectable binding using this assay, including I24S, R28C, R28H, R28L, R28S, and R45S. Mutants exhibiting reduced binding affinity relative to wild-type α0-1 included I24T, R41W, L49F, V31A, R45T, and G46V. Interestingly, the R34W and K48R exhibited binding interactions similar to the wild-type protein.
The initial estimates of binding affinities from HPLC gel filtration assays were used to optimize protein concentrations in subsequent more rigorous measurements of binding affinities using isothermal titration calorimetry (ITC). This method enables determination of stoichiometry, binding affinity, and all thermodynamic parameters for protein-protein interactions in solution without the need to tag or otherwise modify either component. An initial series of ITC experiments using wild-type α0-1 and β16-17 yielded a Kd of 8.7 μM at 37°C and 1.3 μM at 30°C, which are very similar to binding affinities measured using intact spectrin dimers.12,13,58,59 The values obtained by ITC also agree well with previously determined Kd values for the αI domain peptide and β monomer obtained in prior HPLC gel filtration assays (7.1 μM at 37°C and 3.8 μM at 30°C).15 Furthermore, the binding affinity of the wild-type recombinants, as measured by ITC at 23°C in this study, was similar to binding affinities of α0-1 and β16-17 measured at 25°C using surface plasmon resonance.60 Finally, we compared binding affinities in ITC assays of wild-type and several mutants using the α0-1 construct with a longer wild-type α-spectrin recombinant consisting of residues 1 to 584. As these results were comparable, a systematic evaluation of thermodynamic properties for all α0-1 mutants was conducted using 23°C to take advantage of the tighter binding affinity and faster binding kinetics at this temperature compared with physiologic temperature.
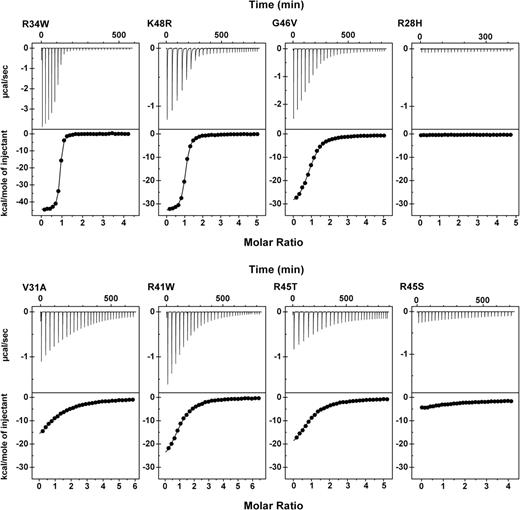
Triplicate titrations were performed for association of each α0-1 mutant with wild-type β16-17 (Table 1). As expected, the stoichiometry for all reactions where binding was observed was 1:1, within experimental error. All quantifiable reactions were strongly exothermic (ie, the observed enthalpy (ΔH) was a large negative value). Only 5 mutants were either incapable of binding to β16-17 or bound with affinities below quantitative limits of this assay. The affinity of the sixth mutant that did not show detectable binding in the HPLC assay, R28L, was slightly above the detection threshold of the ITC assays with an observed Kd of 67 μM. The HE and HPP mutants were grouped based on binding affinities as described in Table 1. Consistent with the HPLC binding assays, K48R and R34W, exhibited tetramer binding affinity similar to wild-type. R34W was particularly intriguing because this mutation subtly perturbed the structure of α0-1 as indicated by decreased thermal stability and increased protease susceptibility, yet this recombinant actually formed tetramers with apparent slightly greater avidity than the wild-type protein (ie, a Kd of 0.085 μM compared with 0.43 μM for the wild-type protein). Titration curves and nonlinear least-squares fits of the data for representative interactions spanning the full range of observed affinities are shown in Figure 3.
Representative isothermal titration calorimetry analyses of tetramerization site complexes. All experiments were conducted at 23°C with β16-17 in the reaction cell and the α0-1 peptide in the titration syringe. Top panels show baseline subtracted titration curve raw data; bottom panels show blank corrected integrated areas of the peaks from the top panel (●) and the best data fit using a nonlinear least-squares method (—). The protein concentrations of α0-1 mutants in the syringe were: R34W = 330 μM; K48R = 560 μM; G46V = 680 μM; R28H = 600 μM; V31A = 600 μM; R41W = 650 μM; R45T = 660 μM; R45S = 600 μM. The protein concentrations of the β16-17 wild-type recombinant in the cell were: 18 μM for R34W and 25 to 30 μM in all other cases.
Representative isothermal titration calorimetry analyses of tetramerization site complexes. All experiments were conducted at 23°C with β16-17 in the reaction cell and the α0-1 peptide in the titration syringe. Top panels show baseline subtracted titration curve raw data; bottom panels show blank corrected integrated areas of the peaks from the top panel (●) and the best data fit using a nonlinear least-squares method (—). The protein concentrations of α0-1 mutants in the syringe were: R34W = 330 μM; K48R = 560 μM; G46V = 680 μM; R28H = 600 μM; V31A = 600 μM; R41W = 650 μM; R45T = 660 μM; R45S = 600 μM. The protein concentrations of the β16-17 wild-type recombinant in the cell were: 18 μM for R34W and 25 to 30 μM in all other cases.
Although ITC requires up to several milligrams of each reactant per experiment, it has the advantage that stoichiometry and all thermodynamic parameters can be determined in a single experiment without modifying or immobilizing either reactant (Table 1). The observed stoichiometry of approximately 1.0 for all mutants where association was detectable indicates that there were not appreciable amounts of inactive protein, such as would be the case if partial aggregation occurred. Comparison of the thermodynamic parameters for wild-type and mutant binding provides useful insights into the relative importance of hydrophilic and hydrophobic interactions as well as shifts in their relative contributions for mutant proteins. As expected, all reactions where head-to-head tetramer-like binding is observed exhibit negative ΔGobs values for the forward (tetramer assembly) direction, indicating that tetramer assembly is favored to various degrees. ΔGobs is the sum of experimentally determined enthalpy (ΔHobs)and an entropy term (−TΔSobs), where T is the reaction temperature in Kelvin. The data in Table 1 show that the wild-type association has a highly favorable enthalpy and a moderately unfavorable entropy component for an overall large negative free energy (ΔGobs = −8.6 kcal/mol). A favorable enthalpy (negative ΔH) involves hydrophilic forces, such as formation of hydrogen-bonds, and protonation events as well as van der Waals interactions resulting primarily from hydrophobic interactions. A favorable entropy term (a negative TΔS) is expected from burial of electrostatic charges or hydrophobic groups. Most HE mutants show a reduced enthalpy contribution and a partially compensatory reduction in the magnitude of the unfavorable entropy component. However, similar affinities are achieved by different mutants through widely differing contributions of these 2 thermodynamic parameters. For example, comparison of the 4 mutations that show approximately 100-fold reduction in binding affinity (category 2 in Table 1) shows that one mutation (V31A) has a nearly wild-type enthalpy and more unfavorable entropy, whereas 2 mutations exhibit greatly reduced enthalpy and partially compensatory reduction in the unfavorable entropy term.
Discussion
The HE/HPP syndromes are the most common inherited abnormalities of erythrocyte shape worldwide. Considering the wide variability in clinical severity and the variable results obtained in prior studies characterizing HE/HPP mutants, we thought it was important to structurally and functionally characterize and compare pathogenic mutations in a system that eliminated confounding effects of modifier alleles or other factors. Because the most common defects identified in HE/HPP are α-spectrin missense mutations in the dimer-tetramer self-association site, the present study systematically compared the 14 known HE/HPP mutations located in the α0 partial spectrin repeat. Minimum size α and β recombinant peptides retaining normal tetramer binding affinity49,51 were used to maximize detection of potential subtle changes in structure and tetramer binding affinities.
None of the α0 mutations induced aggregation or conformational perturbations of the α0-1 complex as indicated by identical Stokes radius and essentially identical secondary structure by CD analysis. This is in contrast to several HE/HPP-associated β17 mutations reported to substantially perturb the conformation of both the β17 partial repeat and the adjacent β16 repeat.49 In the α0 mutants, the only evidence of structural destabilization was a modest, but significant, decrease in the thermal stability of the R34W mutant and slightly increased susceptibility of this mutant to proteolysis when incubated for extended periods at 30°C during the sedimentation equilibrium analyses. Prior analysis of 3 α0 mutations using nuclear magnetic resonance concluded that tetramer destabilization occurred through changes in molecular recognition rather than conformational disruption.48 Our analysis of all 14 α0 mutations using different methods to evaluate structure are consistent with the nuclear magnetic resonance study; as our data indicate, most α0 mutations affect tetramer assembly through perturbation of molecular recognition rather than conformational destabilization. Of course, this conclusion is based on the lack of evidence for perturbed conformations of α0 mutations exhibiting reduced tetramer binding affinity rather than direct measurements of disrupted molecular recognition. Interestingly, the α0 mutants differ markedly from a more distal α-spectrin mutation, Q471P, which is located in the linker region between the α4 and α5 repeats.61 We recently showed this proline mutation, which was located a great distance from the tetramer binding site, was able to bind to β16-17 with normal affinity in ITC head-to-head binding assays. However, this mutation destabilizes the structure of the N-terminal region of α-spectrin at physiologic temperature by disrupting the stabilizing influence of adjacent repeats normally provided by the helical inter-repeat linkers. Presumably, the Q471P mutant disrupts tetramer formation in a more indirect manner through this structural destabilization.
Although all α0 mutants exhibited essentially wild-type structural properties, they exhibited widely various tetramer binding affinities and related thermodynamic properties. The 14 reported HE/HPP missense mutations in α0 could be divided into 4 categories based on approximate order-of-magnitude differences in tetramer binding affinity, as summarized in Table 1. There are several intriguing results. Five mutants were either completely incapable of binding to β16-17 or bound with affinities below the assay's quantitative limits. With 2 intriguing exceptions, K48R and R34W, the other mutants demonstrated substantially reduced binding affinity.
K48R, a very conservative structural substitution, demonstrated Tm, Kd, and thermodynamic parameters essentially identical to wild-type protein. Erythrocytes from the K48R proband exhibited perturbed tetramerization and a diagnostic tryptic αI/74-Kd fragment.37 The K48R mutation could be a polymorphism with the actual pathogenic mutation located elsewhere, presumably in β17, which was not analyzed in the original report.37 No known single nucleotide polymorphisms were detected for this codon in erythroid spectrin when the NCBI single nucleotide polymorphism database was searched, but it could be a rare polymorphism that has not yet been described. Alternative possibilities are that this residue is involved in the lateral inter-strand interaction in tetramers, where it could destabilize formation of the second head-to-head α-β association (tetramer model, Figure 1A) during tetramer formation or it may perturb the closed-open dimer equilibrium (Figure 1A). The univalent head-to-head binding assays used here would not be able to detect either of these potential perturbations.
The R34W mutation, a very nonconservative substitution with a charged basic residue replacing a very large hydrophobic group, exhibits slightly stronger binding than wild-type despite subtle perturbation of its structure as indicated by a significantly reduced Tm and slightly increased proteolytic susceptibility (Figure 3; Table 1). Similar to the K48R mutant, this mutation was characterized in a single family and potential mutations in the β17 region were not evaluated.39 Thus, R34W could also be a polymorphism with an undetected pathogenic mutation in β17 responsible for the observed membrane destabilization. However, this seems less likely than with the K48R mutation for several reasons. The R34W proband had this mutation in trans to the αLELY low expression polymorphism; his peripheral smear had mild HE, and his erythrocyte membranes demonstrated moderate tetramer impairment. The father was also heterozygous for R34W, but he did not have αLELY on either allele; his peripheral smear did not have elliptocytes, but his erythrocyte membranes exhibited moderate tetramer impairment as well. Hence, the membrane disruption and tetramer impairment appeared to track with the α-spectrin allele inherited from the father. Therefore, it is more likely that this mutation is responsible for HE through a mechanism other than the direct α-β interaction probed by the univalent tetramer binding assay used here. It may disrupt lateral interactions with the adjacent strand in the bivalent tetramer, perturb the closed-open dimer equilibrium, or confer greater sensitivity of the tetramer site to tensile stress as a result of its reduced structural stability. A more remote possibility is that moderately increased tetramer affinity may be detrimental to membrane stability by making the membrane too rigid. But such a mechanism is inconsistent with the observed destabilization of tetramers in the proband and the magnitude of this change does seem too modest to account for red cell membrane destabilization unless additional cryptic factors are involved.
It is interesting that 4 of the 9 residues mutated in the α0 C helix are Arg, and 2 of these positions have multiple substituted amino acids (ie, 4 substitutions at R28 and 2 substitutions at R45). The functional effects of substituting Arg at these sites are very diverse, ranging from the tighter than normal binding for R34W to no detectable binding for R45S and 3 of the 4 mutations at R28. In addition, at least some sites are very sensitive to the precise structure of the side chain. For example, the nonconservative substitution of a Thr for either I24 or R45 substantially reduces binding affinity, but substitution with Ser instead of Thr completely abolishes detectable binding, although the difference between Thr and Ser is loss of only a single methyl group.
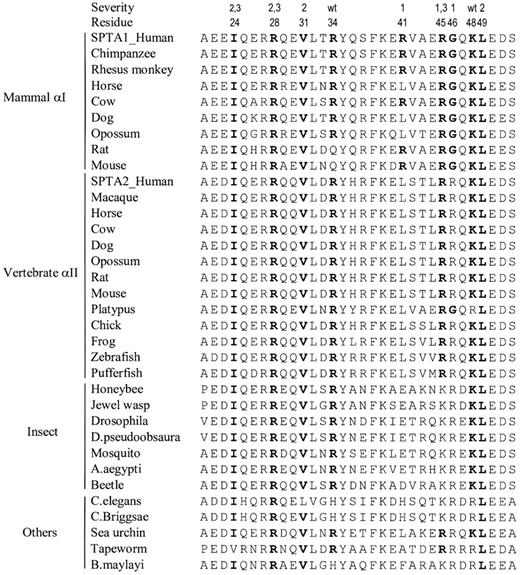
Most amino acid positions where α0 HE/HPP-associated mutations occur are very strongly conserved over great evolutionary distances (Figure 4). For example, when known sequences of the mammalian red cell isoform (αI) are considered, only the R34 and R41 positions show species diversity, and these mutations minimally perturb binding. Even when sequence comparisons are extended to the αII isoform and invertebrate species, critical sites where known mutations reduce binding affinity by at least 100-fold or more (I24, R28, V31, R45, and L49), are very strongly preserved even among lower organisms. Interestingly, reverse comparisons are not true. First, some highly conserved residues in the α0 binding (R27, L32, Y35, F39, K40, and E50) do not have any known mutations associated with HE/HPP. Second, not all highly evolutionarily conserved sites with known HE/HPP mutations result in severe red cell membrane destabilization. Specifically, R34 and K48 are very strongly evolutionarily conserved but observed mutations at these sites are minimally disruptive. Finally, the mildly disruptive mutations at R41 and G46 are relatively conserved among αI spectrin but are in a unique sequence region that distinguishes the αI and αII isoforms.62
Comparison of evolutionary sequence conservation within the spectrin α0 C-helix. Alignments of spectrin sequences encompassing approximately the C-helix of the α0 partial repeat involved in tetramer binding are shown. All known N-terminal region sequences of α-spectrins were extracted from the National Center for Biotechnology Information nonredundant protein sequence database64 using BLASTP 2.2.17 and human red cell spectrin (SPTA1) residues 1 to 158 as the query sequence. The severity of decreased tetramer binding affinity (Table 1) and residue numbers of the 14 known HE/HPP mutations are shown above the sequences. For these positions, residues that match the wild-type human red cell spectrin sequence are shown in bold type.
Comparison of evolutionary sequence conservation within the spectrin α0 C-helix. Alignments of spectrin sequences encompassing approximately the C-helix of the α0 partial repeat involved in tetramer binding are shown. All known N-terminal region sequences of α-spectrins were extracted from the National Center for Biotechnology Information nonredundant protein sequence database64 using BLASTP 2.2.17 and human red cell spectrin (SPTA1) residues 1 to 158 as the query sequence. The severity of decreased tetramer binding affinity (Table 1) and residue numbers of the 14 known HE/HPP mutations are shown above the sequences. For these positions, residues that match the wild-type human red cell spectrin sequence are shown in bold type.
One goal of this study was to correlate binding affinities of the α0 mutants with clinical severity. Grading of clinical severity is confounded by numerous factors, including inheritance of other pathogenic spectrin mutations, some of which may be cryptic, and inheritance of modifier alleles such as αLELY and αLEPRA, that alter spectrin expression. Because some mutations have only been studied in single or a few patients, it is often impossible to assign a clinical phenotype to specific mutations with certainty, as the presence or absence of modifier alleles and other possible spectrin gene variants is sometimes unknown because of incomplete reporting. Thus, the binding affinities of many of the mutations studied here correlate approximately with clinical severity, but correlations cannot be drawn for all mutants. Of note, all 4 known substitutions at R28 either greatly decrease or completely abolish detectable tetramer binding and these mutations are associated with relatively severe clinical symptoms. R41W, R45T, and G46V, associated with only modest impairment in binding affinity, are associated with asymptomatic or mild HE. The asymptomatic K48R variant is discussed above. Single patient or kindred data where lack of modifier allele data made interpretation difficult included I24S, I24T, R34W, L49F, R45S, and L49F.
In an earlier report, homology modeling of the tetramer site hybrid repeat, where the α0 helix C pairs with the A and B helices of the partial β17 repeat (Figure 1), predicted the presence of several key residues and interactions within the contact surface of the tetramer site.63
In this model most of the mutations occur in residues that are expected to stabilize the interaction between the α0 C helix and β17. For example, R28 is predicted to form both hydrogen bonds and salt bridges with β17; therefore, any nonconservative mutation would be expected to be detrimental to tetramer formation. Both I24 and L49 are expected to form hydrophobic interactions with β17; therefore, any mutation that disrupts the hydrophobic effect is expected to destabilize the spectrin tetramer. Analysis of α0 helix C HE/HPP mutations using this model concluded there was good agreement between the locations of mutated residues in the tetramer and clinical severity of the pathogenic mutations.63
We should soon be able to more precisely correlate effects of α0 mutations with structural perturbation of the tetramer interaction site as we have recently obtained a medium resolution crystallographic structure of the tetramer complex. Although structural refinement is not complete, the preliminary structure confirms the general concept of a hybrid α-β repeat tetramer site and demonstrates prior molecular models are reasonable, but incomplete, approximations of this critical membrane skeleton interaction site (S.H., T. Messick, R. Marmorstein, and D. W. Speicher, manuscript in preparation). In this regard, the preliminary structure suggests that the R34W mutant is located within, but near the boundary of the tetramer interface, which is consistent with the observed complex effects on thermodynamic parameters. Furthermore, its location supports the hypothesis that this is a pathogenic mutation that is at least in part destabilized by interstrand interactions in the normal bivalent tetramer. In contrast, the K48R mutant is not located in the tetramer interface, which is consistent with the lack of an effect of this mutant on tetramer binding observed in the current study.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by National Institutes of Health grants HL38794 (D.W.S.) and HL65448 (P.G.G.) and in part by institutional grants to the Wistar Institute, including NCI Cancer Core Grant (CA10815) and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.
National Institutes of Health
Authorship
Contribution: M.G. designed and performed research, analyzed and interpreted data, and drafted the manuscript; S.M. designed and performed research; S.H. assisted with performing research and data analysis/interpretation; P.G.G. and D.W.S. designed and directed research, assisted with data interpretation, and prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David W. Speicher, The Wistar Institute, 3601 Spruce Street, Philadelphia, PA 19104; e-mail: speicher@wistar.org.