Abstract
The genetics of t(11;14)(q13;q32)/cyclin D1–negative mantle cell lymphoma (MCL) is poorly understood. We report here 8 MCL cases lacking t(11;14) or variant CCND1 rearrangement that showed expression of cyclin D1 (2 cases), D2 (2 cases), and D3 (3 cases). One case was cyclin D negative. Cytogenetics and fluorescence in situ hybridization detected t(2;12)(p11;p13)/IGK-CCND2 in one of the cyclin D2-positive cases and t(6;14)(p21;q32)/IGH-CCND3 in one of the cyclin D3-positive cases. Moreover, we identified a novel cryptic t(2;14)(p24;q32) targeting MYCN in 2 blastoid MCLs: one negative for cyclin D and one expressing cyclin D3. Interestingly, both cases showed expression of cyclin E. Notably, all 3 blastoid MCLs showed a monoallelic deletion of RB1 associated with a lack of expression of RB1 protein and monoallelic loss of p16. In sum-mary, this study confirms frequent aberrant expression of cyclin D2 and D3 in t(11;14)-negative MCLs and shows a t(11;14)-independent expression of cy-clin D1 in 25% of present cases. Novel findings include cyclin E expression in 2 t(11;14)-negative MCLs characterized by a cryptic t(2;14)(p24;q32) and identification of MYCN as a new lymphoma oncogene associated with a blastoid MCL. Clinically important is a predisposition of t(11;14)-negative MCLs to the central nervous system involvement.
Introduction
Mantle cell lymphoma (MCL) is an aggressive B-cell lymphoma with distinct pathologic and clinical features.1 The genetic hallmark of MCL is the t(11;14)(q13;q32) that leads to a constitutive expression of CCND1 (11q13) through the action of enhancer elements in the immunoglobulin heavy chain locus (IGH) (14q32). CCND1 encodes cyclin D1 that plays an important role in the G1/S transition of the cell cycle. Although t(11;14)/IGH-CCND1 can be detected in nearly all MCLs, rare t(11;14)-negative or cyclin D1–negative MCL cases have been published.2-5 Recently, Fu et al6 reported 6 such cases with otherwise typical morphology, phenotype, and clinical features of MCL. Importantly, those cases displayed a gene expression signature of CCND1-positive MCLs but they expressed rather CCND2 or CCND3. Their findings not only show that CCND1-negative MCLs are part of the spectrum of MCL but also suggest that aberrant expression of cyclin D2 or D3, 2 other members of the D family of cyclins, can substitute functionally for cyclin D1 in MCL. The molecular mechanisms underlying the pathogenesis of those cases remain largely unknown. We report here 8 t(11;14)-negative MCL cases collected in our institution in the past 15 years that showed the highly variable genetic pathogenesis. So far, this is the largest series of MCL lacking the IGH-CCND1 rearrangement.
Methods
Patients
Patients were selected from the lymphoma database of the Department of Pathology and the Center for Human Genetics, Katholieke Universiteit Leuven. Informed consent was obtained in accordance with the Declaration of Helsinki. Absence of the t(11;14)/IGH-CCND1 rearrangement in these cases was proven by conventional cytogenetics, fluorescence in situ hybridization (FISH), or both with locus-specific identifier (LSI) IGH/CCND1. The clinical features, morphology, and phenotype of these cases were reviewed, and additional immunophenotypic markers were analyzed. All cases were documented with frozen and paraffin-embedded material taken at diagnosis.
Immunohistochemistry
Additional immunohistochemistry (IHC) was performed on paraffin-embedded tissue sections using antibodies against cyclin D1 (Dako Denmark, Glosturp, Denmark), cyclin D2 (Santa Cruz Biotechnology, Santa Cruz, CA), cyclin D3 (Dako Denmark), cyclin E (Novocastra, Newcastle, United Kingdom), p27 (Dako Denmark), and RB1 (Novocastra). Sections were stained according to the manufacturer's recommendations, and staining results were visualized using the EnVision system (Dako Denmark). Negative as well as positive controls were performed. Images were captured with a Leica DMLB microscope (Leica, Wetzlar, Germany) using a Leica PL FLUOTAR objective lens (40×/0.70) and a Leica DC200 camera. Images were imported directly into PowerPoint (Microsoft, Redmond, WA) using the Leica DC200 camera software (version 2.51).
Cytogenetics and FISH
Conventional G-banding chromosomal analysis followed routine methods. In each case, 3 to 11 metaphase cells were analyzed. Chromosomal aberrations are presented in accordance with the International System for Human Cytogenetic Nomenclature.7 FISH was performed on slides prepared from remaining diagnostic or follow-up cytogenetic harvests. Applied probes included LSI IGH/CCND1, LSI IGH, LSI CCND1, LSI N-MYC, LSI p16/CEP9, LSI 13 (RB1 13q14), LSI p53 (Abbott Molecular, Ottignies, Belgium), break-apart probes for CCND2, CCND3, IGK, IGL6,8 and CCNE1 (RP11-372I05 and RP11-354B17), bacterial artificial chromosome (BAC) clones for 2pter, 6p21, 11q13, IGH, 20p selected from Ensembl,9 c123C12 for p27/CDKN1B,10 11q13 cosmids (6.7, 6.22, 3.62, and 3.91),11,12 and 24xCyte mFISH (MetaSystems, Altlussheim, Germany). Noncommercial probes were directly labeled with SpectrumOrange– and SpectrumGreen–d-UTP (Abbott Molecular) using random priming. FISH images were acquired with a 63×/1.4 oil-immersion objective in an Axioplan 2 fluorescence microscope equipped with an Axiophot 2 camera (Carl Zeiss, Jena, Germany) and a MetaSystems Isis imaging system (MetaSystems). One to 6 abnormal metaphase cells, 200 interphase nuclei, or both were evaluated in each FISH experiment.
Comparative expressed sequence hybridization
Total RNA was isolated from eight 20-μm sections of frozen tissue samples of each case by using the RNeasy Mini Kit (Qiagen, Dorking, United Kingdom) following the manufacturer's recommendations. The quality and concentration of the RNA were measured spectrophotometrically. Total RNA (1 μg) was reverse transcribed using random hexamers and superscript II (Invitrogen, Carlsbad, CA).
Comparative expressed sequence hybridization (CESH) was performed according to Lu et al.13 Briefly, test and reference cDNA were differentially labeled with SpectrumGreen- and SpectrumRed-dUTP (Abbott Molecular) during 2 rounds of degenerate oligonucleotide-primed polymerase chain reaction using the degenerate primer UN-1.14 Labeled probes were purified using Qiaquick PCR purification columns (Qiagen). Label incorporation was checked by the Nanodrop spectrophotometer (Isogen Life Science, Ijsselstein, The Netherlands) and varied between 25 and 50 pmol labeled dUTP/μg cDNA. Fluorescently labeled test and reference cDNA were prepared for competitive hybridization onto normal metaphase chromosomes using the CGH kit (Abbott Molecular). Hybridization was performed in a humid chamber for 72 hours at 36°C. Ten good-quality metaphases counterstained with DAPI (4,6-diamidino-2-phenylindole) were analyzed in each experiment. Image acquisition and analysis were performed using a Zeiss Axioplan 2 fluorescence microscope (Carl Zeiss) equipped with a cooled charge-coupled device camera COHU 4910 (Diagnostic Instruments, Sterling Heights, MI) and controlled by Cytovision software v2.81 (Applied Imaging International, Newcastle upon Tyne, United Kingdom). Dynamic standard reference intervals (SRIs) were used as thresholds for determination of relative overexpression and underexpression.15 These SRIs were created in our system as previously described16 and are based on systemic ratio variations seen in normal samples. The acquired CESH data were further interpreted by unsupervised hierarchical cluster analysis. Only those CESH regions registered at the 95% CI were included for this analysis. Data were imported into the Multiexperiment Viewer (The Institute for Genomic Research, http://www.tigr.org/software/), and an average linkage clustering analysis was performed applying the Euclidean distance metric.
Quantitative reverse-transcriptase–polymerase chain reaction
Total RNA used for CESH was subjected to RNase-free DNase treatment performed according to the manufacturer's instructions (Ambion, Austin, TX). DNase-free RNA (1 μg) was reverse transcribed into cDNA using random hexamers and superscript II (Invitrogen) according to the manufacturer's recommendations.
Primers for the MYCN gene within exon2 (forward primer 5′-GGC GTT CCT CCT CCA ACA-3′ and reverse primer 5′-CGT TCT TGG GAC GCA CAG T-3′) were designed with the PrimerExpress software (Applied Biosystems, Foster City, CA) to quantify relative expression levels. Quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) was performed using qPCR Master Mix for SYBR Green (Eurogentec, Seraing, Belgium) on lymph node (LN) samples from cases 1 to 8 and additional bone marrow (BM) and pleural effusion samples from case 8. Normal BM, LN, and t(11;14)-positive MCL were used as negative controls, and one case of neuroblastoma with amplification of MYCN was used as a positive control. Human β-actin gene was used as an internal control to which the threshold cycle (Ct) values of MYCN were normalized. qRT-PCR on 10 ng of total RNA was also performed with the MYCN primer pair to assess the genomic DNA contamination. All samples were run in triplicate, and data were analyzed using the comparative Ct method (ΔΔCt) as previously described.17 The threshold value for overexpression was calculated by the mean fold difference of BM and LN control samples plus 3 times the SD.
Results
Clinical features
The relevant clinical characteristics of the reported cases are shown in Table 1. There were 6 male and 2 female patients aged 42 to 76 years (median age, 57.7 years). The patients showed an advanced stage of disease with BM involvement at presentation and short survival (range, 12-64 months from diagnosis; median, 32 months). Of note, 5 cases showed secondary involvement of the central nervous system (CNS) during the later disease course. Time from initial diagnosis to CNS invasion ranged between 6 and 57 months (median, 31.8 months). The latter 5 patients died within the first year (range, 3 weeks to 9 months; median, 4.3 months) after diagnosis of secondary CNS involvement.
Relevant clinical and pathologic features of t(11;14)–negative MCL cases
| Case no. . | Sex . | Age, y . | Ann Arbor stage . | Extranodal sites . | Initial therapy . | Response . | Follow-up, mo/status . | Pathologic features . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker expression . | Growth pattern/cytology . | ||||||||||||||
| Cyclin . | p27 . | RB1 . | Other reviewed markers . | ||||||||||||
| CD3 . | CD5 . | CD10 . | CD20 . | ||||||||||||
| 1 | F | 43 | IVB | BM, T | CHVmP-BV, CEP, DHAP | PR | 19/D | D1 | +w | + | — | +w | — | + | Vagualy nodular/typical |
| 2 | M | 51 | IVB | BM, S, CNS | CHVmP-BV | PR | 64/D | D1 | + | + | — | + | — | + | Nodular/typical |
| 3 | M | 65 | IVB | BM, PB | CT and RT (no details) | PD | 15/D | D2 | + | + | — | + | — | + | Diffuse/typical |
| 4 | M | 56 | IVA | BM, CNS | CVP, rituximab, fludarabine, RT | PR | 62/D | D2w | + | + | — | + | — | + | Vaguely nodular/typical |
| 5 | F | 76 | NA | BM, PB | NA | NA | NA | D3 | + | + | — | + | — | + | Diffuse/typical |
| 6 | M | 63 | IVB | BM, S, PB, CNS | Chlorambucil, CHOP | CR | 12/D | D3 | + | — | — | + | — | + | Diffuse/blastoid |
| 7 | M | 66 | IVB | BM, CNS | CHVmP-BV | CR | 15/D | D3w/Ew | +w | — | — | — | — | + | Diffuse/blastoid |
| 8 | M | 42 | IVB | BM, CNS | CHOP | CR | 37/D | E | + | — | — | +w | — | + | Diffuse/blastoid |
| Case no. . | Sex . | Age, y . | Ann Arbor stage . | Extranodal sites . | Initial therapy . | Response . | Follow-up, mo/status . | Pathologic features . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker expression . | Growth pattern/cytology . | ||||||||||||||
| Cyclin . | p27 . | RB1 . | Other reviewed markers . | ||||||||||||
| CD3 . | CD5 . | CD10 . | CD20 . | ||||||||||||
| 1 | F | 43 | IVB | BM, T | CHVmP-BV, CEP, DHAP | PR | 19/D | D1 | +w | + | — | +w | — | + | Vagualy nodular/typical |
| 2 | M | 51 | IVB | BM, S, CNS | CHVmP-BV | PR | 64/D | D1 | + | + | — | + | — | + | Nodular/typical |
| 3 | M | 65 | IVB | BM, PB | CT and RT (no details) | PD | 15/D | D2 | + | + | — | + | — | + | Diffuse/typical |
| 4 | M | 56 | IVA | BM, CNS | CVP, rituximab, fludarabine, RT | PR | 62/D | D2w | + | + | — | + | — | + | Vaguely nodular/typical |
| 5 | F | 76 | NA | BM, PB | NA | NA | NA | D3 | + | + | — | + | — | + | Diffuse/typical |
| 6 | M | 63 | IVB | BM, S, PB, CNS | Chlorambucil, CHOP | CR | 12/D | D3 | + | — | — | + | — | + | Diffuse/blastoid |
| 7 | M | 66 | IVB | BM, CNS | CHVmP-BV | CR | 15/D | D3w/Ew | +w | — | — | — | — | + | Diffuse/blastoid |
| 8 | M | 42 | IVB | BM, CNS | CHOP | CR | 37/D | E | + | — | — | +w | — | + | Diffuse/blastoid |
BM indicates bone marrow; T, tonsil; CHVmP-BV, cyclophosphamide, adriamycine, teniposide, methylprednisolone, bleomycine, vincristine; CEP, CCNU, etopsoide, prednisone; DHAP, cisplatinum, cytarabine, dexamethasone; PR, partial response; D, died; w, weak; S, spleen; CNS, central nervous system; PB, peripheral blood; CT, chemotherapy; RT, radiotherapy; PD, progressive disease; CVP, cyclophosphamide, vincristine, prednisone; NA, not available; CHOP, cyclophosphamide, adriamycine, vincrisitine, prednisone; and CR, complete response.
Histologic and immunohistochemical findings
Histology showed a nodular or vaguely nodular growth pattern in 3 cases and a diffuse growth pattern in the others (Table 1). Five lymphomas consisted of tumor cells with typical mantle cell cytology, and blastoid cytology was found in 3 cases. All cases showed a B-cell phenotype with coexpression of CD5 antigen in all but case 7. Nuclear expression of cyclin D1, D2, and D3 was found in, respectively, 2, 2, and 3 cases (Table 1; Figure 1A-C). Expression of these cyclins was observed in almost 90% of the tumor cells. Case 8 did not show expression of any cyclin D. All cases were further analyzed with antibodies against cyclin E, p27, and RB1. Interestingly, cases 7 and 8 showed a positive nuclear expression of cyclin E, with a variable staining intensity in approximately 15% and 30% of the cells, respectively (Figure 1D). Cases 1 to 6 were cyclin E negative. All cases expressed p27. Nuclear RB1 positivity was found in all MCL cases with a typical morphology (cases 1-5), whereas the tumor cells of cases with a blastoid morphology (cases 6-8) were RB1 negative (Table 1).
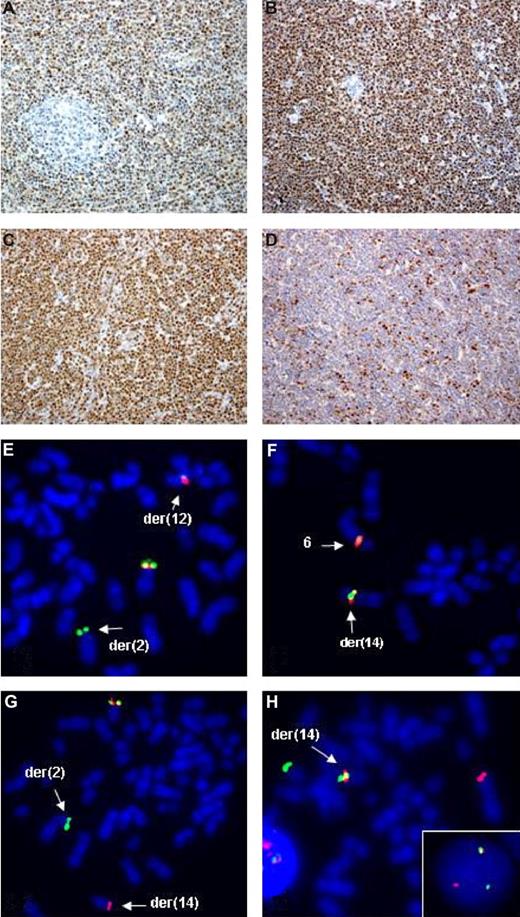
Examples of IHC and FISH analysis on t(11;14)-negative MCL. Upper panels show IHC findings in cases 2 (A), 3 (B), 6 (C), and 8 (D) using antibodies against cyclin D1, D2, D3, and cyclin E, respectively. Note a t(11;14)-independent expression of cyclin D1 in case 2 (A), expression of cyclin D2 in case 3 with a t(2;12)(p11;p13)IGK/CCND2 rearrangement (B), expression of cyclin D3 in case 6 with a t(6;14)(p21;q32)IGH/CCND3 aberration (C), and expression of cyclin E in case 8 negative for D-type cyclins. Lower panels are examples of FISH performed in cases 3 (E), 6 (F), 7 (H, inset) and 8 (G,H). Applied probes were CCND2 break-apart (E), CCND3 break-apart (F), LSI IGH (G), and 3′IGH (SpectrumGreen)/LSI N-MYC (SpectrumOrange) (H). Note a split of signals from probes flanking CCND2 in case 3 (E), translocation of CCND3 on a der(14q) in case 6 (F), split of LSI IGH signals heralding a cryptic t(2;14)(p24;q32) in case 8 (G), a fusion (red LSI N-MYC/2p24 and green 3′IGH/14q32) signal on a der(14) in case 8 with t(2;14) (H), and the same FISH pattern detected in interphase cells from case 7 (H, inset).
Examples of IHC and FISH analysis on t(11;14)-negative MCL. Upper panels show IHC findings in cases 2 (A), 3 (B), 6 (C), and 8 (D) using antibodies against cyclin D1, D2, D3, and cyclin E, respectively. Note a t(11;14)-independent expression of cyclin D1 in case 2 (A), expression of cyclin D2 in case 3 with a t(2;12)(p11;p13)IGK/CCND2 rearrangement (B), expression of cyclin D3 in case 6 with a t(6;14)(p21;q32)IGH/CCND3 aberration (C), and expression of cyclin E in case 8 negative for D-type cyclins. Lower panels are examples of FISH performed in cases 3 (E), 6 (F), 7 (H, inset) and 8 (G,H). Applied probes were CCND2 break-apart (E), CCND3 break-apart (F), LSI IGH (G), and 3′IGH (SpectrumGreen)/LSI N-MYC (SpectrumOrange) (H). Note a split of signals from probes flanking CCND2 in case 3 (E), translocation of CCND3 on a der(14q) in case 6 (F), split of LSI IGH signals heralding a cryptic t(2;14)(p24;q32) in case 8 (G), a fusion (red LSI N-MYC/2p24 and green 3′IGH/14q32) signal on a der(14) in case 8 with t(2;14) (H), and the same FISH pattern detected in interphase cells from case 7 (H, inset).
CESH
CESH techniques give a genomewide view of relative expression patterns within tissues according to chromosomal location in a way similar to that of conventional comparative genomic hybridization, using, however, cDNA but not DNA.13 We have successfully applied CESH for studies of leukemia and lymphoma.16,18
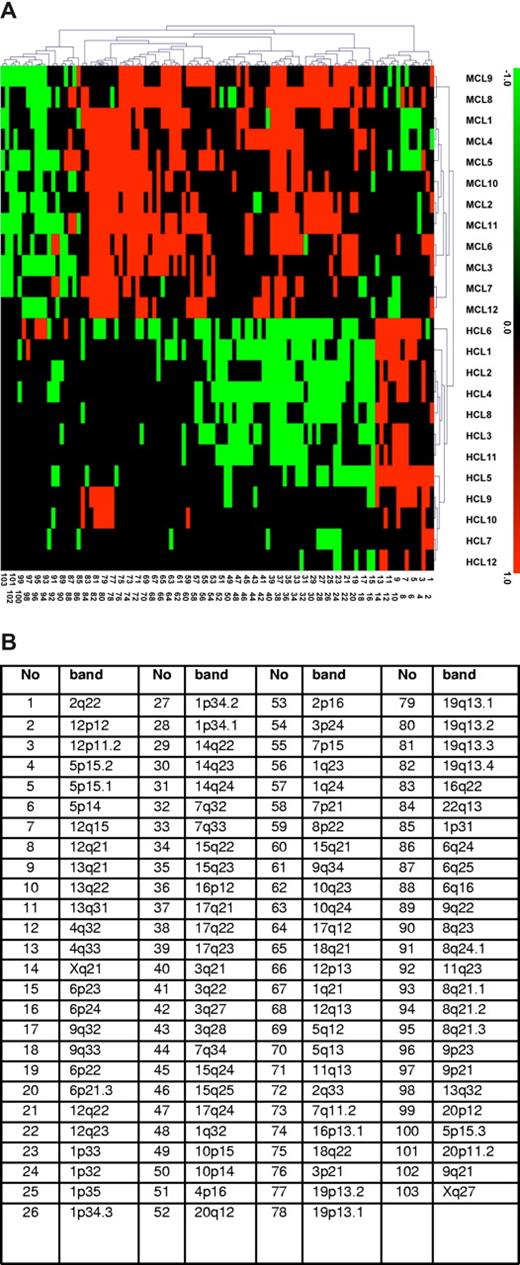
CESH analysis was performed on the 8 t(11;14)-negative MCL cases and 4 control t(11;14)-positive MCLs. For hybridization, differentially labeled test cDNA (MCL) and a reference cDNA (normal LNs) were used. The obtained CESH profiles showed numerous differentially expressed regions involving almost all chromosomes (data not shown). Cases 1 to 5 with a typical cytology had on average 34.6 imbalances per case (range, 27-44 imbalances), cases 6 to 8 with a blastoid cytology showed an average of 37 imbalances (range, 31-40 imbalances), and control t(11;14)-positive MCL displayed on average 31 imbalances per case (range, 19-36 imbalances). There were more overexpressed than underexpressed regions. Chromosomal regions showing a high differential overexpression (↑) and underexpression (↓) in more than 50% of cases were localized at 3p21 (↑), 5q13 (↑), 6p21-22 (↑), 7q11.2 (↑), 7q32-35 (↑), 10q24 (↑), 11q13 (↑), 15q22 (↑), 16p12.1 (↑), 17q21 (↑), 18q22 (↑), 19q13 (↑) and 5p15.2 (↓), 8q21 (↓), 9q21-22 (↓), 20p11 (↓). To compare gene expression profiles of t(11;14)–negative MCL with that of t(11;14)-positive MCL, the acquired CESH data were further interpreted by unsupervised hierarchical cluster analysis. The previously obtained CESH data on 12 cases of hairy cell leukemia (HCL)16 were also included in this analysis. Unsupervised analysis showed the clustering of all 8 t(11;14)-negative cases together with 4 t(11;14)-positive MCLs but separately from the HCL cases (Figure 2).
Gene-expressing profiling by CESH showed clustering t(11;14)-negative and t(11;14)-positive MCLs. (A) Results of unsupervised hierarchical cluster analysis of CESH data obtained in cases 1 to 8 (MCL1-8), 4 control t(11;14)-positive MCL cases (MCL9-12), and 12 additionally included HCL cases (HCL1-12). Each row represents one hybridization experiment and each column represents an informative differentially expressed chromosomal region described in panel B. Relative overexpression and underexpression are depicted as red and green, respectively, whereas black squares represent chromosomal regions showing no differential expression. Only regions recorded in at least 25% of all cases (at least 7 of 26) are displayed in this diagram. Note a clustering of all 8 t(11;14)-negative and 4 typical MCL cases and their separation from 12 HCL cases.
Gene-expressing profiling by CESH showed clustering t(11;14)-negative and t(11;14)-positive MCLs. (A) Results of unsupervised hierarchical cluster analysis of CESH data obtained in cases 1 to 8 (MCL1-8), 4 control t(11;14)-positive MCL cases (MCL9-12), and 12 additionally included HCL cases (HCL1-12). Each row represents one hybridization experiment and each column represents an informative differentially expressed chromosomal region described in panel B. Relative overexpression and underexpression are depicted as red and green, respectively, whereas black squares represent chromosomal regions showing no differential expression. Only regions recorded in at least 25% of all cases (at least 7 of 26) are displayed in this diagram. Note a clustering of all 8 t(11;14)-negative and 4 typical MCL cases and their separation from 12 HCL cases.
Cytogenetic, FISH, and qRT-PCR analyses
Cytogenetics displayed clonal chromosomal aberrations in 6 of the 8 cases (Table 2). Two of them showed a relatively simple karyotype with 2 to 3 aberrations, whereas the remaining 4 cases, including both blastoid cases, displayed complex rearrangements with 6 to 17 aberrations per karyotype. Noteworthy was the presence of t(2;12)(p11;p13) in case 3, involving regions of IGK and CCND2, respectively.
Results of cytogenetic and FISH analysis
| Case no. . | Cytogenetics . | FISH . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample/status . | Results . | Examined loci . | Other probes . | Newly identified aberrations . | ||||||||||
| IGH . | IGK . | IGL . | CCND1 . | CCND2 . | CCND3 . | p16 . | p27 . | RB1 . | TP53 . | |||||
| 1* | LN/D | 46, XY[10] | ||||||||||||
| BM/P | 44, XX, del(1)(p11p31), add(10)(p15),-11, -12,-14, -15,-20,-22,+4mar [3] | 2F | 2F | 2F | 2F | 2F | 2F | ND | ND | ND | ND | |||
| PB/P | 44, idem, t(4;9)(q13; q13)[1][cp3] | |||||||||||||
| 2* | S/P | 46-47, XY, del(3)(q21)[8], add(5)(p15),-6 [3], add(6) (q25)[8], der(14)t(7;14) (p11;p11),+mar [cp11] | 2F | 2F | 2F | 2F | 2F | 2F | 2s | 2s | 2s | 2s | ||
| 3* | LN/P | 48, XY, t(2;12)(p11;p13),+3, +15 [6] | 2F | 1F1R1G (25%) | 2F | 2F | 1F1R1G (26%) | 2F | 1s (28%) | 2s | 2s | 2s | del(9)(p21p21) | |
| 4 | LN/D | 46, XY, del(2)(q31), t(9;11) (q34;q13)[5] | 2F | 2F | 2F | 2F | 2F | 2F | 2s | 2s | 2s | 2s | ish der(9)t(9;11)(q34;q13) (CCND1+, RP5-901A4+) | |
| 5 | LN/P | No mitosis | 2F | 2F | 2F | 2F | 2F | 2F | 2s | 2s | 2s | 2s | ||
| 6 | LN/D | 46, XY[5] | ||||||||||||
| PB/D | 45-47, XY,+3, del(4)(q31), del(5)(q21q33),-6[4], del(6) (q14q24), add(6)(q12)[2], +8, t(8;11) ((q21;q23),-9,-10, -12, add(14)(q32), der(15) t(10;15)(q21;q14),-17,+19, del(20)(p11), +1-2mar [cp6] | 1F1R1G (76%) | 2F | 2F | 2F | 2F | 2F | 1s (74%) | 2s | 1s (78%) | 1s (78%) | ish add(14)(q32)(3′IGH+, 5′IGH-, wcp6+, CCND3+, RP11-321B9+);ish del(20) (p11)(5′IGH+, CEP20+, RP11-329D4-, CTB- 530N10-, CTP-81F12+) | der(14)t(6;14)(p21;q32), der(20) t(14;20)(q32;p11), del(13)(q14q14) | |
| 7 | LN/D | No mitosis | 1F1R1G (80%) | 2F | 2F | 2F | 3F (30%) | 2F | 1s (80%) | 1s (34%) | 1s (78%) | 2s | nuc ish(MYCNx2),(3′IGHx2), (MYCN con 3′IGHx1); (RP11-282G6x2, RP11- 542H15x2)(RP11-282G6 sep RP11-542H15x1) | t(2;14)(p24;q32), del(9)(p21p21), del(12)(p13p13), del(13)(q14q14), |
| 8 | BM/D | 49, XY,+3, del(6)(q23q25), del(9)(p21), del(13) (q14q22), -15,+18,+add(18)(p11), +19 [5] | 1F1R1G (75%) | 2F | 2F | 2F | 2F | 2F | 1s (22%) | 2s | 1s (79%) | 2s | ish der(14)t(2;14)(p24;q32) (3′IGH+,5′IGH-, RP11- 417P24+, MYCN+, RP11 -282G6+); ish der(2)t(2;14) (p24;q32) (MYCN-, RP11- 282G6-, RP11-542H15+, 5′IGH+, RP11-417P24+); nuc ish (CCNE1x3) | t(2;14)(p24;q32) |
| Case no. . | Cytogenetics . | FISH . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample/status . | Results . | Examined loci . | Other probes . | Newly identified aberrations . | ||||||||||
| IGH . | IGK . | IGL . | CCND1 . | CCND2 . | CCND3 . | p16 . | p27 . | RB1 . | TP53 . | |||||
| 1* | LN/D | 46, XY[10] | ||||||||||||
| BM/P | 44, XX, del(1)(p11p31), add(10)(p15),-11, -12,-14, -15,-20,-22,+4mar [3] | 2F | 2F | 2F | 2F | 2F | 2F | ND | ND | ND | ND | |||
| PB/P | 44, idem, t(4;9)(q13; q13)[1][cp3] | |||||||||||||
| 2* | S/P | 46-47, XY, del(3)(q21)[8], add(5)(p15),-6 [3], add(6) (q25)[8], der(14)t(7;14) (p11;p11),+mar [cp11] | 2F | 2F | 2F | 2F | 2F | 2F | 2s | 2s | 2s | 2s | ||
| 3* | LN/P | 48, XY, t(2;12)(p11;p13),+3, +15 [6] | 2F | 1F1R1G (25%) | 2F | 2F | 1F1R1G (26%) | 2F | 1s (28%) | 2s | 2s | 2s | del(9)(p21p21) | |
| 4 | LN/D | 46, XY, del(2)(q31), t(9;11) (q34;q13)[5] | 2F | 2F | 2F | 2F | 2F | 2F | 2s | 2s | 2s | 2s | ish der(9)t(9;11)(q34;q13) (CCND1+, RP5-901A4+) | |
| 5 | LN/P | No mitosis | 2F | 2F | 2F | 2F | 2F | 2F | 2s | 2s | 2s | 2s | ||
| 6 | LN/D | 46, XY[5] | ||||||||||||
| PB/D | 45-47, XY,+3, del(4)(q31), del(5)(q21q33),-6[4], del(6) (q14q24), add(6)(q12)[2], +8, t(8;11) ((q21;q23),-9,-10, -12, add(14)(q32), der(15) t(10;15)(q21;q14),-17,+19, del(20)(p11), +1-2mar [cp6] | 1F1R1G (76%) | 2F | 2F | 2F | 2F | 2F | 1s (74%) | 2s | 1s (78%) | 1s (78%) | ish add(14)(q32)(3′IGH+, 5′IGH-, wcp6+, CCND3+, RP11-321B9+);ish del(20) (p11)(5′IGH+, CEP20+, RP11-329D4-, CTB- 530N10-, CTP-81F12+) | der(14)t(6;14)(p21;q32), der(20) t(14;20)(q32;p11), del(13)(q14q14) | |
| 7 | LN/D | No mitosis | 1F1R1G (80%) | 2F | 2F | 2F | 3F (30%) | 2F | 1s (80%) | 1s (34%) | 1s (78%) | 2s | nuc ish(MYCNx2),(3′IGHx2), (MYCN con 3′IGHx1); (RP11-282G6x2, RP11- 542H15x2)(RP11-282G6 sep RP11-542H15x1) | t(2;14)(p24;q32), del(9)(p21p21), del(12)(p13p13), del(13)(q14q14), |
| 8 | BM/D | 49, XY,+3, del(6)(q23q25), del(9)(p21), del(13) (q14q22), -15,+18,+add(18)(p11), +19 [5] | 1F1R1G (75%) | 2F | 2F | 2F | 2F | 2F | 1s (22%) | 2s | 1s (79%) | 2s | ish der(14)t(2;14)(p24;q32) (3′IGH+,5′IGH-, RP11- 417P24+, MYCN+, RP11 -282G6+); ish der(2)t(2;14) (p24;q32) (MYCN-, RP11- 282G6-, RP11-542H15+, 5′IGH+, RP11-417P24+); nuc ish (CCNE1x3) | t(2;14)(p24;q32) |
LN, lymph node; D, diagnosis; BM, bone marrow; P, progression; F, fused signal; ND, not done; PB, peripheral blood; S, spleen; s, signal; R, red signal; G, green signal.
Previously published case.2
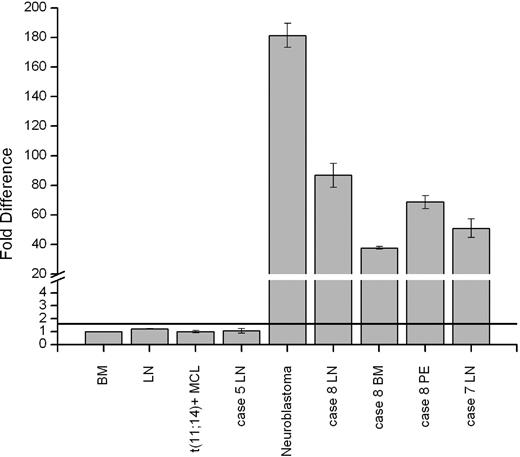
All 8 cases were further analyzed by FISH with break-apart probes for CCND1, CCND2, CCND3, IGH, IGK, and IGL. FISH aberrations were detected in 4 cases. As expected, the t(2;12)(p11;p13) found in cyclin D2–expressing case 3 showed rearrangements of IGK and CCND2 (Figure 1E). In case 6 with add(14)(q32), translocation of the 5′IGH region (green signal) on del(20)(p11) was seen. Further multicolor FISH analysis identified the add(14)(q32) as a der(14)t(6;14) and pointed to an unbalanced 3-way t(6;14;20)(p21;q32;p11) associated with loss of the der(6). CCND3 break-apart probes hybridized with the der(14) (Figure 1F), and, eventually, the 6p21 breakpoint was mapped approximately 0.5 Mb centromeric to CCND3 (RP1-321B9 < - > RP1-242G1). The resulting IGH-CCND3 aberration correlated with the expression of cyclin D3 in this lymphoma. Additional FISH analysis of the der(20)t(14;20) mapped the 20p11 breakpoint in the near-centromeric, gene-negative region distally flanked by RP13-329D4. Case 7, with unsuccessful cytogenetic analysis, showed a split of LSI IGH signals, heralding t(14q32). The CCND1 to CCND3 loci examined in this case, however, displayed a normal FISH pattern. Particularly interesting were FISH findings in case 8. Hybridization with LSI IGH showed a red signal on a normal-looking chromosome 14 and a green signal at 2pter, indicating a cryptic t(2;14) (Figure 1G). The IGH breakpoint was further mapped in the IGCH region covered by RP11-417P24. Using a set of BACs from 2p23-p25, the breakpoint on chromosome 2 was mapped in the 2p24.3 region flanked by RP11-282G6 and RP11-542H15. The first gene located approximately 545 kilobases (kb) telomeric from the breakpoint was MYCN. To prove that t(2;14)(p24;q32) targets MYCN, we applied qRT-PCR to analyze the expression of MYCN mRNA in LN, BM, and pleural effusion (PE) samples from this case. In comparison with normal BM, LN, and 2 typical MCLs, qRT-PCR detected 86.7-, 37.7-, and 68.6-fold up-regulation of MYCN mRNA in these samples, respectively (Figure 3). An 180-fold expression of MYCN mRNA was seen in a neuroblastoma case with amplification of MYCN. Expression of MYCN was further analyzed in all the remaining t(11;14)–negative cases. Only case 7 with a presumed t(14q32) showed an up-regulated expression of MYCN (51-fold; Figure 3). FISH with LSI N-MYC (SpectrumOrange) and 3 pooled BACs from the 3′ region of IGH (SpectrumGreen-labeled RP11-1087P08, RP11-346I20, RP11-675H01) showed one red, one green, and one fusion signal in interphase cells confirming t(2;14)(p24;q32) (Figure 1H inset). A rearrangement of the MYCN region was further proven by FISH with LSI N-MYC combined with RP11-542H15 that bordered the centromeric 2p24 breakpoint in case 8. The observed 1 fusion signal and 2 separated red and green signals are evidence of the 2p24 rearrangement also in this case. Given that further IHC studies showed expression of cyclin E in both cases with t(2;14), we additionally checked FISH status of CCNE1 with probes flanking the gene. Case 7 showed 2-fusion signals and case 8 showed 3-fusion signals (trisomy 19) in 78% of interphase nuclei but no break-apart rearrangement of CCNE1.
MYCN is up-regulated in MCL cases with t(2;14)(p24;q32). Expression of MYCN in 3 samples (LN, BM, and PE) from case 8 and LN samples from cases 5 and 7. Note up to 90-fold overexpression of MYCN in the analyzed samples from case 8 and case 7 with t(2,14)(p24;q32)/IGH-MYCN. The negative controls and the LN sample from case 5 did not exceed the threshold level (black line) of 1.6. The neuroblastoma case that was used as positive control showed up to 180-fold overexpression of MYCN mRNA. The qRT-PCR data are representative of three independent experiments. Measurements were performed in triplicate and results are means (± SEM).
MYCN is up-regulated in MCL cases with t(2;14)(p24;q32). Expression of MYCN in 3 samples (LN, BM, and PE) from case 8 and LN samples from cases 5 and 7. Note up to 90-fold overexpression of MYCN in the analyzed samples from case 8 and case 7 with t(2,14)(p24;q32)/IGH-MYCN. The negative controls and the LN sample from case 5 did not exceed the threshold level (black line) of 1.6. The neuroblastoma case that was used as positive control showed up to 180-fold overexpression of MYCN mRNA. The qRT-PCR data are representative of three independent experiments. Measurements were performed in triplicate and results are means (± SEM).
The remaining 4 t(11;14)-negative cases (1, 2, 4, and 5) did not show any FISH aberration of the examined loci. Lack of CCND1 rearrangement in cases 1 and 2 expressing cyclin D1 was further proved by FISH with 11q13 cosmid probes covering and flanking CCND1.11 Case 4 with t(9;11)(q34;q13) showed an 11q13 breakpoint at least 2 Mb distal from CCND1. Expression of cyclin D3 (but not D1) by this lymphoma additionally proved that CCND1 was not targeted by the t(9;11)(q34;q13) (see Table 1).
The status of p16/CDKN2A (9p21), p27/CDKN1B (12p13), RB1 (13q14), and TP53 (17p13) was analyzed by interphase FISH in 7 available cases. We detected monoallelic loss of 1 to 3 examined loci in 4 cases, including the 3 blastoid variants of MCL (Table 2). Briefly, case 3 lost one p16 allele; case 6 lost p16 (monosomy 9), RB1, and TP53 (monosomy 17); case 7 showed single signals of p16, p27 (subclonal), and RB1; and case 8 lost p16 (subclonal) and RB1 (del(13)).
Discussion
We report 8 MCL cases negative for t(11;14)/IGH-CCND1 or variant CCND1 rearrangements with otherwise classical histologic and immunophenotypic features of MCL collected in our institution in the past 15 years. Three of them displayed a blastoid cytology. One of the latter cases (case 7) was CD5 negative. Although expression of this glycoprotein is characteristic for MCL, sporadic lack of CD5 reactivity has been recurrently observed in MCL, particularly in a blastoid variant of MCL (BV-MCL).18,19 Clinical features of t(11;14)-negative MCLs, including male predominance, advanced stage of disease at presentation with BM involvement, and a short survival, are reminiscent of t(11;14)-positive MCL. Of note, 5 cases, including 2 typical and all 3 BV-MCLs, showed secondary involvement of the CNS during the course of the disease. The high incidence of this complication in the present series (62.5%) contrasts with CNS invasion in t(11;14)-positive MCL which in our institution was registered in 6.8% of the cases. As cerebrospinal fluid examination, CNS imaging, or both were performed only in case of neurologic symptoms, data of occult CNS involvement are lacking. The reported CNS involvement in MCL varied between 4% and 22% in retrospective analyses.19,21,22 In a recent study of Ferrer et al,23 CNS involvement was detected in 13% of patients (11 of 83) with MCL; that is, 6 (8%) of 62 typical MCLs and 5 (23%) of 21 blastoid MCLs. Our findings indicate that CNS infiltration in t(11;14)-negative MCL heralds a poor prognosis, similar to that in t(11;14)-positive MCL. To the best of our knowledge, this is the first report of secondary CNS involvement in t(11;14)-negative MCL.
Gene expression profiling by CESH further underscores the relation between t(11;14)-negative and t(11;14)-positive MCL. Unsupervised hierarchical cluster analysis showed clustering of all 8 t(11;14)-negative MCL cases together with 4 t(11;14)-positive MCL cases but separately from HCL (Figure 2). These findings further support the suggestion of Fu et al6 that both MCL subtypes belong to the same entity.
Our initial IHC studies showed that t(11;14)-negative MCLs express cyclin D1 (2 cases), D2 (2 cases), and D3 (3 cases; Table 1), consistent with published data.6 However, a novel finding was the identification of one case that did not express any cyclin D. The pattern of cyclin D expression in t(11;14)-negative MCL resembles that in multiple myeloma (MM). In the latter neoplasm, dysregulation of cyclins D, driven or not driven by chromosomal translocations, was recognized as an early and common pathogenic event documented in 98% of cases.24 However, approximately 2% of MMs do not express any cyclin D.
To check whether ectopic expression of cyclins D in t(11;14)-negative cases is caused by chromosomal aberrations of the respective loci, all 8 cases were analyzed by FISH. These studies showed an IGK-CCND2 rearrangement in one of the cyclin D2–positive MCL cases with t(2;12)(p11;p13). Interestingly, this translocation was shown to be recurrent in cyclin D1–negative MCL because 2 similar cases with t(2;12)(p11;p13) were recently reported by Gesk et al.25 We further identified a t(6,14)(p21;q32)/IGH-CCND3 aberration in one of the cyclin D3–positive cases. The latter case is the first reported MCL with t(6;14)(p21;q32), although this translocation has been recurrently observed in MM and various other mature B-cell malignancies.26,27 Finally, we identified a novel IGH-mediated t(2;14)(p24;q32) targeting MYCN in 2 of the 3 blastoid MCL cases. This submicroscopic translocation resulted in up-regulation of MYCN mRNA, as proven by qRT-PCR, seems to be associated with a poor clinical outcome. MYCN is a member of the MYC family of helix-loop-helix transcription factors, well known for their potent oncogenic activities in a large number of human cancers.28 MYC proteins affect a variety of cellular functions, including cell-cycle regulation, apoptosis, metabolism, cell adhesion, and cellular differentiation.29,30 Genetic abnormalities underlying up-regulation of MYC oncogenes in tumors include chromosomal translocations, eg, t(8,14)(q24;q32)/IGH-MYC in Burkitt lymphoma,31 amplification of MYCL in lung carcinomas,32 and amplification of MYCN in neuroblastoma and a variety of neuroectodermal tumors.33 So far, translocations targeting MYCN have not been reported in lymphoma.
To investigate the status of some other components of the cell-cycle pathway in t(11;14)-negative MCL, we performed IHC studies of cyclin E, p27, and RB1 and FISH analysis of p16, p27, RB1, and TP53 in all available cases. Particularly interesting was the finding of cyclin E expression in 2 blastoid MCLs: one negative for cyclin D (case 8) and one displaying a weak expression of cyclin D3 (case 7). Notably, both these cases showed a cryptic t(2;14)(p24;q32), resulting in overexpression of MYCN. Cyclin E, a member of another class of mammalian G1 cyclins,34 is believed to control G1/S phase progression downstream of cyclin D and pRB. It associates with cyclin-dependent kinase 2 (CDK2), activates its kinase activity shortly before entry into the S phase, and phosphorylates proteins involved in DNA replication.35 An aberrant up-regulation of cyclin E has been detected in a variety of human tumors, including lymphoma (reviewed by Malumbres and Barbacid36 ). In MCL, an increased expression of cyclin E was found in blastoid cases but not in cases with a classic histology.37 Whether aberrant expression of cyclin E detected exclusively in 2 t(11;14)-negative cases with t(2;14) is functionally associated with up-regulation of MYCN remains unknown. So far, the functional consequences of MYCN overexpression in tumors and the exact pathways critically influenced by this oncogene are poorly understood. However, several lines of evidence indicate a role of MYCN in deregulation of the G1/S transition. It has been shown that overexpression of MYCN reduces the G1 phase of the neuroblastoma cell cycle,38 induces the reentry of quiescent cells into the cell cycle,39 stimulates cyclin E-CDK2 activity, and decreases activity of p27.40 In support of our findings are data showing that rodent neurons overexpressing N-myc have increased expression levels of cyclin E and CDK2 proteins.41 The finding of t(2;14) in 2 BV-MCLs expressing either cyclin D3 or any cyclin D suggests that this translocation is a secondary event in MCL, similar to that of t(8q24/MYC) sporadically observed in t(11;14)-positive MCL.42-44
Finally, we could show that p16/CDKN2, p27/CDKN1B, RB1, and TP53, 4 tumor suppressor genes of potential relevance for development and for progression of MCL,45-48 were commonly monoallelically deleted in all 3 t(11;14)-negative blastoid MCL cases. Particularly interesting was a correlation between loss of one copy of RB1 and lack of expression of RB1 protein in these cases. The latter finding probably heralds inactivation of RB1 in all 3 BV-MCLs, possibly because of mutations of the second not-deleted RB1 allele. RB1 is a key element in the cell-cycle machinery frequently affected by inactivated mutations in various human cancers.49 Approximately 50% of MCL harbors hemizygous deletions of RB1,50 ; however, inactivation of pRB1 is a rare event.51 Recently, Pinyol et al52 identified inactivating intragenic deletions of RB1 in 1 (UPN-1) of 4 studied MCL cell lines (UPN-1) and in 1 of 32 analyzed primary MCL cases comprising 22 typical and 10 blastoid variants. Of note, the original lymphoma from which UPN-1 was established and the latter case were diagnosed as a blastoid variant. The low incidence (10%) of RB1 inactivation events detected by this group in typical BV-MCL contrasts with our finding of loss of pRB1 expression in all 3 t(11;14)-negative BV-MCL.
In summary, our study further supports the existence of t(11;14)-negative MCL. These lymphomas predominantly express cyclin D2 or cyclin D3, the expression of which may be driven by IG-mediated translocations. However, t(11;14)-negative MCL cases that do express cyclin D1 in a translocation-independent manner or not express any cyclin D have been also identified in our series. The novel findings included identification of MYCN as a lymphoma oncogene targeted by a cryptic t(2;14)(p24;q32) in 2 MCLs with a blastoid cytology, expression of cyclin E in both cases with t(2;14), and loss of pRB1 in all 3 blastoid MCLs. A clinically distinctive feature of t(11;14)-negative MCLs appears to be their predisposition to CNS involvement. Altogether, our study sheds new light on the highly variable genetic pathogenesis underlying t(11;14)-negative MCL and underscores the diagnostic problem recognizing t(11;14)-negative MCLs.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank U. Pluys and M. Vanherck for their excellent technical assistance, I. Vanden Bempt for help in IHC studies, and R. Logist for editorial assistance. PV is a senior clinical investigator of Fonds voor Wetenschappelijk Onderzoek (FWO) Vlaanderen.
Authorship
Contribution: I.W. designed the study, interpreted data, and wrote the paper; D.D., G.V., and J.T provided and analyzed clinical data; V.V., K.V.R., and H.P. performed research; F.M. collected and reviewed cases; P.V. provided cytogenetic data and contributed to the paper; C.D.W-P. designed the study, reviewed cases, interpreted data, and contributed to the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Iwona Wlodarska, Center for Human Genetics, KU Leuven, Gasthuisberg, Herestraat 49, Box 602, B-3000 Leuven, Belgium; e-mail: iwona.wlodarska@uz.kuleuven.ac.be.