Hematopoietic stem cells (HSCs) are a key paradigm system for stem cell therapy applications. How these cells change from their multipotent to lineage-restricted cell states is still not fully understood. In this issue of Blood, McKinney-Freeman and colleagues show that members of a single transcription factor gene family are used in diverse and dynamic ways to implement this critical organogenesis program.
Organ development from multipotent precursor cells is a complex and opaque process even in perhaps the best-characterized mammalian stem cell system, the blood. These so-called hematopoietic stem cells (HSCs) are remarkable for their ability to reconstitute nearly the entire blood system when transplanted into a patient. Dramatic success in the treatment of innate and acquired blood disorders has demonstrated the full value of stem cell therapies. However, despite significant traction in this area, we still do not understand the differentiation cascade that leads to HSCs, or how HSCs make important decisions regarding proliferation versus differentiation. Arguably, the difficulty in obtaining full knowledge of the biology of this highly tractable system suggests that the broader impact of other, less well-studied and defined stem cells will be similarly limited in practical utility. The hope is that the paradigm developed for HSCs will not only have an impact on how these stem cells form for their organ (the blood), but will also provide key insight into how other stem cells operate in both the normal and disease state.
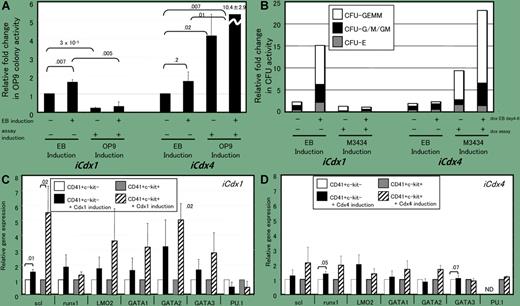
In the article by McKinley-Freeman and colleagues, these authors merge biological insight from diverse model systems. First, the caudal gene from the fruit fly (Drosophilia melanogaster) is identified as an important transcription factor in development. Second, in vivo data from the zebrafish demonstrate that several vertebrate orthologs of these genes are critical for blood development.1,2 Third, these authors deploy in vitro blood differentiation models based on embryonic stem cells that can be cultured to generate hematopoietic lineages. By the addition of 3 different orthologs of the caudal gene family from mammals, the authors show that these genes encode biochemically distinct protein forms that affect multiple aspects of HSC biology (see figure). Early in the life of an HSC lineage, cdx1 and cdx4 genes are capable of inducing differentiation, while cdx2 inhibits differentiation. However, in the hematopoietic progenitor differentiation step, cdx1 and cdx2 inhibit while cdx4 can enhance. This is critical, suggesting that the regulation of these genes provides a potential mechanism in HSC developmental decision-making processes. These genes do not appear to be the actual trigger, but they do appear to be important in implementing this key process in stem cell biology for blood lineages.
Cdx genes selectively affect the hematopoietic potential and hematopoietic-specific gene expression of CD41+c-kit+ EB-derived cells. See the complete figure in the article beginning on page 4944.
Cdx genes selectively affect the hematopoietic potential and hematopoietic-specific gene expression of CD41+c-kit+ EB-derived cells. See the complete figure in the article beginning on page 4944.
What are the broader implications of this work? First, assuming the in vitro system recapitulates the in vivo scenario, it implies that a similar focus on key transcription factor families is likely to yield important insight into other stem cell pathways. Secondly, this work suggests that these important differences among bioinformatically indistinguishable members of a protein family will need to be deduced using functional assay systems. The use of an in vitro stem cell–based model is one such tool, demonstrating a second and important role for in vitro culture systems for other organs. Will this be a common mechanism for other stem cell systems? That is not yet clear. But the HSC system is providing an important reference paradigm against which other systems can be compared.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal