Thrombin-activable fibrinolysis inhibitor (TAFI) is a plasma zymogen that acts as a molecular link between coagulation and fibrinolysis. Numerous single nucleotide polymorphisms (SNPs) have been identified in CPB2, the gene encoding TAFI, and are located in the 5′-flanking region, in the coding sequences, and in the 3′-untranslated region (UTR) of the CPB2 mRNA transcript. Associations between CPB2 SNPs and variation in plasma TAFI antigen concentrations have been described, but the identity of SNPs that are causally linked to this variation is not known. In the current study, we investigated the effect of the SNPs in the 5′-flanking region on CPB2 promoter activity and SNPs in the 3′-UTR on CPB2 mRNA stability. Whereas the 5′-flanking region SNPs (with 2 exceptions) did not have a significant effect on promoter activity, either alone or in haplotypic combinations seen in the human population, all of the 3′-UTR SNPs substantially affected mRNA stability. We speculate that these SNPs, in part, contribute to variation in plasma TAFI concentrations via modulation of CPB2 gene expression through an effect on mRNA stability.
Introduction
Thrombin-activable fibrinolysis inhibitor (TAFI), also known as procarboxypeptidase U or R or plasma procarboxypeptidase B, is a human plasma zymogen that may play a role in mediating the balance between blood coagulation and fibrinolysis.1 Upon cleavage of TAFI by thrombin,2 thrombin-thrombomodulin,3 or plasmin,4 an enzyme is formed (TAFIa) that possesses basic carboxypeptidase activity. TAFIa has been demonstrated to attenuate plasminogen activation, and thus fibinolysis, by removing from partially degraded fibrin the carboxyl-terminal lysine residues that mediate positive feedback in the fibrinolytic cascade.5 In addition, TAFIa has been shown to remove the carboxyl-terminal arginine residues from bradykinin and the anaphylatoxins C3a and C5a, thereby implicating the TAFI pathway as a link between coagulation and inflammation.6,,–9
Plasma concentrations of TAFI vary significantly in the human population.10,11 The vast majority of individuals have TAFI antigen levels between 50% and 150% of the mean population value,12,13 thereby ranging from approximately 100 to 200 nM. Importantly, these concentrations of TAFI fall below the Km for activation of TAFI by thrombin or thrombomodulin (1 μM),3 indicating that individuals with higher plasma TAFI concentrations would exhibit a higher rate of TAFIa production following a procoagulant stimulus. Indeed, variation in “functional” TAFI concentrations has been observed using a clot lysis assay of plasma samples.14 Therefore, it is reasonable to consider the gene encoding TAFI (CPB2) as a candidate gene for thrombotic disorders. Indeed, elevated concentrations of TAFI have been shown to be a mild risk factor for first venous thrombosis15 as well as recurrent venous thrombosis,16 and to be more common in carriers of Factor V Leiden with venous thromboembolism than in asymptomatic carriers.17 High functional TAFI concentrations have been found to be associated with an increased risk for ischemic stroke.18 While a possible association between plasma TAFI concentrations and coronary artery disease and coronary events has been a point of controversy,19,,,,–24 an association between plasma TAFI antigen concentrations and restenosis following percutaneous coronary interventions has been reported.25
It has been demonstrated that there is virtually no association between plasma TAFI concentrations and conventional cardiovascular risk factors,26 which led to the suggestion that plasma TAFI concentrations are largely under genetic control. Further studies identified numerous single nucleotide polymorphisms (SNPs) in the 5′-flanking, protein-coding, and 3′-untranslated coding regions of CPB2.27,28 Early studies determined that virtually all of these SNPs were strongly associated with plasma TAFI antigen concentrations,27,28 and that 2 SNPs in CPB2 accounted for approximately 60% of variation in plasma TAFI antigen concentrations.27 However, it was subsequently determined that the enzyme-linked immunosorbent assays (ELISAs) used in many of these studies were sensitive to the Thr/Ile polymorphism at position 325 and thus the extent of variation in plasma TAFI antigen concentrations, the association between individual SNPs and TAFI levels, and the extent to which the SNPs explained TAFI antigen variation was overestimated.12,13,29 The most recent analyses indicate that less than 20% of the variation in plasma TAFI antigen concentrations can be attributed to SNPs.13,30 However, there is extensive linkage disequilibrium between all of the SNPs,27,28,30 complicating the identification of SNPs that directly influence plasma TAFI concentrations, such as by altering expression of the gene encoding TAFI. Therefore, in the current work, we have assessed the effect of polymorphisms in the CPB2 5′-flanking region and 3′-untranslated region (UTR) on promoter activity and mRNA stability, respectively.
Methods
Work on human DNA and plasma samples was conducted at the University of Vermont and the University of Sao Paolo. Informed consent was obtained from patients in accordance with the Declaration of Helsinki for donation of blood samples and the protocols were approved by the institutional review boards at these respective locations.
Reporter plasmids and mutagenesis
The parental luciferase reporter plasmid used for the mutagenesis was TAFI[−2699]-luc, described by Boffa and coworkers.31 This construct consists of the CPB2 5′-flanking region spanning the EcoRI site at −2699 to the HindIII site at +21 inserted into pGL3 Basic (Promega, Madison, WI). The initiator methionine codon immediately upstream of the HindIII site was mutated to TTG. Nucleotide numbering is as described,31 where the +1 nucleotide is the first nucleotide of the cDNA reported by Eaton and coworkers.32 The sequence of the genomic insert is identical to that described by Boffa et al.31
All of the reported SNPs27,28 as well as an additional novel SNP −298G/A in the CPB2 5′-flanking region were introduced into the TAFI[−2699]-luc plasmid by site directed mutagenesis using either the GeneEditor (Promega) or QuikChange (Stratagene, La Jolla, CA) kits according to the manufacturers' recommendations. Nucleotide numbering is as in the corresponding first reports of the SNPs27,28 and in the original description of the sequence of the CPB2 5′-flanking region.31 The corresponding rs numbers for each SNP are presented in Table 1.33 Oligonucleotides used in the mutagenesis reactions were purchased from Cortec DNA Service Laboratories (Kingston, ON). The presence of the appropriate mutations was verified by DNA sequence analysis. Where multiple substitutions were necessary, as in the construction of the different haplotypes, mutagenesis reactions were carried out sequentially. Reporter plasmid DNA was purified using QIAGEN Maxi-Prep kits (Valencia, CA). The DNA concentration was determined by UV absorbance spectroscopy. All preparations were subjected to restriction enzyme digestion followed by agarose gel electrophoresis to assess their quality.
SNPs investigated in this study
| SNP . | rs no. . |
|---|---|
| 5′-flanking region SNPs | |
| −2599 C/G | rs34813434 |
| −2345 1G/2G | rs35814191 |
| −1925 T/C | rs17844145 |
| −1690 A/G | rs9526146 |
| −1102 G/T | rs7999168 |
| −1053 C/T | rs9526144 |
| −530 C/T | rs11574977 |
| −438 G/A | rs2146881 |
| −298 G/A | rs17843980 |
| −152 A/G | rs11574980 |
| 3′-UTR SNPs | |
| +1344 G/A | rs1049669 |
| +1542 C/G | rs940 |
| +1583 A/T | rs1087 |
| SNP . | rs no. . |
|---|---|
| 5′-flanking region SNPs | |
| −2599 C/G | rs34813434 |
| −2345 1G/2G | rs35814191 |
| −1925 T/C | rs17844145 |
| −1690 A/G | rs9526146 |
| −1102 G/T | rs7999168 |
| −1053 C/T | rs9526144 |
| −530 C/T | rs11574977 |
| −438 G/A | rs2146881 |
| −298 G/A | rs17843980 |
| −152 A/G | rs11574980 |
| 3′-UTR SNPs | |
| +1344 G/A | rs1049669 |
| +1542 C/G | rs940 |
| +1583 A/T | rs1087 |
The parental fusion mRNA reporter plasmid used for mutagenesis was βG-TAFI/1273-1819,34 which was constructed in the context of the pC7βG vector described by Wilson and Deeley.35 This plasmid features a 460-bp segment of the rabbit β-globin cDNA whose expression is driven by the cytomegalovirus promoter, and also contains Epstein-Barr virus genomic sequences and a hygromycin resistance cassette to allow for stable episomal maintenance of the plasmid in mammalian cells.35 A segment of the CPB2 cDNA between +1273 (the first nucleotide after the stop codon) and +1819 (the 3′-most polyadenylation site) was inserted downstream of the β-globin sequences. Using site-directed mutagenesis, substitutions corresponding to the SNPs in the CPB2 3′-UTR were incorporated either alone or in the combinations potentially observed in the human population. Site-directed mutagenesis was carried out using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's specifications. In addition to the +1542 C/G and +1583 T/A SNPs identified by Henry and coworkers,27 we also considered a novel SNP +1344 G/A. The corresponding rs numbers for each SNP are presented in Table 1.
Luciferase reporter gene assays
HepG2 cells (human hepatocellular carcinoma)36 were grown in modified Eagle medium (MEM) containing 10% fetal calf serum (MP Biomedicals, Solon, OH) and 1% penicillin-streptomycin-Fungizone (PSF; Invitrogen, Carlsbad, CA). Cells were maintained in a humidified 37°C incubator under a 95% air/5% CO2 atmosphere. Transient transfection of the cells was performed in 6-well tissue culture dishes (Corning, Corning, NY) using the method of calcium phosphate coprecipitation.37 Cells were seeded into the wells approximately 18 hours prior to transfection at a density of 20% confluence. Each well received 5 μg of luciferase reporter plasmid and 2 μg of the internal control plasmid RSV-βgal which contains the Escherichia coli β-galactosidase gene under the control of the Rous sarcoma virus promoter.38 After a 6-hour exposure to the precipitate, the cells were washed 3 times with phosphate-buffered saline (PBS), and fresh complete medium was added to each well. At 48 hours later, cytoplasmic extracts were prepared by first washing each well 3 times with ice-cold PBS, then lysing the cells using ice-cold lysis buffer (25 mM glycylglycine [pH 7.8], 15 mM MgSO4, 4 mM EGTA, and 1% [vol/vol] Triton X-100). Extracts were clarified by centrifugation at 6000g for 5 minutes at room temperature.
Luciferase and β-galactosidase activities in the cell extracts were measured using a MicroLumat Plus plate-reading luminometer (EG&G Berthold, Bad Wildbad, Germany) running WinGlow version 1.24 software and a SpectraMax Plus plate reading spectrophotometer (Molecular Devices, Sunnyvale, CA) running SoftMax Pro version 3.1.1 software, respectively. To assay luciferase activity, 10 μL of extract was placed in the wells of a white 96-well plate and 50 μL luciferase assay buffer (Promega) was injected sequentially; relative light units were measured 0.1 seconds after injection. To assay β-galactosidase activity, 10 μL extract was placed in the wells of a 96-well plate and 40 μL water and 50 μL β-galactosidase assay buffer (1.33 mg/mL O-nitrophenyl-β-D-galactopyranoside [ONPG]; as substrate), 200 mM sodium phosphate (pH 7.3), 2 mM MgCl2, and 100 mM β-mercaptoethanol. Absorbance at 405 nm was measured every 30 seconds at 37°C; the β-galactosidase activity was taken to be the rate of increase of absorbance at 405 nm. Relative luciferase activity was calculated as the luciferase activity per unit β-galactosidase activity per unit volume of extract.
For each reporter plasmid, 4 independent transfection experiments were performed. In each independent experiment, triplicate wells were transfected, and luciferase and β-galactosidase activities for each well were determined in duplicate. The promoter activity of constructs containing polymorphisms was compared with that of the wild-type construct using paired t tests.
mRNA stability assays
In order to construct HepG2 cells lines stably expressing the β-globin/TAFI 3′-UTR fusion mRNAs, dishes of cells were stably transfected with the respective plasmids using FuGENE 6 Transfection Reagent (Roche Diagnostics, Indianapolis, IN). After a 48-hour recovery period, selection of positive clones was performed by adding hygromycin B (Roche Diagnostics) to a final concentration of 300 μg/mL and changing the media every 2 to 3 days to remove dead cells. After approximately 2 weeks, hygromycin-resistant colonies were pooled and grown in MEM containing 10% fetal calf serum and 1% PSF, while maintaining hygromycin B at 300 μg/mL.
For the mRNA stability assays, actinomycin D (Sigma, St Louis, MO) was added to 90% confluent stably-transfected HepG2 cells, to a final concentration of 5 μg/mL. Cells were maintained without hygromycin B during the course of these experiments. Incubation was continued for different times up to 8 hours, at which point total RNA was harvested using TriZOL reagent (Invitrogen) as recommended by the manufacturer. Poly(A)+RNA was prepared using Nucleotrap mRNA purification kits (Clontech, Palo Alto, CA). Poly(A)+ RNA (approximately 2 μg/lane; in 50% [vol/vol] formamide, 10 mM MOPS [pH 7.0], and 2.2 M formaldehyde) was incubated at 65°C for 15 minutes, quenched on ice, and then fractionated on a 1% (wt/vol) agarose gel containing 10 mM MOPS (pH 7.0) and 2.2 M formaldehyde for Northern blot analysis. The RNA was blotted onto a nylon membrane (Hybond-XL; Amersham Pharmacia Biotech, Piscataway, NJ) via capillary action in 20× SSC (1× SSC is 15 mM trisodium citrate [pH 7] and 150 mM NaCl). RNA was crosslinked to the membrane using the Spectrolinker XL-1000 UV Crosslinker (Stratagene), and blots were hybridized with radiolabeled probes corresponding to the coding region of the rabbit β-globin cDNA. In order to correct for differences in RNA loading and transfer, blots were stripped with boiling 0.5% SDS and hybridized with radiolabeled probes corresponding to the glyceraldehyde-6-phosphate dehydrogenase (GAPDH) cDNA. All probes were prepared using [α-32P]-dATP and the Prime-It II random primer labeling kit (Stratagene). Hybridization was carried out at 68°C for 1 hour in ExpressHyb solution (Clontech). The blots were then washed at room temperature in 1× SSC and 0.1% (wt/vol) SDS, and then at 50°C in 0.2× SSC and 0.1% (wt/vol) SDS. Blots were exposed to a phosphor screen (Kodak, Eastman, NY) and band intensities were quantitated using a Molecular Imager FX phosphorimager (Bio-Rad, Hercules, CA). The amount of fusion RNA present at each time point was calculated as previously described by Wilson and Deeley,35 with the half-life of GAPDH mRNA assumed to be 8 hours.39 The results shown are the mean of 3 independent actinomycin D time courses for each construct.
Results
Effect of 5′-flanking region SNPs on CPB2 promoter activity
To date, 9 SNPs in the CPB2 5′-flanking region have been described (Figure 1).27,28,40 In order to assess the effect of each SNP on CBP2 promoter activity, SNPs were individually introduced, using site-directed mutagenesis, into the 5′-flanking region in the context of luciferase reporter plasmids. All reporter plasmids contained genomic sequence spanning from −2699 to +21 of CPB2; the sequence of the genomic DNA in the parental reporter plasmid was that reported by Boffa and coworkers.31 The resultant reporter plasmids were then transiently transfected into HepG2 cells, which are an excellent model for CPB2 gene expression as the gene is expressed endogenously in these cells.41 An internal control plasmid containing the β-galactosidase gene under the control of the RSV promoter was included in each transfection in order to control for differences in transfection and harvesting efficiency. When evaluated individually, none of the SNPs had a significant impact on CPB2 promoter activity, with the exception of the −298 A and −152 G substitutions; each of these only increased promoter activity by less than 20% (Figure 2).
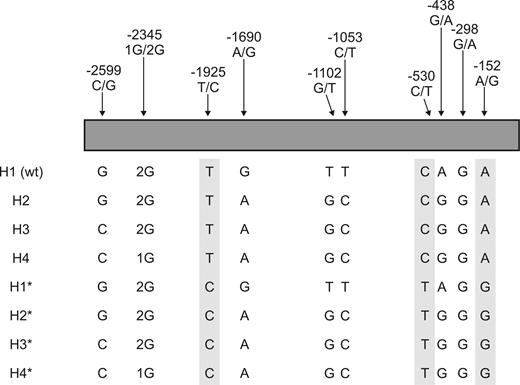
CPB2 5′-flanking region SNPs and haplotypes examined in this study. The H1 to H4 haplotypes are those described by Henry and coworkers.27 The haplotypes denoted H1* to H4* have the substitutions corresponding to a second block of SNPs (shaded) reported by Franco and coworkers.28 The linkage relationship of the relatively infrequent −298 G/A SNP to the other SNPs is not known. The H1 haplotype (wt) corresponds to the sequence of the 5′-flanking region originally reported by Boffa and coworkers.31
CPB2 5′-flanking region SNPs and haplotypes examined in this study. The H1 to H4 haplotypes are those described by Henry and coworkers.27 The haplotypes denoted H1* to H4* have the substitutions corresponding to a second block of SNPs (shaded) reported by Franco and coworkers.28 The linkage relationship of the relatively infrequent −298 G/A SNP to the other SNPs is not known. The H1 haplotype (wt) corresponds to the sequence of the 5′-flanking region originally reported by Boffa and coworkers.31
Effect of individual SNPs on CPB2 promoter activity. The indicated substitutions were introduced into the wild-type (wt; H1 haplotype) sequence31 by site-directed mutagenesis, in the context of the TAFI[−2699]-luc luciferase reporter plasmid.31 Reporter plasmids were transiently transfected into HepG2 cells along with an internal control plasmid (RSV-βgal) to control for differences in transfection and harvesting efficiency. At 48 hours after transfection, cytoplasmic extracts were prepared for assay of luciferase and β-galactosidase activities. The relative luciferase activity is defined as the luciferase activity per unit β-glacatosidase activity per unit volume of extract, and is expressed at a percentage of the relative luciferase acivity of the wt plasmid. The data shown are the means plus or minus SEM of 4 independent experiments. *P < .05 compared with the wt plasmid.
Effect of individual SNPs on CPB2 promoter activity. The indicated substitutions were introduced into the wild-type (wt; H1 haplotype) sequence31 by site-directed mutagenesis, in the context of the TAFI[−2699]-luc luciferase reporter plasmid.31 Reporter plasmids were transiently transfected into HepG2 cells along with an internal control plasmid (RSV-βgal) to control for differences in transfection and harvesting efficiency. At 48 hours after transfection, cytoplasmic extracts were prepared for assay of luciferase and β-galactosidase activities. The relative luciferase activity is defined as the luciferase activity per unit β-glacatosidase activity per unit volume of extract, and is expressed at a percentage of the relative luciferase acivity of the wt plasmid. The data shown are the means plus or minus SEM of 4 independent experiments. *P < .05 compared with the wt plasmid.
There is substantial linkage disequilibrium between all CBP2 SNPs identified to date.27,28 As such, there is a limited number of haplotypes observable in the population. Henry and coworkers described 4 main haplotypes (H1-4) involving −2599 C/G, −2345 1G/2G, −1690 A/G, −1102 G/T, −1053 C/T, and −438 G/A in the 5′-flanking region.27 Accordingly, we constructed haplotypes H2, H3, and H4 by mutagenesis of the wild-type (H1) reporter plasmid (Figure 1). When the promoter activity of these 4 constructs was compared in HepG2 cells, we observed no significant differences between the haplotypes (Figure 3).
Promoter activity of different CPB2 5′-flanking region haplotypes. Substitutions were introduced into the wild-type (wt; H1 haplotype) sequence31 in the context of the TAFI[−2699]-luc luciferase reporter plasmid in the haplotypic combinations shown in Figure 1. Transfections and reporter gene assays were performed as described in the legend to Figure 2. The data shown are the means plus or minus SEM of 4 independent experiments.
Promoter activity of different CPB2 5′-flanking region haplotypes. Substitutions were introduced into the wild-type (wt; H1 haplotype) sequence31 in the context of the TAFI[−2699]-luc luciferase reporter plasmid in the haplotypic combinations shown in Figure 1. Transfections and reporter gene assays were performed as described in the legend to Figure 2. The data shown are the means plus or minus SEM of 4 independent experiments.
Franco and coworkers reported a separate block of 3 SNPs in the CPB2 5′-flanking region (−1925 T/C, −530 C/T, and −152 A/G).28 These SNPs are in virtually complete linkage disequilibrium, and so the corresponding substitutions were introduced as a unit into the 4 main haplotypes to construct H1*, H2*, H3*, and H4* (Figure 1). Once again, there was no difference in promoter activity between any of the haplotypes (Figure 3).
Effect of 3′-UTR SNPs on CPB2 mRNA stability
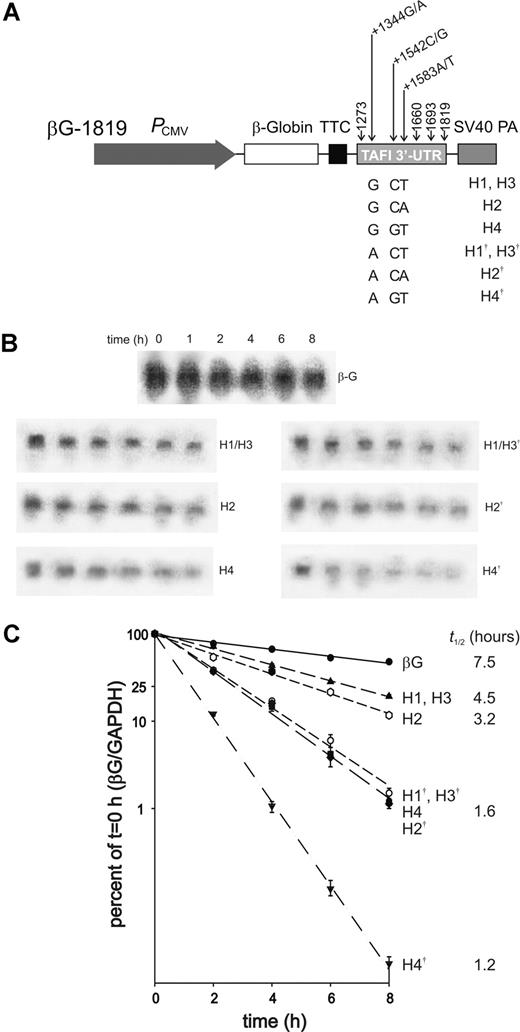
To investigate whether SNPs in the 3′-UTR can act to modulate CPB2 gene expression through an effect on mRNA stability, we used a β-globin reporter mRNA system in which HepG2 cells are stably transfected with plasmids expressing fusion mRNA species consisting of the CPB2 3′-UTR fused to rabbit β-globin mRNA.34 These RNA species can be accurately quantitated (and distinguished from the endogenous CPB2 mRNA in these cells) by Northern blot analysis using β-globin as a probe. The parental plasmid (Figure 4A) represents the 3′-UTR sequence originally reported by Eaton and coworkers,32 and contains CPB2 cDNA sequence downstream of the stop codon and encompasses all 3 potential polyadenylation sites identified in the 3′-UTR.31,34 Using this system, we have found that cis-acting sequences specifically localized to the 3′-UTR determine the stability of the CPB2 mRNA.34
Effect of CPB2 SNPs on mRNA stability. (A) The parental construct consists of the CPB2 3′-UTR (nucleotides +1273 to +1819) inserted into pC7βG downstream of the rabbit β-globin cDNA.33 The construct also contains the cytomegalovirus promoter (PCMV), translation termination cassette (TTC), and simian virus 40 polyadenylation sequences (SV40 PA). Using site-directed mutagenesis, SNPs in the CPB2 3′-UTR, at positions +1344, +1542, and +1583, were introduced either alone or in the haplotypic combinations observed in the human population. The H1 to H4 haplotypes are those described by Henry and coworkers.27 The haplotypes denoted H1† to H4† contain the +1344 A substitution. Also shown are the locations of the 3 polyadenylation signal sequences present in the 3′-UTR at +1660, +1693, and +1819. (B) Representative Northern blots for each construct are shown, along with a representative blot for the parental pC7βG plasmid (βG). In each case, the lanes correspond to mRNA harvested at 0, 1, 2, 4, 6, and 8 hours after the addition of actinomycin D. The full collection of blots is available as Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). (C) HepG2 cells stably transfected with the β-globin fusion mRNA reporter plasmids indicated to the right of the graph, or with a reporter plasmid lacking CPB2 3′-UTR sequences (βG), were treated with actinomycin D. RNA was harvested at various times after the addition of the drug and subjected to Northern blot analysis using a probe specific for rabbit β-globin. To control for differences in RNA loading and transfer, the blots were stripped and hybridized with a probe specific for human GAPDH. Corrected fusion transcript abundance was normalized to that observed before the addition of actinomycin D to obtain the decay curves shown. The data shown are the means plus or minus SEM of 3 independent experiments. The mean half-lives of the respective fusion transcripts are shown to the right of the graph.
Effect of CPB2 SNPs on mRNA stability. (A) The parental construct consists of the CPB2 3′-UTR (nucleotides +1273 to +1819) inserted into pC7βG downstream of the rabbit β-globin cDNA.33 The construct also contains the cytomegalovirus promoter (PCMV), translation termination cassette (TTC), and simian virus 40 polyadenylation sequences (SV40 PA). Using site-directed mutagenesis, SNPs in the CPB2 3′-UTR, at positions +1344, +1542, and +1583, were introduced either alone or in the haplotypic combinations observed in the human population. The H1 to H4 haplotypes are those described by Henry and coworkers.27 The haplotypes denoted H1† to H4† contain the +1344 A substitution. Also shown are the locations of the 3 polyadenylation signal sequences present in the 3′-UTR at +1660, +1693, and +1819. (B) Representative Northern blots for each construct are shown, along with a representative blot for the parental pC7βG plasmid (βG). In each case, the lanes correspond to mRNA harvested at 0, 1, 2, 4, 6, and 8 hours after the addition of actinomycin D. The full collection of blots is available as Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). (C) HepG2 cells stably transfected with the β-globin fusion mRNA reporter plasmids indicated to the right of the graph, or with a reporter plasmid lacking CPB2 3′-UTR sequences (βG), were treated with actinomycin D. RNA was harvested at various times after the addition of the drug and subjected to Northern blot analysis using a probe specific for rabbit β-globin. To control for differences in RNA loading and transfer, the blots were stripped and hybridized with a probe specific for human GAPDH. Corrected fusion transcript abundance was normalized to that observed before the addition of actinomycin D to obtain the decay curves shown. The data shown are the means plus or minus SEM of 3 independent experiments. The mean half-lives of the respective fusion transcripts are shown to the right of the graph.
Nucleotide substitutions corresponding to the SNPs were incorporated into the cDNA segment encoding the 3′-UTR in combinations potentially observed in the human population. To determine whether the SNPs in the 3′-UTR have an effect on CPB2 mRNA stability, we carried out mRNA decay assays using HepG2 cell lines stably transfected with the respective fusion mRNA reporter plasmids. Transcription was arrested using actinomycin D, and RNA was harvested at different times after addition of this drug. The remaining fusion mRNA at each time point was quantified by Northern blot analysis (Figure 4B,C). Compared with the “wild-type” haplotype (H2), mRNA corresponding to the H1/H3 haplotype has an increased stability (half-life of 4.5 hours vs 3.2 hours), while the H4 haplotype has an approximately 2-fold decreased stability (1.6 hours). The +1344 A substitution has a general destabilizing effect, as each haplotype containing this substitution (denoted with the daggers in Figure 4) had a decreased stability relative to the variant containing a G at this position.
Discussion
While it is clear that SNPs in CPB2 (the human gene encoding TAFI) influence plasma concentrations of TAFI antigen, the molecular basis of this relationship remains to be defined, and the identity of the particular SNP(s) directly responsible is not known.
SNPs could influence plasma TAFI concentrations through a variety of mechanisms. SNPs in the 5′-flanking region could influence gene transcription by altering the binding of transcription factors to promoter or enhancer elements, or by altering the local chromatin architecture. SNPs in the 3′-UTR could affect CPB2 mRNA abundance by influencing the stability of the CPB2 transcript or polyadenylation site selection. SNPs in the coding regions of the gene could influence the residence time of the TAFI polypeptide in plasma. Furthermore, the functional SNPs could be outside the CPB2 locus itself, albeit strongly linked to certain of the SNPs. Thus, analysis of the direct effect of individual SNPs on factors that influence CPB2 gene expression is a key aspect of the search for the functional SNPs.
The high degree of linkage disequilibrium that exists between the different SNPs complicates identification of the SNPs that directly influence plasma concentrations of TAFI. Genome-wide scans have confirmed that the CPB2 locus itself (or a locus in close proximity to CPB2) explains virtually all the genetic variation in plasma TAFI antigen levels.42 A recent study examining the association between CPB2 genotype and plasma TAFI antigen concentrations using assays insensitive to the Thr-Ile polymorphism at position 325 suggest that 2 SNPs (possibly −1102 G/T and +1583 T/A) underlie the variability in concentrations.13 That these SNPs are in fact functional could not be ascertained from this type of analysis, and their linkage disequlibrium with other SNPs prevented an entirely conclusive demonstration of their direct effects on variability. A recent trans-ethnic haplotype analysis suggested the presence of 3 quantitative trait nucleotides (QTNs; functional SNPs): +1583 T/A, and possibly −2599 C/G and −2345 1G/2G.30
The data presented here indicate that, with 2 exceptions, none of the SNPs in the CPB2 5′-flanking region influence promoter activity when examined individually. Furthermore, none of the haplotypic combinations examined influenced promoter activity. Collectively, these data suggest against a significant direct functional role of 5′-flanking region SNPs in determining CPB2 gene expression and thus plasma concentrations of TAFI. It is possible that the 2 putative 5′-flanking region QTNs described here are in linkage disequilibrium with the true QTNs30 ; indeed, the sample size in this study may have been too small to unequivocally demonstrate an independent association between the presence of these SNPs and plasma TAFI concentrations.30
Both the −298 A and −152 G substitutions resulted in a modest but significant increase in promoter activity. The −298 G/A SNP is relatively rare in the population (allele frequency of approximately 0.017),43 and in a sample of 359 healthy elderly individuals, no difference in plasma TAFI antigen concentrations was observed between the −298 GG and −298 GA genotypes (data not shown). No difference in plasma TAFI antigen levels was observed in individuals with the −1925 TT/−530 CC/−152 AA versus the −1925 TC/−530 CT/−152 AG genotype.28 A recent transethic study has shown that the −1925 T/C, −1053 C/T, −152 A/G block of SNPs is present in African subjects, but not in Europeans, and is in complete linkage disequilibrium in the former population.30 However, the 2 haplotypes that differ at those positions did not have significantly different plasma TAFI concentrations.30 It is possible that these particular SNPs are too rare for their effects on CPB2 gene expression to be observed in the relatively small samples analyzed thus far. Interestingly, in recent studies from our laboratory which used DNase I footprinting to identify transcription factor binding sites in the CPB2 promoter, both the −298 G/A and −152 A/G SNPs were located between, rather than within, transcription factor binding sites, in keeping with their relatively modest effect on CPB2 promoter activity.44
In contrast to the results observed for the 5′-flanking region SNPs, all of the SNPs in the CPB2 3′-UTR influenced mRNA stability. The combinations of SNPs introduced into the fusion transcripts (Figure 4) are based on the pattern of major haplotypes reported by Henry and coworkers.27 Accordingly, there are 3 combinations with respect to +1542 C/G and +1583 T/A, specifically +1542 C/+1583 T (H1/H3 haplotype), +1542 C/+1583 A (H2), and +1542 G/+1583 T (H4), with the H2 haplotype corresponding to the cDNA cloned by Eaton and coworkers.32 It has very recently been demonstrated that an allele containing +1542 G/+1583 A exists in an African population, albeit at the low frequency of 0.04.30 The +1344 G/A SNP is relatively rare, with an allele frequency of approximately 0.05. Inspection of genotyping data from 127 unrelated individuals (data not shown) reveals the presence of either +1344 A or +1344 G with 2 of the 3 combinations; the +1344 A/+1542 C/+1583 A haplotype could not be conclusively identified in this small sample. Thus, the panel of 3′-UTR model alleles that we have constructed likely corresponds to all the major haplotypes present in the population.
The +1583 T SNP is associated with lower plasma TAFI antigen levels.13,19,30 However, the +1344 G/+1542 C/+1583 T (H1/H3 haplotypes)13,30 transcript was more stable than the +1344 G/+1542 C/+1583 A (H2 haplotype) transcript (Figure 4C). On the other hand, when combined with +1542 G (+1344 G/+1542 G/+1583 T; H3 haplotype), the +1583 T SNP resulted in a 2-fold decrease in transcript stability. These data are consistent with recent studies in which the +1542 G/+1583 T haplotype was associated with the lowest plasma TAFI antigen concentrations and the +1542 C/+1583 A haplotype with the highest.19,30 However, it is clear that additional factors beside an effect of the 3′-UTR SNPs on mRNA stability must be at play, since the H1 and H3 haplotypes feature the same mRNA stability yet are associated with different plasma TAFI antigen levels.19,30
The +1344 A SNP had a general destabilizing effect, with the +1344 A/+1542 G/+1583 T (H4†) transcript being the least stable of all the transcripts tested (Figure 4C). Moreover, the +1344 A/+1542 C/+1583 T (H1†/H3†) transcript was less stable than the +1344 G/+1542 C/+1583 T (H1/H3) transcript. Despite the marked effect of this SNP on mRNA stability, there was no significant association between the novel +1344 G/A SNP and TAFI antigen concentrations in a sample of 127 unrelated individuals, although there was a trend toward lower TAFI antigen concentrations with the A allele (Joost C. M. Meijers, Academic Medical Center, University of Amsterdam, the Netherlands; written communication, July 2004). The relatively low frequency of this SNP indicates that larger sample sizes would be required to detect associations with TAFI concentrations. Indeed, power calculations reveal that in order to have an 80% chance of rejecting the null hypothesis of no association between the +1344 G/A SNP and TAFI concentrations (SD = 10%; α = 0.05), the sample size would have to increase to almost 1000.
The intrinsic stability of mRNA species is determined by cis-acting sequences located within the mRNA (often in the 3′-UTR), as well as trans-acting RNA-binding proteins.45 As such, the nucleotide substitutions in the 3′-UTR that arise from the SNPs may alter the binding of these trans-acting factors, and thus influence mRNA half-life. One possibility is that the SNPs are located within the binding sites for these factors. Alternatively, the SNPs may alter the secondary structure of the 3′-UTR, which is an important determinant of protein binding site formation. Alterations in secondary structure may also influence the rate of endonucleolytic cleavage of the mRNA. Secondary structure analysis using RNA folding software (MFOLD; Michael Zuker, Rensselaer Polytechnic Institute, Troy, NY) predicts that the region spanning the 3 SNPs in the CPB2 3′-UTR can form extensive stem loop structures through internal base pairing. More specifically, in the H2 sequence (+1344 G/+1542 C/+1583 A), +1344 G is involved in base pairing at the base of an internal loop, +1542 C is located in a terminal loop, and +1583 A is involved in base pairing. The +1344 A variant results in disruption of secondary structure in the surrounding region, placing +1344 A in a relatively large terminal loop consisting of approximately 20 bases. Therefore, it is no longer involved in base pairing, perhaps rendering the transcript more susceptible to endonucleolytic cleavage in this region. In addition, +1344 is in a region that is A/U rich, and it is possible that substitution of a G for an A at this position results in targeting the transcript for rapid degradation, which is commonly observed in transcripts with A/U rich sequence elements.46,47 Substitution of a C for a G at position +1542 results in the involvement of this nucleotide in base pairing; the nucleotide at position +1583 remains involved in base pairing when an A is substituted for a T at this position. Finally, the H4† (+1344 A/+1542 G/+1583 T) haplotype, which resulted in a profound effect on mRNA stability (Figure 4C), corresponds to a marked change in predicted secondary structure. In this context, there are a greater number of relatively large terminal loops formed that may increase the potential for degradation of this transcript.
A precedent for the effect of a 3′-UTR SNP on mRNA abundance and thus plasma concentrations of a hemostatic factor can be found in the +20210 G/A SNP in the F2 gene encoding human prothrombin. This SNP results in more efficient processing of the 3′-end of the mRNA through increased endocytic cleavage site recognition, with the effect of increasing the abundance of the mRNA encoding prothrombin.48 The F2 SNP is located downstream of the polyA signal sequence but upstream of the U-rich region; since none of the CPB2 SNPs investigated here are located in the regions where processing of the pre-mRNA occurs,49 we would argue that an analogous mechanism is not at work in the case of the CPB2 mRNA. Although our fusion transcript system is not appropriate for examining the effects of SNPs on the efficiency of polyA site selection or mRNA 3′-end processing, owing to the presence of the strong SV40 polyA signal in the plasmid constructions,34 it must be pointed out that the SNPs are in the region common to all 3 polyadenylated forms, and thus the effects of the SNPs on mRNA stability would impact all 3 forms of the transcript. We have previously demonstrated that our fusion mRNA reporter system accurately models the effect of instability elements in the CPB2 3′-UTR.34 In fact, our previous studies had shown that the CPB2 3′-UTR acts as an autonomous regulatory element that strongly specifies transcript stability, even in the presence of the SV40 sequences.
Our studies represent the first demonstration of a direct effect of SNPs on CPB2 gene expression. These findings will assist in the interpretation of association studies aimed at identifying functional quantitative trait loci both within and outside of the CPB2 gene. Moreover, our findings underscore the idea that such analyses, coupled with epidemiological studies aimed at uncovering associations between CPB2 genotype and the occurrence of vascular disease, should take into account the haplotypic combinations of the 3′-UTR SNPs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Joost Meijers (Academic Medical Center, University of Amsterdam) for performing ELISA assays and Dr Roger Deeley (Queen's University) for providing the pC7βG plasmid.
This work was supported by a grant (to M.L.K.) from the Canadian Institutes for Health Research (no. MOP- 36491). M.L.K. is a Career Investigator of the Heart and Stroke Foundation of Ontario.
Authorship
Contribution: M.B.B. designed research, analyzed data, and wrote the paper; D.M., J.D.H., N.B., P.C., N.S.J., Z.T., E.M.M, and R.F.F. performed research; R.P.T., M.E.N., M.L.K. supervised research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael B. Boffa, Department of Biochemistry, Rm 253, Botterell Hall, Queen's University, Kingston, ON, K7L 3N6 Canada; e-mail: boffam@post.queensu.ca.

![Figure 2. Effect of individual SNPs on CPB2 promoter activity. The indicated substitutions were introduced into the wild-type (wt; H1 haplotype) sequence31 by site-directed mutagenesis, in the context of the TAFI[−2699]-luc luciferase reporter plasmid.31 Reporter plasmids were transiently transfected into HepG2 cells along with an internal control plasmid (RSV-βgal) to control for differences in transfection and harvesting efficiency. At 48 hours after transfection, cytoplasmic extracts were prepared for assay of luciferase and β-galactosidase activities. The relative luciferase activity is defined as the luciferase activity per unit β-glacatosidase activity per unit volume of extract, and is expressed at a percentage of the relative luciferase acivity of the wt plasmid. The data shown are the means plus or minus SEM of 4 independent experiments. *P < .05 compared with the wt plasmid.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/1/10.1182_blood-2007-03-078543/3/m_zh80010811060002.jpeg?Expires=1769236484&Signature=GSxXzUobrBPuZjzv-t3TvuSwB0yNAs3WBRTgePNoLzU9-SCudywA~lHX3MSm~2ogd3ZXJZa-YwyJYvUliCDx4GlEYH3PGM9~aVd8lLHNYJDG580ZsHYj~BhyKpGBOR3ByxYyGu1-nKCxB933ypKgECiSXxAJzJSmDlxbBmfDIPUjACud3Dij3CY67BEHavkP62Nf4zb6mo9HO5k4vTZPBAtranQW0IKL-hHhFK9s6B4S1l5hlQdtfgs-ZFEqM7MF4U~WOw4gmLNZBElJRXEa7Wgbjrb5p5YZyzZMRU~jojN-4wdwtyUmdASS6ldfmA1R8PjhWAI0LZyPQR1E4I80bg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Promoter activity of different CPB2 5′-flanking region haplotypes. Substitutions were introduced into the wild-type (wt; H1 haplotype) sequence31 in the context of the TAFI[−2699]-luc luciferase reporter plasmid in the haplotypic combinations shown in Figure 1. Transfections and reporter gene assays were performed as described in the legend to Figure 2. The data shown are the means plus or minus SEM of 4 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/1/10.1182_blood-2007-03-078543/3/m_zh80010811060003.jpeg?Expires=1769236484&Signature=z5p39exWD2NCN9~75-B9S0Lxi9n4VXgX20AaUjOThOs8pJuhXzEmw83c1gHrGjB7f5bc-HC~En4rV35TnRKP3Ao3phpZpTJVfv7rJQdYZ-YHNnshALLLvqdmWtsnntkwIFle~LAbtPSTfHPa2XyvY9JTDsSSxVmEjoUT-jpr8nS8pF2A488O9BNJ4e-n3ea4XKkniGvns3gOURaJIH6N-c046kFHwXexWR57pTXd78IYO3hTd4m71vve2vp9hfwdU4KsoZd~ap5t1rSLGmtAzSKmFQziq5fYs2HlhKHzQd6GeOtqkSDHfjzyZDTiBZxhre91kgHiZYvZfIfNwt6ATA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)