Abstract
Butyrate is a prototype of histone deacetylase inhibitors that is believed to reactivate silent genes by inducing epigenetic modifications. Although butyrate was shown to induce fetal hemoglobin (HbF) production in patients with hemoglobin disorders, the mechanism of this induction has not been fully elucidated. Our studies of the epigenetic configuration of the β-globin cluster suggest that DNA methylation and histone H3 acetylation are important for the regulation of developmental stage-specific expression of the β-like globin genes, whereas acetylation of both histones H3 and H4 seem to be important for the regulation of tissue-specific expression. These studies suggest that DNA methylation may be important for the silencing of the β-like globin genes in nonerythroid hematopoietic cells but may not be necessary for their silencing in nonhematopoietic cells. Furthermore, our studies demonstrate that butyrate exposure results in a true reversal of the normal developmental switch from γ- to β-globin expression. This is associated with increased histone acetylation and decreased DNA methylation of the γ-globin genes, with opposite changes in the β-globin gene. These studies provide strong support for the role of epigenetic modifications in the normal developmental and tissue-specific regulation of globin gene expression and in the butyrate-mediated pharmacologic induction of HbF production.
Introduction
The human β-like globin genes are expressed in vivo in a tissue- and developmental stage–specific manner. The mechanisms of regulation of globin gene expression have been the subject of intense investigation for several decades.1,2 These studies have led to the identification of a repertoire of cis elements and trans-acting factors that are critically important for regulation of the expression of the different genes of the β-globin cluster. Moreover, these studies led to an appreciation of the role of epigenetic modifications such as DNA methylation and histone acetylation in the regulation of globin gene expression.1-10 As a result, considerable efforts have been focused on the pharmacologic induction of fetal hemoglobin (HbF) by agents that target DNA methylation and histone acetylation. Several pharmacologic agents including 5-azacytidine,11 decitabine,12,13 butyrate, and other histone deacetylase (HDAC) inhibitors14-16 were shown to induce HbF in vivo in patients with hemoglobin disorders. However, the mechanisms responsible for the reactivation of γ-globin gene expression by these pharmacologic agents have not been elucidated yet.17,18
Our previous studies have demonstrated that butyrate can induce high levels of HbF in a majority of patients with sickle cell disease (SCD).14 Patients who responded to intermittent arginine butyrate that was administrated as 4-day cycles every 4 weeks maintained high levels of HbF as long as they continued to receive the drug, even during the 3 weeks between cycles of butyrate therapy.14 Moreover, the peak HbF levels achieved in patients with SCD were similar after therapy with butyrate continuously or intermittently.14 These observations suggest that the induction of HbF by butyrate, a well-known HDAC inhibitor, may be a result of heritable epigenetic modifications that are maintained for several weeks after butyrate exposure. The major goal of the studies described in this report was to investigate the role of epigenetic modifications in the normal development stage– and tissue-specific regulation of globin gene expression and in the butyrate-mediated induction of HbF.
Patients, materials, and methods
Cell cultures
Peripheral blood was obtained from 9 patients with SCD after they had signed the informed consent, which was in accordance with the Declaration of Helsinki and which was approved by the institutional review board at the Mount Sinai School of Medicine. Three of these patients were enrolled at the time of this study in a trial of intermittent arginine butyrate therapy.14,19 Peripheral blood was obtained from these patients before and 24 hours after the initiation of a cycle of butyrate therapy (500 mg/d for 4 days/month). Mononuclear cells were isolated from peripheral blood after separation on Ficoll-Hypaque gradient and cultured into burst-forming unit-erythroid (BFU-E) and colony-forming unit-granulocyte, macrophage (CFU-GM) colonies as described previously20 in the absence and presence of butyrate (B-5887; Sigma, St Louis, MO) at 50 and 150 μM final concentration; 50 μM butyrate was the lowest concentration that induced the γ-globin expression, and 150 μM was the highest nontoxic concentration. K562 cells were cultured in RPMI 1640 medium, and HeLa cells were cultured in Dulbecco modified Eagle medium containing 10% fetal bovine serum, 105 U/L penicillin, and 100 mg/L streptomycin. Cells were cultured at 37°C in a 5% CO2 humidified atmosphere.
Chromatin immunoprecipitation assay
Analysis of histone modification was carried out as recommended by the manufacturer (Upstate Biotechnology, Lake Placid, NY). Briefly, protein–DNA cross-linking was performed by incubating 106 cells at 37°C for 10 minutes in 1% formaldehyde. After cell lysis, the lysate was sonicated to result in DNA fragments of 0.2 to 1.0 kb in size. DNA was coimmunoprecipitated in duplicate with acetylated histones using either a polyclonal antibody against diacetyl-histone H3 (acH3, 06-599; Upstate Biotechnology) or antiserum against tetra-acetyl-histone H4 (acH4, 06-598; Upstate Biotechnology). The immunoprecipitated DNA was purified by phenol/chloroform extraction and dissolved in 50 μL of distilled water. At the same time, 50-μL aliquots of the sonicated lysate (total DNA) were directly subjected to DNA extraction before immunoprecipitation and dissolved in 100 μL of distilled water.
Quantitative real-time polymerase chain reaction analysis
Three microliters of immunoprecipitated DNA and 1 μL of total DNA were analyzed in duplicate by quantitative real-time polymerase chain reaction (PCR) using 200 nM TaqMan probes and primers in a 25-μL iQ-Supermix (Bio-Rad, Hercules, CA) reaction volume. To generate a standard curve, serial dilutions of genomic DNA were used. Experimental data were collected relative to the standard curve at the threshold at which amplification was linear. To adjust for the DNA amount used in each amplification, the immunoprecipitated DNA and the total DNA values for each gene were normalized to the constitutively expressed β-actin gene promoter. The relative unit for each primer pair was determined by dividing the amount of target sequence in immunoprecipitated DNA by the amount of the same target sequence in total DNA. The regions of the β-globin gene cluster that were subjected to PCR amplification are represented schematically in Figure 2A. We selected α-actin, a gene that is not expressed in K562, BFU-E–, or CFU-GM–derived cells, as a negative control. The primers and TaqMan probes (Table 1) were designed using Beacon Designer software (Premier Biosoft International, Palo Alto, CA).
Sequences of primers and TaqMan probes used in quantitative real-time PCR for the ChIP assay
| PCR primer . | Sequence . |
|---|---|
| ϵ-globin | |
| Forward | AGAGAGGCAGCAGCACATATCTG |
| Reverse | CATCTTGCTCCACAGGCTAGTGA |
| Probe | FAM-AGCTGCAATCACTAGCAAGCTCTCAGGCC-BHQ1 |
| γ-globin | |
| Forward | GGCTGGCTAGGGATGAAGAATAAA |
| Reverse | TGGCGTCTGGACTAGGAGCTTA |
| Probe | TexRd-XN-CCTTCAGCAGTTCCACACACTCGCTTCTGG-BHQ2 |
| β-globin | |
| Forward | AGGGAGGGCTGAGGGTTTGA |
| Reverse | CAGGGTGAGGTCTAAGTGATGACA |
| Probe | FAM-TCCAACTCCTAAGCCAGTGCCAGAAGAGCC-BHQ1 |
| α-actin | |
| Forward | GTCTCCCTGTCCTTGCAGAAACTA |
| Reverse | CCCACGATGGACGGGAACAC |
| Probe | FAM-ACAATGTGCGACGAAGACGAGACCACCG-BHQ1 |
| β-actin | |
| Forward | CCCTGGCGGCCTAAGGACTC |
| Reverse | CACATGCCGGAGCCGTTGTC |
| Probe | HEX-CGACGAGCGCGGCGATATCATCATCCATG-BHQ2 |
| PCR primer . | Sequence . |
|---|---|
| ϵ-globin | |
| Forward | AGAGAGGCAGCAGCACATATCTG |
| Reverse | CATCTTGCTCCACAGGCTAGTGA |
| Probe | FAM-AGCTGCAATCACTAGCAAGCTCTCAGGCC-BHQ1 |
| γ-globin | |
| Forward | GGCTGGCTAGGGATGAAGAATAAA |
| Reverse | TGGCGTCTGGACTAGGAGCTTA |
| Probe | TexRd-XN-CCTTCAGCAGTTCCACACACTCGCTTCTGG-BHQ2 |
| β-globin | |
| Forward | AGGGAGGGCTGAGGGTTTGA |
| Reverse | CAGGGTGAGGTCTAAGTGATGACA |
| Probe | FAM-TCCAACTCCTAAGCCAGTGCCAGAAGAGCC-BHQ1 |
| α-actin | |
| Forward | GTCTCCCTGTCCTTGCAGAAACTA |
| Reverse | CCCACGATGGACGGGAACAC |
| Probe | FAM-ACAATGTGCGACGAAGACGAGACCACCG-BHQ1 |
| β-actin | |
| Forward | CCCTGGCGGCCTAAGGACTC |
| Reverse | CACATGCCGGAGCCGTTGTC |
| Probe | HEX-CGACGAGCGCGGCGATATCATCATCCATG-BHQ2 |
FAM indicates 6-carboxyfluorescein; BHQ1, black hole quencher 1; TexRd-XN, Texas red-X NHS ester; BHQ2, black hole quencher 2; HEX, hexachlorofluorescein.
Bisulfite treatment and methylation analysis
Methylation at the promoters of the γ- and β-globin genes was analyzed by bisulfite treatment of DNA followed by pyrosequence analysis, a powerful quantitative method that measures methylation at all cytosine-phosphate-guanosine (CpG) sites within a region of interest. Genomic DNA was prepared from cells using a DNA-purification kit (Gentra Systems, Minneapolis, MN). Sodium bisulfite conversion of 1 μg of DNA was carried out in duplicate using a CpG genome DNA-modification kit (Chemicon International, Temecula, CA). A fragment of the promoter of the γ-globin gene (−107 to 73) that includes the 5 CpG dinucleotides at positions −54, −51, +5, +16, and +49 was amplified from an aliquot of converted DNA with primer set 1 (Table 2). An aliquot of the first PCR amplification was subjected to a second amplification with primer set 2 or 3 (Table 2). Methylation of 3 CpG dinucleotides at positions −306, −265, and −125 of the β-globin gene was assessed by amplifying an aliquot of converted DNA with primer set 4 or 5 (Table 2). The PCR products then were prepared for pyrosequence analysis in a Vacuum Prep workstation (Biotage, Uppsala, Sweden), and the sequencing reactions were performed on a PSQ 96HS system (Biotage). For each CpG site, the mean methylation level of duplicate samples is presented as percent methylation. The mean methylation level under each experimental condition is presented as the average of the methylation level of all CpG sites for each gene. Primer set 1 was designed using Beacon Designer software, and primer sets 2 through 5 and the sequencing primers were designed using PSQ Assay Design 1.0 software (Biotage) (Table 2).
PCR and sequencing primers used in pyrosequence analysis for the DNA-methylation assay
| . | Sequence . | CpG dinucleotides . |
|---|---|---|
| γ-globin | ||
| Set 1 | ||
| PCR primer1 | TTGATAAGGTAAATTTGATTAATAGTTTTA | na |
| PCR primer2 | CCTCCTCTATAAAATAACCCATAAC | na |
| Set 2 | ||
| PCR primer1 | GAGTATTTAGTGAGGTTAGGGG | na |
| PCR primer2 | Biotin-ATACTTCCTTTTATTCTTCATCCC | na |
| Sequencing primer | ATTTAGTGAGGTTAGGGG | −54 and −51 |
| Set 3 | ||
| PCR primer1 | Biotin-TTGGTTAGGGATGAAGAATAAAAG | na |
| PCR primer2 | CCTCCTCTATAAAATAACCCATAA | na |
| Sequencing primer1 | AAAACTTATTAATAACCTCA | +5 and +16 |
| Sequencing primer2 | TCCTCTATAAAATAACCCAT | +49 |
| β-globin | ||
| Set 4 | ||
| PCR primer1 | GTGTAATAAGAAAATTGGGAAAA | na |
| PCR primer2 | Biotin-ATATCTCTTAACCCCATACCATCA | na |
| Sequencing primer1 | TGTAATAAGAAAATTGGGAA | −306 |
| Sequencing primer2 | AGTTGTGATTTTAAATATTA | −265 |
| Set 5 | ||
| Forward | GGGTTGAGGGTTTGAAGTTTAATT | na |
| Reverse | Biotin-CCAACCCTAAAATATAACTCCACA | na |
| Sequencing primer | AGAAGAGTTAAGGATAGGTA | −125 |
| . | Sequence . | CpG dinucleotides . |
|---|---|---|
| γ-globin | ||
| Set 1 | ||
| PCR primer1 | TTGATAAGGTAAATTTGATTAATAGTTTTA | na |
| PCR primer2 | CCTCCTCTATAAAATAACCCATAAC | na |
| Set 2 | ||
| PCR primer1 | GAGTATTTAGTGAGGTTAGGGG | na |
| PCR primer2 | Biotin-ATACTTCCTTTTATTCTTCATCCC | na |
| Sequencing primer | ATTTAGTGAGGTTAGGGG | −54 and −51 |
| Set 3 | ||
| PCR primer1 | Biotin-TTGGTTAGGGATGAAGAATAAAAG | na |
| PCR primer2 | CCTCCTCTATAAAATAACCCATAA | na |
| Sequencing primer1 | AAAACTTATTAATAACCTCA | +5 and +16 |
| Sequencing primer2 | TCCTCTATAAAATAACCCAT | +49 |
| β-globin | ||
| Set 4 | ||
| PCR primer1 | GTGTAATAAGAAAATTGGGAAAA | na |
| PCR primer2 | Biotin-ATATCTCTTAACCCCATACCATCA | na |
| Sequencing primer1 | TGTAATAAGAAAATTGGGAA | −306 |
| Sequencing primer2 | AGTTGTGATTTTAAATATTA | −265 |
| Set 5 | ||
| Forward | GGGTTGAGGGTTTGAAGTTTAATT | na |
| Reverse | Biotin-CCAACCCTAAAATATAACTCCACA | na |
| Sequencing primer | AGAAGAGTTAAGGATAGGTA | −125 |
na indicates not applicable.
Quantitative real-time reverse-transcription–PCR analysis
Total RNA was extracted from cells using a TRIzol kit (Invitrogen, Carlsbad, CA) and treated with RNase-free DNase I (Ambion, Austin, TX) for 30 minutes at 37°C, followed by 5 minutes at 75°C. cDNA synthesis was performed in duplicate in a 25-μL reaction volume using oligo(dT) primers and an Omniscript reverse-transcription (RT) kit (QIAGEN, Valencia, CA). Aliquots of cDNA were amplified in a 25-μL iQ-Supermix reaction volume using 200 nM TaqMan probes and primers for the different globin genes. The amplification efficiencies of the γ-and β-globin messenger RNAs (mRNAs) were very similar in the exponential phase of amplification (data not shown), which made it possible for us to compare the relative levels of γ- and β-globin mRNA in the same sample. Data were collected relative to a standard curve at the threshold at which amplification was linear. The levels of expression of the different globin genes were normalized to the level of expression of 18S rRNA (Applied Biosystems, Foster City, CA), and the data are expressed as percentage γ/(β + γ) and percentage β/(β + γ). The primers and TaqMan probes (Table 3) were designed using Beacon Designer software.
Sequences of primers and TaqMan probes used in quantitative real-time RT-PCR
| PCR primer . | Sequence . |
|---|---|
| γ-globin | |
| Forward | AATGTGGAAGATGCTGGAGGAGAA |
| Reverse | CTTCTTGCCATGTGCCTTGACTT |
| Probe | TexRd-XN-CCCTGGGAAGGCTCCTGGTTGTCTACCC-BHQ2 |
| β-globin | |
| Forward | CCTGAGGAGAAGTCTGCCGTTAC |
| Reverse | TAGACCACCAGCAGCCTGCC |
| Probe | TexRd-XN-ACCACCAACTTCATCCACGTTCACCTTGCC-BHQ2 |
| PCR primer . | Sequence . |
|---|---|
| γ-globin | |
| Forward | AATGTGGAAGATGCTGGAGGAGAA |
| Reverse | CTTCTTGCCATGTGCCTTGACTT |
| Probe | TexRd-XN-CCCTGGGAAGGCTCCTGGTTGTCTACCC-BHQ2 |
| β-globin | |
| Forward | CCTGAGGAGAAGTCTGCCGTTAC |
| Reverse | TAGACCACCAGCAGCCTGCC |
| Probe | TexRd-XN-ACCACCAACTTCATCCACGTTCACCTTGCC-BHQ2 |
TexRd-XN indicates Texas red-XN; BHQ2, black hole quencher 2.
Globin chain synthesis
To assess γ-globin chain synthesis in BFU-E–derived colonies, cells were incubated with [3H]leucine for 24 hours. Lysates were prepared and resolved on urea-polyacrylamide gels as described previously.20 After fluorography, autoradiograms were scanned, analyzed with the Scion image program (Scion, Frederick, MD), and used to determine the percentage γ-globin chain synthesis (ie, % γ/(γ + β)).
Statistical analysis
Student's t test was used to determine statistical significance. P equal to or less than .05 was considered statistically significant. Data are presented as means (± SD).
Results
Induction of HbF by butyrate is heritable
Before the initiation of butyrate infusions, the mean γ-globin production in BFU-E–derived cells from 3 patients with SCD was 23% (± 1%). The level of γ-globin increased to 42% (± 7%) when BFU-E cells from these same patients were cultured in vitro in the presence of butyrate (P =.02) (Figure 1A). When BFU-E cells from blood harvested 24 hours after butyrate infusion were cultured in vitro in the absence of butyrate, γ-globin production levels (45% ± 5%) were significantly higher than the baseline levels in the same patients before the initiation of butyrate infusions (ie, 23% ± 1%; P =.01, Figure 1B). Interestingly, exposure to butyrate in vivo (45% ± 5%) and in vitro (42% ± 7%) induced similar levels of γ-globin production in BFU-E–derived cells (P =.12) (Figure 1A,B). Finally, γ-globin production in BFU-E–derived cells from blood harvested 24 hours after the initiation of butyrate infusions was similar in the presence (48% ± 10%) or absence (45% ± 5%) of butyrate (P = .27) (Figure 1C). Thus, our in vitro experimental system for the induction of HbF by butyrate recapitulates the in vivo observations and provides an excellent model for the study of the molecular mechanisms of induction of γ-globin expression by butyrate.
Effect of butyrate on γ-globin chain synthesis. Mononuclear cells from 3 patients with SCD (□, ▵, ◇) were cultured into BFU-E–derived colonies. (A) Effects of butyrate exposure in vitro on γ-globin chain synthesis in BFU-E cultured before initiation of butyrate infusions. (B) γ-Globin chain synthesis in BFU-E colonies cultured (without in vitro butyrate) from the same patients before and 24 hours after butyrate infusions. (C) Effects of butyrate exposure in vitro on γ-globin chain synthesis in BFU-E cultures initiated 24 hours after butyrate infusions in vivo. Data are expressed as percentageof γ-globin chain synthesis (ie, % γ/[γ + β]). Each data point represents the mean plus or minus 1 SD of triplicate cultures. Solid horizontal lines represent the mean value for every condition.
Effect of butyrate on γ-globin chain synthesis. Mononuclear cells from 3 patients with SCD (□, ▵, ◇) were cultured into BFU-E–derived colonies. (A) Effects of butyrate exposure in vitro on γ-globin chain synthesis in BFU-E cultured before initiation of butyrate infusions. (B) γ-Globin chain synthesis in BFU-E colonies cultured (without in vitro butyrate) from the same patients before and 24 hours after butyrate infusions. (C) Effects of butyrate exposure in vitro on γ-globin chain synthesis in BFU-E cultures initiated 24 hours after butyrate infusions in vivo. Data are expressed as percentageof γ-globin chain synthesis (ie, % γ/[γ + β]). Each data point represents the mean plus or minus 1 SD of triplicate cultures. Solid horizontal lines represent the mean value for every condition.
Developmental and tissue-specific regulation of histone acetylation at the β-globin cluster
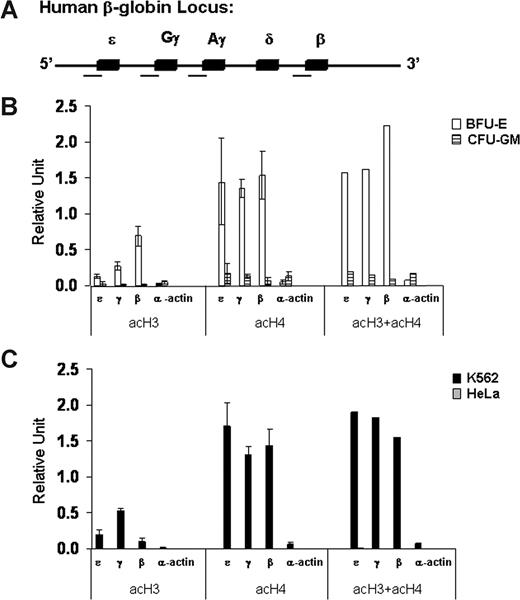
As a first step in our investigation of the role of epigenetic modifications in globin gene regulation, we analyzed the patterns of histone acetylation and DNA methylation at the promoters of the fetal γ-globin and adult β-globin genes in erythroid cells at different developmental stages (K562 and BFU-E) and in nonerythroid cells of different origins (HeLa and CFU-GM). We developed a quantitative real-time PCR-based chromatin immunoprecipitation (ChIP) assay to determine the level of acetylation of histones H3 and H4 at the promoters of the β-like globin genes (Figure 2A). We found high levels of acH3 and acH4 at the promoters of the embryonic ϵ-, γ-, and β-globin genes in erythroid cells in which the globin genes are expressed (Figure 2B,C). In marked contrast, histones H3 and H4 were hypoacetylated at the promoters of the same globin genes in nonerythroid cells in which the globin genes are silent (Figure 2B,C). Interestingly, acH4 levels were high at all β-like globin gene promoters in erythroid cells regardless of their relative levels of expression. In contrast, the levels of acH3 correlated well with the transcriptional activity of the different β-like genes. In other words, in the adult erythroid environment of BFU-E–derived cells, the high-level of acH3 at the promoter of the adult β-globin gene correlated with its high-level expression, and the lower level of acH3 at the promoter of the fetal γ-globin genes correlated with their lower level of expression. In contrast, in the embryonic/fetal environment of K562 cells, a high level of acH3 was found at the promoters of the highly expressed γ-globin genes, an intermediate level was found at the promoter of the expressed ϵ-globin gene, and a low level was found at the promoter of the nonexpressed β-globin gene. Finally, in erythroid and nonerythroid cells, the levels of acetylation of histones H3 and/or H4 were very low at the promoter of the transcriptionally silent α-actin gene.
Patterns of histone acetylation at the promoters of the β-like globin genes in erythroid (K562 and BFU-E) and nonerythroid (HeLa and CFU-GM) cells. (A) Schematic map of the human β-globin gene cluster in which the 5 genes are represented by solid boxes. The regions analyzed for histone acetylation are marked by a horizontal line under the map. (B,C) Levels of acetylation of histones H3 (acH3) and H4 (acH4) and their sum (acH3 + acH4) at the promoters of ϵ-, γ-, and β-globin and α-actin genes assessed by quantitative real-time PCR-based ChIP analysis. The data are expressed in relative units. Error bars are SD. This experiment is representative of 3 independent experiments.
Patterns of histone acetylation at the promoters of the β-like globin genes in erythroid (K562 and BFU-E) and nonerythroid (HeLa and CFU-GM) cells. (A) Schematic map of the human β-globin gene cluster in which the 5 genes are represented by solid boxes. The regions analyzed for histone acetylation are marked by a horizontal line under the map. (B,C) Levels of acetylation of histones H3 (acH3) and H4 (acH4) and their sum (acH3 + acH4) at the promoters of ϵ-, γ-, and β-globin and α-actin genes assessed by quantitative real-time PCR-based ChIP analysis. The data are expressed in relative units. Error bars are SD. This experiment is representative of 3 independent experiments.
Developmental regulation of DNA methylation at the β-globin cluster
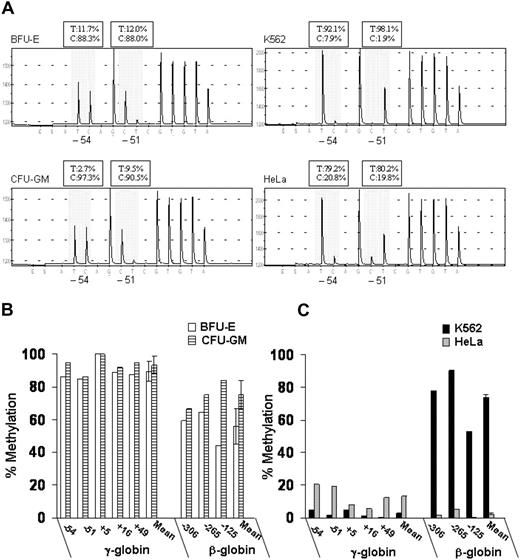
We also measured DNA methylation at the promoters of the γ- and β-globin genes using DNA bisulfite treatment followed by pyrosequence analysis. This new method provides quantitative data on the level of methylation at all CpG sites in a region of interest, as shown in Figure 3A. To simplify the presentation of the methylation data, we expressed overall methylation at the γ-globin promoters as the average of methylation at the 5 CpG dinucleotides located at positions −54, −51, +5, +16, and +49 relative to the transcription-initiation site. Murray and Grosveld21 had previously shown that methylation at these CpG sites leads to suppression of transcription from the human γ-globin promoters. We also measured DNA methylation of 3 CpG dinucleotides at the promoter of the β-globin gene (positions −306, −265, and −125 relative to the transcription-initiation site). In BFU-E–derived cells that express predominantly adult hemoglobin, the methylation levels at the γ-globin promoters ranged from 84% to 100% (mean, 89% ± 6%), whereas the methylation levels at the β-globin promoter ranged from 44% to 65% (mean, 56% ± 11%) (Figure 3B). In contrast, in K562 cells that express predominantly embryonic and fetal hemoglobin, the methylation levels at the γ-globin promoters ranged from 0% to 5% (mean, 3.0% ± 0.7%), whereas the methylation levels at the β-globin promoter ranged from 53% to 78% (mean, 74% ± 2%) (Figure 3C). Thus, in erythroid cells, the level of DNA methylation at the γ- and β-globin promoters is inversely correlated with their level of expression.
Patterns of DNA methylation at the promoters of the γ- and β-globin genes in erythroid (K562 and BFU-E) and nonerythroid (HeLa and CFU-GM) cells. (A) Example of pyrograms generated from pyrosequencing reactions at CpG sites −54 and −51 of the γ-globin promoters (shadowed area). (B,C) Levels of DNA methylation at 5 CpG dinucleotides at γ-globin promoters and 3 CpG dinucleotides at the β-globin promoter were determined by pyrosequencing. The data are expressed as percent methylation. Error bars represent SD. This experiment is representative of 3 independent experiments. T indicates thymidine; C, cytosine.
Patterns of DNA methylation at the promoters of the γ- and β-globin genes in erythroid (K562 and BFU-E) and nonerythroid (HeLa and CFU-GM) cells. (A) Example of pyrograms generated from pyrosequencing reactions at CpG sites −54 and −51 of the γ-globin promoters (shadowed area). (B,C) Levels of DNA methylation at 5 CpG dinucleotides at γ-globin promoters and 3 CpG dinucleotides at the β-globin promoter were determined by pyrosequencing. The data are expressed as percent methylation. Error bars represent SD. This experiment is representative of 3 independent experiments. T indicates thymidine; C, cytosine.
Interestingly, in nonerythroid cells, the patterns of DNA methylation were markedly different between hematopoietic cells (ie, CFU-GM) and epithelial cervical cancer cells (ie, HeLa). In CFU-GM–derived cells, the levels of methylation at the γ-globin promoters ranged from 86% to 100% (mean, 93% ± 5%) and at the β-globin promoter from 66% to 84% (mean, 75% ± 9%) (Figure 3B). In contrast, in HeLa cells, the levels of methylated CpG dinucleotides ranged from 6% to 20% (mean, 12.6% ± 0.6%) at the γ-globin promoters and from 0.5% to 5.0% (mean, 2.3% ± 0.4%) at the β-globin promoter (Figure 3C).
Butyrate induces an “open” chromatin structure at the γ-globin gene and a “closed” chromatin structure at the β-globin gene
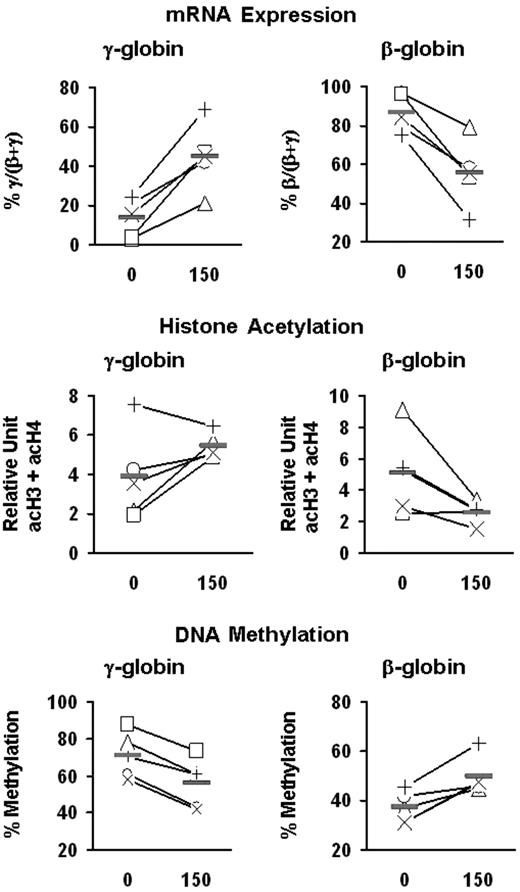
To investigate the potential role of epigenetic modifications in butyrate-induced activation of γ-globin expression, we examined the effect of butyrate on histone acetylation and DNA methylation at the γ-globin promoters in erythroid cells. BFU-E colonies from patients with SCD were cultured in the absence or presence of butyrate at 2 different concentrations (50 and 150 μM). These concentrations of butyrate seem to be nontoxic, because they did not inhibit colony formation (Figure 4A) or decrease the number of cells per colony (mean, 51 000 ± 3000 and 49 000 ± 6000 cells per colony in the absence and presence of butyrate, respectively). Exposure of BFU-E–derived cells to butyrate resulted in a dose-dependent augmentation of γ-globin mRNA levels and γ-globin chain synthesis (Figure 4B-D). Although at the lower concentration of butyrate, the increase in the levels of acetylated histone were relatively small, at a high concentration of butyrate, γ-globin induction was associated with a marked increase in the total level of acetylation of histones H3 and H4 (expressed as acH3 + acH4) at the γ-globin promoters (Figure 4E). Interestingly, butyrate exposure was also associated with a dose-dependent decrease in DNA methylation at the promoters of the γ-globin genes (Figure 4F). We then investigated the effect of 150 μM butyrate on the γ- and β-globin genes in these patients. Exposure of BFU-E–derived cells to butyrate in vitro resulted in an increase in percentage γ/(β + γ) mRNA levels and a decrease in percentage β/(β + γ) mRNA levels (Figure 5). Interestingly, the magnitude of the decrease in the level of β/(β + γ) mRNA was essentially identical to the magnitude of the increase in the level of γ/(β + γ) mRNA. Butyrate exposure was associated with increased histone acetylation at the γ-globin promoters and decreased histone acetylation at the β-globin promoter (Figure 5). Moreover, butyrate exposure was associated with decreased DNA methylation at the γ-globin gene promoters and increased DNA methylation at the β-globin promoter in all 5 patients (Figure 5).
Effects of different concentrations of butyrate on the expression and the epigenetic modifications of γ-globin genes. (A) Mononuclear cells from a patient with SCD were cultured at 50 and 150 μM concentrations of butyrate. BFU-E–derived colonies were counted and expressed as number of colonies per 106 nucleated cells (colonies/106 NC). Error bars are SD. (B) γ-Globin chain synthesis was assessed by polyacrylamide gel electrophoresis and fluorography and expressed as percentage of γ-globin chain synthesis (ie, % γ/[γ + β]). (C,D) mRNA levels were measured by quantitative real-time RT-PCR and expressed as fold change relative to the control (0 μM) and as percent γ/(β + γ). (E) Acetylation of histones H3 and H4 at the γ-globin promoters was measured by quantitative real-time PCR-based ChIP assay and is expressed in relative units and is presented as the sum of acH3 and acH4 (acH3 + acH4). (F) DNA methylation at 5 CpG sites in the γ-globin promoters was determined by pyrosequencing and is expressed as the mean of all the methylation measurements at all 5 sites. Medium and large symbols indicate that the changes relative to the control and to the 50 μM butyrate were statistically significant, respectively. This experiment is representative of 3 independent experiments.
Effects of different concentrations of butyrate on the expression and the epigenetic modifications of γ-globin genes. (A) Mononuclear cells from a patient with SCD were cultured at 50 and 150 μM concentrations of butyrate. BFU-E–derived colonies were counted and expressed as number of colonies per 106 nucleated cells (colonies/106 NC). Error bars are SD. (B) γ-Globin chain synthesis was assessed by polyacrylamide gel electrophoresis and fluorography and expressed as percentage of γ-globin chain synthesis (ie, % γ/[γ + β]). (C,D) mRNA levels were measured by quantitative real-time RT-PCR and expressed as fold change relative to the control (0 μM) and as percent γ/(β + γ). (E) Acetylation of histones H3 and H4 at the γ-globin promoters was measured by quantitative real-time PCR-based ChIP assay and is expressed in relative units and is presented as the sum of acH3 and acH4 (acH3 + acH4). (F) DNA methylation at 5 CpG sites in the γ-globin promoters was determined by pyrosequencing and is expressed as the mean of all the methylation measurements at all 5 sites. Medium and large symbols indicate that the changes relative to the control and to the 50 μM butyrate were statistically significant, respectively. This experiment is representative of 3 independent experiments.
Effects of butyrate on the expression and epigenetic modifications of the γ- and β-globin genes. Mononuclear cells from 5 patients with SCD (+, ×, □, ▵, ○) were cultured into BFU-E–derived colonies in the absence and presence of butyrate. mRNA levels were measured by quantitative real-time RT-PCR and expressed as percentage γ/(β + γ) and percent β/(β + γ). The levels of acetylation of histones H3 and H4 are expressed in relative units and are presented as the sum of acH3 and acH4 (acH3 + acH4). The levels of DNA methylation at the promoters of γ- and β-globin genes are expressed as the mean of methylation levels at all CpG sites (ie, 5 sites for γ-globin and 3 sites for β-globin). Solid horizontal lines represent the mean values for every condition.
Effects of butyrate on the expression and epigenetic modifications of the γ- and β-globin genes. Mononuclear cells from 5 patients with SCD (+, ×, □, ▵, ○) were cultured into BFU-E–derived colonies in the absence and presence of butyrate. mRNA levels were measured by quantitative real-time RT-PCR and expressed as percentage γ/(β + γ) and percent β/(β + γ). The levels of acetylation of histones H3 and H4 are expressed in relative units and are presented as the sum of acH3 and acH4 (acH3 + acH4). The levels of DNA methylation at the promoters of γ- and β-globin genes are expressed as the mean of methylation levels at all CpG sites (ie, 5 sites for γ-globin and 3 sites for β-globin). Solid horizontal lines represent the mean values for every condition.
Discussion
Several previous studies have demonstrated the presence of hypomethylated DNA and hyperacetylated histones at some of the promoters of the expressed globin genes and the locus control region (LCR) in erythroid cells.5-10 However, the exact role of these epigenetic modifications in the regulation of human γ- to β-globin gene switching was not known. In other words, it was not clear whether these epigenetic modifications are responsible for or are a result of the developmental changes in globin gene expression.22 It was proposed that in the fetal stage, interactions between transcription factors and chromatin-remodeling complexes at the relevant regulatory elements result in a chromatin structure that favors expression of the human γ-globin genes. Reciprocal interactions in adult erythroid cells result in a chromatin structure that favors expression of the β-globin gene.1,2
We have conducted a comprehensive analysis of histone acetylation and DNA methylation in the β-globin cluster in erythroid cells and nonerythroid cells. Our epigenetic analysis of BFU-E–derived cells in which the γ- and β-globin genes are transcriptionally active showed that histones H3 and H4 are highly acetylated at the promoters of both γ- and β-globin genes (Figure 2). Interestingly, the levels of acetylation of histone H3 (but not histone H4) at the promoters of the β-like globin genes seem to correlate with their relative levels of expression. Moreover, methylation analysis in BFU-E–derived cells showed that the promoter of the more active β-globin gene is hypomethylated, whereas the promoters of the less active fetal γ-globin genes are relatively hypermethylated (Figure 3). We found a similar correlation between epigenetic modifications and expression of the genes of the β-globin cluster in the embryonic-fetal environment of K562. In these cells, the promoters of the more actively expressed γ-globin genes were characterized by high levels of acetylated H3 and low levels of methylated DNA (Figures 2,3). Thus, DNA methylation and histone H3 acetylation at the globin promoters seem to be important for regulating the developmental stage-specific expression of the different globin genes. Moreover, in agreement with other studies,7,8,10 our data show that acetylation of both histones H3 and H4 and DNA methylation are important for the regulation of tissue-specific expression. Surprisingly, DNA methylation levels are high at the promoters of β-like globin genes in nonerythroid hematopoietic cells and low in nonhematopoietic cells. Thus, DNA methylation seems to be necessary for the maintenance of the silent state of the β-like globin in nonerythroid hematopoietic cells but not in nonhematopoietic cells.
When patients with SCD were treated with an intermittent butyrate regimen, the peak HbF levels that were achieved were similar to those achieved in patients who received butyrate continuously.14 Thus, it seemed that short-term exposure to butyrate resulted in long-term activation of the HbF production program, which suggests that the effects of butyrate on γ-globin gene expression are heritable. Our data presented here show that when erythroid progenitors are exposed to butyrate for only 8 hours in vivo, the colonies that are generated from these cells in vitro after 15 days in culture in the absence of butyrate produce high levels of HbF (Figure 1). This experiment demonstrates that the effect of butyrate on γ-globin gene expression is heritable after multiple cell divisions, which, in turn, suggests that the induction of HbF by butyrate might be mediated by epigenetic modifications that persist after cell division. The fact that additional exposure to butyrate in vitro after the progenitor cells were exposed to butyrate in vivo did not result in an additional increase in γ-globin chain synthesis suggests that the activation of the γ-globin genes by butyrate is maximal after a relatively short exposure in vivo (Figure 1).
We used this experimental system to investigate the potential role of histone acetylation and DNA methylation in the pharmacologically induced activation of HbF by butyrate. Our analysis showed that induction of γ-globin gene expression in adult progenitor-derived cells is associated with an increase in histone acetylation and a decrease in DNA methylation at the γ-globin gene promoters (Figures 4,5). These observations suggest that the silencing of the fetal globin genes is maintained by a suppressive chromatin structure and that butyrate-induced epigenetic changes may be responsible, at least in part, for the activation of γ-globin genes in adult life by opening the chromatin structure at the γ-globin promoters and making it more accessible to the transcriptional machinery. Interestingly, butyrate exposure resulted in an increase in histone acetylation at the promoter of the nonexpressed ϵ-globin gene. However, the increased histone acetylation of the ϵ-globin gene was not associated with reactivation of its expression in adult BFU-E–derived cells (data not shown). Thus, the butyrate-induced chromatin modifications at the ϵ-globin gene may not be sufficient to induce its expression, which suggests that reactivation of the ϵ-globin gene may require additional factors such as the availability of transcription factors or other chromatin modifications, including epigenetic changes in other regulatory domains such as the LCR.
The changes in DNA methylation that we observed at the γ-globin promoters were unexpected. It is not clear how exposure to an agent that inhibits histone deacetylation results in changes in DNA methylation. Xiong et al23 recently described a novel mechanism by which HDAC inhibitors could modulate DNA methylation by down-regulating DNMT3B, a methyltransferase that is responsible for de novo DNA methylation. This mechanism might explain the observed butyrate-mediated hypomethylation of DNA at the γ-globin gene promoters. Interestingly, Lavelle et al24 recently made a reciprocal observation that exposure of baboon erythroid progenitors to decitabine, a DNA hypomethylating agent, results in decreased methylation and increased histone acetylation at the promoters of the γ-globin genes. 5-Methylcytosine can recruit silencing complexes that consist of methyl-DNA–binding proteins and HDAC in regulatory elements of specific genes and interfere with the DNA binding of complexes that activate transcription,25 which suggests that exposure of cells to a DNA hypomethylating agent might displace the HDAC-containing protein complex and result in increased histone acetylation. Thus, there is clearly cross-talk between histone acetylation and DNA methylation, and pharmacologic agents that modulate one type of epigenetic modifications directly may have indirect effects on other types of epigenetic modifications. This would lead to further activation of γ-globin expression by making the DNA structure at the γ-globin gene promoters more favorable for transcription.
The experiments described in this paper show that the activation of γ-globin gene expression by butyrate is associated with suppression of β-globin gene expression. Thus, butyrate exposure results in a reversal of the normal developmental switch from fetal to adult hemoglobin. Interestingly, the effect of the butyrate-induced activation of γ-globin gene expression on the HbF level (ie, γ/(β + γ) ratio) is enhanced further by the butyrate-mediated suppression of β-globin gene expression. As a result, butyrate treatment results in higher levels of HbF in circulating red blood cells of patients with SCD than would be expected from the activation of γ-globin gene expression. Moreover, this butyrate-mediated suppression of β-globin gene expression will also result in a decrease in the effective concentration of sickle hemoglobin, which will further reduce the tendency of the red blood cell to sickle. Interestingly, our studies show that the inhibition of β-globin gene expression by butyrate is also associated with a change in its chromatin configuration. In contrast to its effects on the γ-globin gene promoters, butyrate exposure is associated with hypoacetylation of histones and hypermethylation of DNA at the promoter of the β-globin gene, which gives rise to a less accessible chromatin structure that would limit the access of the transcription factors to the β-globin promoter.
The studies described in this report demonstrate not only specific but opposite effects of butyrate on the activity of 2 different genes within the same cluster. These studies show that butyrate exposure results in differential sequence-specific modifications of histone acetylation and DNA methylation associated with activation or suppression of different genes within the same locus. Our studies did not allow us to conclude whether the effects of butyrate on the chromatin configuration of the β-globin gene cluster were direct or indirect. We speculate that the effects of butyrate on the γ-globin gene may be direct, and its effects on the β-globin gene may be indirect. The most widely accepted model for globin switching is based on competition between the fetal and adult globin genes for interaction with LCR.26 The activation of the γ-globin gene by butyrate may favor its interaction with the LCR and interfere with the interaction of the LCR with the β-globin genes. Thus, the changes that we observed in the β-globin gene may be a reflection of its decreased interaction with the LCR.
Finally, because epigenetic modifications are known to be heritable after cell division, our observations of the butyrate-induced changes in histone acetylation and DNA methylation may explain the clinical observation that intermittent exposure of bone marrow cells to butyrate is sufficient to activate γ-globin gene expression for several weeks after drug exposure. Several other HDAC inhibitors are being used in clinical trials in patients with cancer, genetic diseases, and metabolic diseases. The globin gene system continues to provide an excellent model for elucidating the therapeutic potential and the mechanism(s) of action of these agents because the target genes are known and the end point of the treatment is well defined.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We acknowledge Dr William Davis for facilitating the collection of blood samples from patients with SCD for this study.
This work was supported by National Institutes of Health grant HL-073438 (G.F.A.).
National Institutes of Health
Authorship
Contribution: H.F. and Y.G. performed all experiments; H.F., R.S.W., and G.F.A. designed the experiments and the overall research project; M.S. conducted the clinical research study and provided the blood samples for the experiments described in the article; and H.F. and G.F.A. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hassana Fathallah, Mount Sinai School of Medicine, Division of Hematology/Medical Oncology, Box 1079, One Gustave L. Levy Place, New York, NY 10029; e-mail: hassana.fathallah@mssm.edu.

![Figure 1. Effect of butyrate on γ-globin chain synthesis. Mononuclear cells from 3 patients with SCD (□, ▵, ◇) were cultured into BFU-E–derived colonies. (A) Effects of butyrate exposure in vitro on γ-globin chain synthesis in BFU-E cultured before initiation of butyrate infusions. (B) γ-Globin chain synthesis in BFU-E colonies cultured (without in vitro butyrate) from the same patients before and 24 hours after butyrate infusions. (C) Effects of butyrate exposure in vitro on γ-globin chain synthesis in BFU-E cultures initiated 24 hours after butyrate infusions in vivo. Data are expressed as percentageof γ-globin chain synthesis (ie, % γ/[γ + β]). Each data point represents the mean plus or minus 1 SD of triplicate cultures. Solid horizontal lines represent the mean value for every condition.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/9/10.1182_blood-2007-02-076091/7/m_zh80210708850001.jpeg?Expires=1767704269&Signature=HgnGbDXw1nBjs99Hl1zFKsqn83YAIm8jmSfcUXNH61PoJjvqt9MSydFo90cw5ZBdHdfKJeQu2ouXgtWUS4-lzanpgDjZwf96gLI8wNTAgEg5u2FHOC9vf0MgInLgZ-5FTcZpAIis0hhhKo4U1auc4XErzZF63g2GCHm7b0WrRmad-EQICk7ZW0dEOEAAsUpRMcMfsaL4iVE~dHlT6fd2UMAGP~XbSs3lH2zoEvomxjXBa3C21UR8~8eKiwM5xWpai0oFwduuP~JS~G2eVu~xIr6umM28-NsAK6l3aKfaw8R1vu4bZUhh786nMvZu0eAeHb-LxG8SkMkW1HII-iewrw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Effects of different concentrations of butyrate on the expression and the epigenetic modifications of γ-globin genes. (A) Mononuclear cells from a patient with SCD were cultured at 50 and 150 μM concentrations of butyrate. BFU-E–derived colonies were counted and expressed as number of colonies per 106 nucleated cells (colonies/106 NC). Error bars are SD. (B) γ-Globin chain synthesis was assessed by polyacrylamide gel electrophoresis and fluorography and expressed as percentage of γ-globin chain synthesis (ie, % γ/[γ + β]). (C,D) mRNA levels were measured by quantitative real-time RT-PCR and expressed as fold change relative to the control (0 μM) and as percent γ/(β + γ). (E) Acetylation of histones H3 and H4 at the γ-globin promoters was measured by quantitative real-time PCR-based ChIP assay and is expressed in relative units and is presented as the sum of acH3 and acH4 (acH3 + acH4). (F) DNA methylation at 5 CpG sites in the γ-globin promoters was determined by pyrosequencing and is expressed as the mean of all the methylation measurements at all 5 sites. Medium and large symbols indicate that the changes relative to the control and to the 50 μM butyrate were statistically significant, respectively. This experiment is representative of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/9/10.1182_blood-2007-02-076091/7/m_zh80210708850004.jpeg?Expires=1767704269&Signature=NbUV5CEsihusRM9HX-0fzivwd6Wg4lE02Wy-0hhms7CoTlepoWk6UVJaYXYX2uOWtUYvrFIEkbh2L3LIPGdbxiaV83ww-XTGxuyYDd-nPe5cdaC-xBNcEUS~AofZB5t~eROHF7iu-K9hVlq0qOVtH0J1DfDw9OZaVO-bTOtiY0xxtsKVGdJZ-MGJJDulMUG0is3hF5H0Jq-I3JoB-vpF7bgG99FNeu0rkFT~2cJD33ho8h9J1zOfhQLVrMvy28Efh~5O3kiwtPg5THHQV9VtP35yTQxSLwaqcuaNl~M~TanjsA-VG8y575h4fNtR77mm8ckh5qzHjZMh45shijK3JA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal