Abstract
Aberrant activities of JAK/STAT signaling pathways have been observed in several hematologic malignancies. Here, we show high expression of JAK2 in the tumor cells of lymphocyte-predominant Hodgkin lymphoma in 85% of cases and activation of JAK2 in 39% of cases. STAT6, which is a target of JAK2, was activated in 50% of the cases. SOCS1 controls JAK2 activity and degradation. Mutations in SOCS1 of either somatic or germ-line origin were observed in micromanipulated tumor cells of 50% of cases. Most mutations truncated SOCS1 or caused replacement of amino acids in functional important regions. Activating mutations in exon 12 of JAK2, which are frequent in myeloproliferative diseases, were not observed. In lymphocyte-predominant Hodgkin lymphoma SOCS1 function may thus be frequently impaired by mutations, and this may contribute to high JAK2 expression and activation of the JAK2/STAT6 pathway.
Introduction
Hodgkin lymphoma (HL) is distinguished into the classical (c) and nodular lymphocyte-predominant (lp) forms. While the derivation of the lymphocytic and histiocytic (L&H) tumor cells of lpHL from germinal center (GC) B cells is well established,1 knowledge about their pathogenesis is limited. Besides BCL6-IGH translocations in fractions of cases,2 aberrant somatic hypermutation (SHM) of PIM1, PAX5, RHOH, and c-MYC has been observed.3
JAK/STAT signaling pathways are constitutively activated in several hematologic malignancies.4 In a survey of JAK2 expression in lymphomas, we observed high JAK2 expression in lpHL and analyzed the causes and consequences of the high JAK2 expression in L&H cells.
Materials and methods
Approval was obtained from the University of Frankfurt School of Medicine Institutional Review Board for these studies. Informed consent was obtained in accordance with the Declaration of Helsinki.
Tissue samples
Forty-seven formalin-fixed paraffin-embedded and 12 frozen lpHL biopsies were used for immunohistochemistry (IHC) and micromanipulation, respectively.
Immunohistochemistry
IHC for JAK2 was performed as previously described and verified for a fraction of cases with an additional JAK2-specific antibody (24B11; Cell Signaling, Beverly, MA).5 P-STAT3 and p-STAT5 IHC was performed as recommended by the supplier (no. 9131 and no. 9359; Cell Signaling). For p-STAT6 (no. 611566; B&D Biosciences, San Diego, CA) and p-JAK2 (no. 3771; Cell Signaling) IHC sections were cooked for 15 minutes in a microwave oven in citrate buffer (10 mM, pH 6) and incubated at room temperature overnight with 1:1000 dilutions of the antibodies. All p-STAT and p-JAK2 IHCs were developed with the CSAII system (Dako, Hamburg, Germany). For specificity tests of antibodies, see Data Supplements (available on the Blood website; see the Supplemental Materials link at the top of the online article). For picture acquisition, an Olympus BX40 (Olympus, Hamburg, Germany) microscope equipped with UPLANApo objective lenses (10×/0.4, 20×/0.7, 40×/0.65) and a Nikon Coolpix 4500 (Nikon, Düsseldorf, Germany) digital camera were used. All pictures were processed with Photoshop 7.0d (Adobe, San Jose, CA).
Western blot analysis
Cells were lysed in Lämmli buffer (8 minutes, 100°C). Lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), blotted onto PDVF membranes (BioRad, Munich, Germany), and incubated overnight with 1:1000 dilutions of primary antibodies.
Micromanipulation, PCR, and sequence analysis
Single CD20+ L&H cells were micromanipulated as previously described.1 For every 10 cells, 4 aliquots of buffer covering the sections were used as negative controls. The complete coding region of SOCS1 (a single exon of 633 bp) was amplified in 3 overlapping fragments and exon 12 of JAK2 as a single PCR product from genomic DNA of single cells in seminested 2-round PCRs. For each case and SOCS1 fragment, usually 5 to 6 PCR products from different cells and for JAK2 amplificates from at least 4 different cells were analyzed. Primer sequences and PCR details are given in Table S1. All PCR products were directly sequenced.
Results and discussion
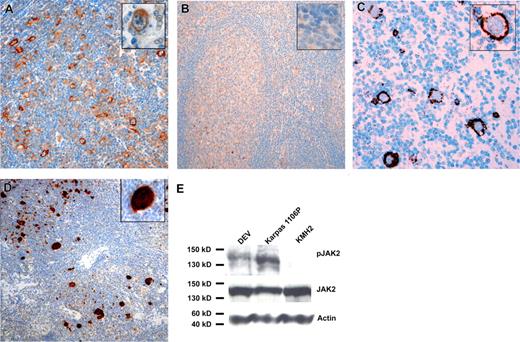
IHC revealed high JAK2 expression in the majority of L&H cells in most lpHL cases (40/47 cases, 85%) (Figure 1). In GC B cells, from which L&H cells are derived, and other normal B cells JAK2 protein amounts were below the detection limit of IHC. Besides the previously reported high JAK2 expression in cHL and mediastinal large B-cell lymphoma (MLBCL),5 JAK2 expression was only occasionally observed in other B-cell lymphomas (3/20 diffuse large B-cell, 0/10 mantle cell, and 2/16 follicular lymphomas and 1/9 chronic lymphocytic leukemias). With a p-JAK2–specific antibody, JAK2 activation was observed in 39% (13/33) of cases (Figure 1). Western blot analysis with the lpHL-derived cell line DEV revealed that JAK2 was expressed and also activated (Figure 1).
Immunohistochemistry for JAK2 and p-STAT6. IHC for JAK2 with an lpHL case (A) and a tonsil (B) is shown. Inserts show higher magnifications of an L&H cell in panel A and the mantle zone/GC border in panel B. While JAK2 amounts in normal B cells were below the detection limit of IHC, 40 of 47 cases of lpHL showed positivity in L&H cells. (C) IHC with an lpHL case and a p-JAK2 specific antibody is shown. Fractions of L&H cells were positive in 13 of 33 analyzed cases. (D) IHC for p-STAT6 with an lpHL case is shown. Twenty-one of 43 analyzed lpHL cases showed positivity for p-STAT6. All cases were from the files of the Department of Pathology of the University of Frankfurt. Bound antibodies were detected using the Envision system (Dako, Hamburg, Germany) for anti-JAK2 and the CSAII signal amplification system for anti–p-JAK2 and anti–p-STAT6 with horseradish peroxidase and 3.3-diaminobenzidine as substrate. Specificity of antibodies was verified by comparison of IHC with Western blot analysis (JAK2 and p-STAT6; Figures S1,S2) and/or by the use of 2 different antibodies directed against the same antigen (JAK2 and p-JAK2). Specificity of anti–p-JAK2 and anti–p-STAT6 antibody for phosphorylated JAK2 and STAT6 was tested by pretreatment of several cases with T-cell phosphatase,5 which abolished staining completely (Figure S3). (E) Western blot analysis of JAK2 expression and activation in the lpHL-derived cell line DEV is shown. Whereas JAK2 was expressed in the MLBCL-derived cell line Karpas1106P, the cHL-derived cell line KMH2, and DEV, activation of JAK2 was observed only in Karpas1106P and DEV, in line with the presence of SOCS1 mutation only in Karpas1106P and DEV6 .
Immunohistochemistry for JAK2 and p-STAT6. IHC for JAK2 with an lpHL case (A) and a tonsil (B) is shown. Inserts show higher magnifications of an L&H cell in panel A and the mantle zone/GC border in panel B. While JAK2 amounts in normal B cells were below the detection limit of IHC, 40 of 47 cases of lpHL showed positivity in L&H cells. (C) IHC with an lpHL case and a p-JAK2 specific antibody is shown. Fractions of L&H cells were positive in 13 of 33 analyzed cases. (D) IHC for p-STAT6 with an lpHL case is shown. Twenty-one of 43 analyzed lpHL cases showed positivity for p-STAT6. All cases were from the files of the Department of Pathology of the University of Frankfurt. Bound antibodies were detected using the Envision system (Dako, Hamburg, Germany) for anti-JAK2 and the CSAII signal amplification system for anti–p-JAK2 and anti–p-STAT6 with horseradish peroxidase and 3.3-diaminobenzidine as substrate. Specificity of antibodies was verified by comparison of IHC with Western blot analysis (JAK2 and p-STAT6; Figures S1,S2) and/or by the use of 2 different antibodies directed against the same antigen (JAK2 and p-JAK2). Specificity of anti–p-JAK2 and anti–p-STAT6 antibody for phosphorylated JAK2 and STAT6 was tested by pretreatment of several cases with T-cell phosphatase,5 which abolished staining completely (Figure S3). (E) Western blot analysis of JAK2 expression and activation in the lpHL-derived cell line DEV is shown. Whereas JAK2 was expressed in the MLBCL-derived cell line Karpas1106P, the cHL-derived cell line KMH2, and DEV, activation of JAK2 was observed only in Karpas1106P and DEV, in line with the presence of SOCS1 mutation only in Karpas1106P and DEV6 .
JAK2 phosphorylates STAT transcription factors. We thus investigated the activation status of STAT3, STAT5, and STAT6, which are constitutively phosphorylated in cHL and MLBCL,6-8 in lpHL. While no activation of STAT3 and STAT5 was observed, STAT6 was phosphorylated in 49% of cases (21/43). Twenty of the 21 p-STAT6–positive cases showed high JAK2 expression and 9 of 17 p-STAT6–positive cases were also p-JAK2 positive (Figure 1). Taking the technical problems performing p-Y–specific IHC into account,9 it is likely that JAK2 and STAT6 are activated in most cases of lpHL.
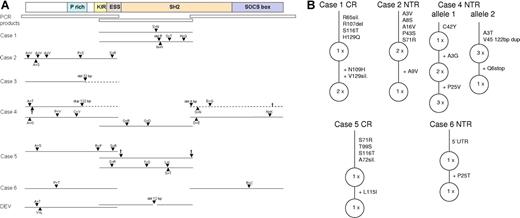
JAK2 activity and protein amount are regulated by a negative-feedback loop. P-STATs induce SOCS1 expression, which binds via its SH2 domain to p-JAK2, thereby inhibiting JAK2 kinase activity and initiating JAK2 degradation by recruitment of a E3-ubiquitin ligase.10 In cHL and MLBCL, this negative-feedback loop is disturbed by inactivating mutations in SOCS1 with subsequent accumulation of JAK2 protein.6,11 We thus analyzed the SOCS1 gene in L&H cells for presence of mutations. Three overlapping fragments encompassing the complete coding region of SOCS1 were amplified from genomic DNA of single micromanipulated L&H cells and directly sequenced (Figure 2A; Table S2).
Schematic presentation of SOCS1 mutations in L&H cells. The complete coding region of SOCS1 was amplified and sequenced from genomic DNA of single micromanipulated L&H cells of 12 cases and the lpHL-derived cell line DEV. (A) A scheme of the SOCS1 protein with functionally important regions and the 3 PCR fragments amplified are shown at top (P rich indicates proline rich; KIR, kinase inhibitory region; and ESS, extended SH2 domain). For each case with mutations in SOCS1, only regions and alleles that carried mutations are presented. Mutations are indicated by ▴. The deletion in case 3 is of germ-line origin. Protein sequences translated out of frame due to deletions or duplications are shown in − − − and stop codons, by †. In addition to the mutated alleles, unmutated alleles were amplified for the central region (CR) of case 1 (7×), the N-terminal region (NTR) of case 2 (3×), the NTR of case 3 (1×), the CR of case 4 (1×), the NTR and CR of case 5 (1× and 2×), and the NTR and C-terminal region (CTR) of case 6 (2× and 3×). Amplification of only 1 unmutated allele for a fragment of a case (NTR of case 3, CR of case 4, and NTR of case 5) is most likely due to cellular contamination during micromanipulation. Apart from the replacement mutations shown, altogether 7 silent mutations were observed (not shown; see Table S2 for details). For alleles with ongoing somatic mutation, the variant carrying the most mutations is always presented. (B) Stepwise accumulation of mutations for alleles with intraclonal diversity is shown. Presence of truncating and replacement mutations in functional important regions of SOCS1 suggest that SOCS1 function is impaired in L&H cells.
Schematic presentation of SOCS1 mutations in L&H cells. The complete coding region of SOCS1 was amplified and sequenced from genomic DNA of single micromanipulated L&H cells of 12 cases and the lpHL-derived cell line DEV. (A) A scheme of the SOCS1 protein with functionally important regions and the 3 PCR fragments amplified are shown at top (P rich indicates proline rich; KIR, kinase inhibitory region; and ESS, extended SH2 domain). For each case with mutations in SOCS1, only regions and alleles that carried mutations are presented. Mutations are indicated by ▴. The deletion in case 3 is of germ-line origin. Protein sequences translated out of frame due to deletions or duplications are shown in − − − and stop codons, by †. In addition to the mutated alleles, unmutated alleles were amplified for the central region (CR) of case 1 (7×), the N-terminal region (NTR) of case 2 (3×), the NTR of case 3 (1×), the CR of case 4 (1×), the NTR and CR of case 5 (1× and 2×), and the NTR and C-terminal region (CTR) of case 6 (2× and 3×). Amplification of only 1 unmutated allele for a fragment of a case (NTR of case 3, CR of case 4, and NTR of case 5) is most likely due to cellular contamination during micromanipulation. Apart from the replacement mutations shown, altogether 7 silent mutations were observed (not shown; see Table S2 for details). For alleles with ongoing somatic mutation, the variant carrying the most mutations is always presented. (B) Stepwise accumulation of mutations for alleles with intraclonal diversity is shown. Presence of truncating and replacement mutations in functional important regions of SOCS1 suggest that SOCS1 function is impaired in L&H cells.
Mutations were found in 6 of 12 cases and the lpHL cell line DEV. Comparisons with SOCS1 from corresponding nontumor cells revealed that a deletion in one case was of germ-line origin, while all other mutations were of somatic origin. In 3 cases, inactivating mutations were observed, and presence of several replacement mutations in functionally important regions in the 3 other cases suggest that SOCS1 function was also impaired in these cases.
In 3 cases (cases 1, 4, and 5) alleles with completely different mutation patterns and in 4 of 6 cases (cases 1, 2, 5, and 6) with mutated alleles, unmutated alleles were also repeatedly amplified (Figure 2A; Table S2). This could be due to the presence of different alleles in all L&H cells of a case or due to presence of different alleles only in subclones of L&H cells. In the former case, the different alleles should be occasionally coamplified from single cells. This was observed only for both mutated N-terminal fragments of case 4 and mutated and unmutated N-terminal, central, and C-terminal fragments of case 4 and the central fragment of case 5. For the other differing alleles, it remained thus unclear whether they were present in all L&H cells of the respective cases or restricted to different subclones of tumor cells.
In all 5 cases with somatic mutations, intraclonal diversity of individual SOCS1 alleles with stepwise accumulation of mutations was observed (Figure 2B). Hotspots of SHM (RGYW) were 4.5 times more frequently affected than expected (Table S2), and single base pair substitutions were much more frequent than deletions or insertions (39 versus 3, respectively). The somatic mutations are thus likely due to ongoing activity of SHM usually active in GC B cells.
In myeloproliferative disorders, JAK2 is frequently activated by a replacement mutation in exon 12.12 In micromanipulated L&H cells of 11 lpHLs, no such mutations were observed.
Taken together, these data suggest that in L&H cells SOCS1 function might be impaired by mutations that are in most cases likely due to activity of SHM. This may cause increases in JAK2 protein amount and activity and contribute to constitutive activation of STAT6, although SOCS1 mutations are not necessarily accompanied by JAK2 phosphorylation, and JAK2 and STAT6 phosphorylations did not correlate at the level of individual cases.13 P-STAT6 promotes expression of AID and SOCS1.14,15 As SHM is dependent on transcription of target genes,16 the constitutive p-STAT6 activity in L&H cells may drive further acquisition of mutations in SOCS1 and thus cause the high mutation frequency of SOCS1 compared with other genes targeted by normal or aberrant SHM (average mutation frequency in mutated alleles: 1.2% for SOCS1; 0.7% for BCL6; 0.25% for CD95 in GC B cells; and 0.1%-0.2% for aberrant SHM of PAX5, PIM1, RHOH, and c-MYC).3,17,18
Although SOCS1 mutations may in some cases be subclonal and thus a late event in tumor cell development, the selection of replacement mutations (observed ratio of replacement–silent mutations: 4.6 (32/7); expected 3)19 indicates that potential impairment of SOCS1 function is advantageous for L&H cells. In addition to the observed mutations, SOCS1 may be affected by genomic deletions, as losses of the region encompassing SOCS1 are frequent in lpHL,20 and inactivation by hypermethylation, which has been observed in several other malignancies.21,22
STAT6 is also constitutively activated in cHL and MLBCL.7,8 However, STAT6 activation in the context of a GC B-cell expression program as in L&H cells may have different consequences. Thus, BCL6 is expressed in L&H cells but not cHL and MLBCL, and BCL6 may interfere with STAT6 on several promotors.23 Finally, potential impairment of SOCS1 function could in addition to JAK2 also influence the expression of other proteins such as NFκB and VAV1 in lpHL.24,25
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Deutsche Forschungs Gemeinschaft (BR1238/6–2) and the Deutsche Krebshilfe (102362).
We would like to thank Ralf Küppers for discussion and critical reading of the paper and Sabine Albrecht, Christiane Kehm, and Ekaterini Hadzoglou for excellent technical assistance.
Authorship
Contribution: A.M. and C.R. designed and performed research and analyzed and interpreted data; K.W. performed research and analyzed data; M.-L.H. analyzed and interpreted data; A.B. designed research, analyzed and interpreted data, and drafted the paper. A.M. and C.R. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andreas Bräuninger, Senckenberg Institute of Pathology, University of Frankfurt, Theodor-Stern-Kai 7, 60590 Frankfurt, Germany; e-mail: braeuninger@em.uni-frankfurt.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal