Abstract
We performed fluorescent in situ hybridization (FISH) for 16q23 abnormalities in 861 patients with newly diagnosed multiple myeloma and identified deletion of 16q [del(16q)] in 19.5%. In 467 cases in which demographic and survival data were available, del(16q) was associated with a worse overall survival (OS). It was an independent prognostic marker and conferred additional adverse survival impact in cases with the known poor-risk cytogenetic factors t(4;14) and del(17p). Gene expression profiling and gene mapping using 500K single-nucleotide polymorphism (SNP) mapping arrays revealed loss of heterozygosity (LOH) involving 3 regions: the whole of 16q, a region centered on 16q12 (the location of CYLD), and a region centered on 16q23 (the location of the WW domain-containing oxidoreductase gene WWOX). CYLD is a negative regulator of the NF-κB pathway, and cases with low expression of CYLD were used to define a “low-CYLD signature.” Cases with 16q LOH or t(14;16) had significantly reduced WWOX expression. WWOX, the site of the translocation breakpoint in t(14;16) cases, is a known tumor suppressor gene involved in apoptosis, and we were able to generate a “low-WWOX signature” defined by WWOX expression. These 2 genes and their corresponding pathways provide an important insight into the potential mechanisms by which 16q LOH confers poor prognosis.
Introduction
Recurrent structural and numerical genomic abnormalities are characteristic features of multiple myeloma. The most common primary translocations are found in both the premalignant monoclonal gammopathy of undetermined significance (MGUS) and smoldering myeloma (SMM), and in multiple myeloma itself, and involve the immunoglobulin heavy chain (IgH) gene at 14q32: t(4;14), t(6;14), t(11;14), t(14;16), and t(14;20).1 Trisomies of chromosomes 3, 5, 7, 9, 11, 15, 19, and 21 plus gains of 1q are the most frequent gains along with deletions of 13q, 6q, 8p, 16q, and 17p.2-4 It is difficult to determine the precise significance of large areas of gain or loss; therefore, we have previously used single-nucleotide polymorphism (SNP)-based arrays,5 and others have used array-based comparative genomic hybridization (CGH),6,7 to define with greater precision the size and location of regions of deletion or gain. The ability to integrate these data with gene expression analysis has allowed us to map potential genes of interest back to these genomic regions. SNP-based arrays have allowed identification of copy neutral loss of heterozygosity (LOH), known as uniparental disomy (UPD), as another mechanism for gene inactivation in myeloma.5
Deletion of all or part of 16q has been reported in 14% to 18% of myeloma cases by conventional cytogenetics or CGH.8-10 Using 50K SNP arrays, we identified LOH of 16q in 17% of newly diagnosed myeloma cases, all of which appeared to be whole-arm deletions.5 The prognostic impact of deletion of 16q [del(16q)] has not been clearly established, although deletions of 3 regions of 16q identified by array CGH were reported to be associated with adverse prognosis.6 Deletions of all or part of 16q have also been described in a large number of nonhematologic cancers, including breast and prostate carcinomas and Wilms tumors.11-13 Specifically, a WW domain-containing oxidoreductase gene (WWOX) has been identified at 16q23.1 at the common fragile site FRA16D as being the likely tumor suppressor gene deleted in this region.14-20 The primary IgH translocation t(14;16) occurs at WWOX21 and juxtaposes the oncogene MAF into the IgH locus, leading to MAF overexpression.22 It has been identified in 25% to 5% of myeloma cases and has been associated with a poor prognosis.23,24
In this study, we sought to determine the clinical consequences of del(16q) by interphase fluorescent in situ hybridization (FISH) in newly diagnosed myeloma cases and to determine its genetic and clinical associations. We also have used high-density gene mapping to define accurately abnormalities of 16q in myeloma and integrated gene expression profiling to identify potential critical genes and to determine the pathways dysregulated as a consequence of 16q abnormalities.
Patients, materials, and methods
Patients
This study was performed on 861 bone marrow (BM) samples from newly diagnosed myeloma patients (within 6 months of diagnosis) with informed consent obtained in accordance with the Declaration of Helsinki for cytogenetic/FISH analysis received by the Leukaemia Research Fund UK Myeloma Forum Cytogenetic Database between March 2001 and December 2005. Institutional Review Board approval for these studies was obtained from the United Kingdom Multicentre Research Ethics committee. A proportion of these cases has previously been described.25,26 Baseline clinical data were available in 501 cases, of which survival data were available in 467. The median age was 65 years (range, 33-92 years) with 49% older than 65 years and 19% older than 75 years. Maximum follow-up was 68 months with a median of 19 months. Overall survival (OS) was calculated from date of diagnosis to December 2006. The 467 patients evaluable for survival analysis were treated using local induction chemotherapy protocols in accordance with British Committee for Standards in Hematology (BCSH) Guidelines on the Diagnosis and Management of Multiple Myeloma 2005.27 In a subset of 131 patients, adequate tumor RNA was available for gene expression analysis, and in 55 of these, adequate tumor and germ-line DNA was available for additional gene-mapping investigations.
Human myeloma cell lines
Nine human myeloma cell lines (HMCLs) were obtained from the DSMZ cell culture database, Brunswick, Germany (LP1, H929) or ATCC, Manassas, VA (RPMI8226, U266) or kindly provided by Professor T. Otsuki, Kawasaki Medical School, Kawasaki, Japan (KMS11, KMS12BM, KMS26) or Birmingham University, Birmingham, United Kingdom (JIM1, JIM3).
FISH studies
FISH analysis was performed on CD138 selected plasma cells from 861 patients using the micro-FISH technique as described.26 These represent the 94% of successfully purified cases received within the time period and had a median plasma cell percentage of 95%. Probes were a mixture of those commercially available and those grown and labeled in the laboratory using standard techniques.25 Cases were classified according to the following abnormalities: the presence of any IgH translocation (including unknown partner chromosome); the common translocations t(4;14), t(6;14), t(11;14), t(14;16), t(14;20), and t(8;14); deletions of 13q14, 14q32, 16q23, and 17p13; and hyperdiploid status using the iFISH (interphase fluorescence in situ hybridization) ploidy classification previously described.25 Deletion of 14q was deduced from loss of the whole IgH probe or loss of the proximal signal where there was a split, and del(16q) was deduced from loss of the MAF part of the Abbott IgH/MAF probe combination. When scoring fusion probes, the number of copies of each probe was carefully noted as were the presence or absence and number of fusions.
DNA/RNA extraction
Cells for DNA and RNA extraction from 55 patients were frozen in RLT buffer (Qiagen, Valencia, CA) immediately after CD138 selection and stored at −80°C until extraction. HMCL and plasma cell DNA and RNA along with matched germ-line DNA from peripheral white cells were extracted using commercially available kits as described.5
Genome mapping and expression analysis
Mapping analysis was performed using 500 ng of tumor and germ-line DNA from each patient. DNA was prepared according to manufacturer's instructions using the GeneChip mapping 500K assay protocol for hybridization to GeneChip Mapping 250K Nsp and Sty arrays (Affymetrix, Santa Clara, CA). Briefly, genomic DNA was digested in parallel with restriction endonucleases NspI and StyI, ligated to an adaptor, and subjected to polymerase chain reaction (PCR) amplification with adaptor-specific primers. The PCR products were digested with DNaseI and labeled with a biotinylated nucleotide analog. The labeled DNA fragments were hybridized to the microarray, stained by streptavidin–phycoerythrin conjugates, and washed using the Affymetrix Fluidics Station 450 then scanned with a GeneChip scanner 3000 7G. For expression arrays, 100 ng of total tumor RNA was amplified using the 2-cycle target labeling kit (Affymetrix) and hybridized to Human Genome U133 Plus 2.0 arrays according to the manufacturer's instructions, washed, and scanned as described.5
Statistical analysis of FISH and survival data
SPSS (SPSS, Chicago, IL) and Statistica (Statsoft, Tulsa, OK) were used for statistical analysis. Comparisons of frequencies were carried out with Fisher exact test. OS was calculated using the Kaplan-Meier method with the log rank test used to calculate significance. Regression analysis to determine which variables were predictive of survival was carried out, and those attaining a significance of P less than or equal to .05 were included in a stepwise Cox proportional hazards model.
Copy number and LOH analysis
SNP genotypes were obtained using Affymetrix GCOS software (version 1.4) to obtain raw feature intensity and Affymetrix GTYPE software (version 4.0) using the Dynamic Model algorithm with a call threshold of 0.26 to derive SNP genotypes. Samples were analyzed using CNAG 2.0 (http://plaza.umin.ac.jp/genome/)28 using paired tumor (test) samples with the self-reference control (reference) samples to determine copy number and LOH caused by imbalance. The position of regions of LOH and gain were identified using the University of California Santa Cruz (UCSC) Genome Browser, May 2004 Assembly (http://genome.ucsc.edu/cgi-bin/hgGateway). All samples were also analyzed with dChip 2006 (www.dchip.org)29,30 to verify findings obtained with CNAG.
Integration of expression and SNP mapping array data
The samples were grouped into classes based on the presence or absence of LOH at the region of interest. Supervised hierarchical clustering was then performed using dChip, based on these classes, to determine genes that were differentially expressed between the classes. Samples without LOH were used as the baseline (B) and compared with those with LOH, the experimental group (E). Comparison criteria used were lower bound-fold change E/B more than 1.2 or B/E more than 1.2, mean difference E − B more than 100 or B − E more than 100, t test P less than .05. Gene lists were generated containing only genes within the region with LOH.
Generation of signatures defined by CYLD and WWOX
Gene signatures were generated from 131 cases, including the 55 cases in which mapping analysis had also been performed. Expression of a set of 640 NF-κB-related and apoptosis-related probe sets was defined using KEGG pathway information (www.genome.ad.jp/kegg/pathway.html). Cases were classified into quartiles based on CYLD and WWOX expression. A 2-tailed t test (P < .05) was performed to determine the probe sets significantly differentially expressed in cases with low-quartile vs. high-quartile expression for both CYLD and WWOX. A Monte Carlo simulation was performed on 10 000 samples of real expression data to determine the likelihood of finding a given number of probe sets by chance alone. For validation, the analysis was repeated in an independent dataset of 414 newly diagnosed myeloma cases publicly available from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO).31
Results
Correlation between del(16q) and other FISH abnormalities
Deletion of 16q23 [del(16q)] was identified using FISH in 168 of 861 cases (19.5%). The correlations between other FISH abnormalities and cytogenetic abnormalities have been reported previously.25 Del(16q) was significantly associated with deletion of 13q (P = .009), deletion of the IgH probe on 14q (P < .001), deletion of p53 on 17p (P < .001), and with nonhyperdiploid status (P = .043; Table 1).
Correlation between FISH abnormalities according to the presence of del(16q)
| FISH abnormality . | All cases, n (%) . | No del(16q), n (%) . | Del(16q), n (%) . | P . |
|---|---|---|---|---|
| del(13q) | 387/847 (45.7) | 296/681 (43.5) | 91/166 (54.8) | .009* |
| del(14q) | 93/857 (10.9) | 60/689 (8.7) | 33/168 (19.6) | <.001* |
| del(16q) | 168/861 (19.5) | 0/693 (0) | 168/168 (100) | NA |
| del(17p) | 75/764 (9.8) | 44/614 (7.2) | 31/150 (20.7) | <.001* |
| Any IgHt | 393/857 (45.9) | 325/689 (47.2) | 68/168 (40.5) | .121 |
| t(4;14) | 102/859 (11.9) | 89/691 (12.9) | 13/168 (7.7) | .083 |
| t(6;14) | 14/848 (1.7) | 11/683 (1.6) | 3/165 (1.8) | .742 |
| t(11;14) | 132/861 (15.3) | 106/693 (15.3) | 26/168 (15.5) | 1.000 |
| t(14;16) | 33/861 (3.8) | 30/693 (4.3) | 3/168 (1.8) | .177 |
| t(14;20) | 16/850 (1.9) | 14/685 (2.0) | 2/165 (1.2) | .750 |
| t(8;14) | 22/845 (2.6) | 20/681 (2.9) | 2/164 (1.2) | .282 |
| nonhrd | 329/781 (42.1) | 254/630 (40.3) | 75/151 (47.9) | .043* |
| FISH abnormality . | All cases, n (%) . | No del(16q), n (%) . | Del(16q), n (%) . | P . |
|---|---|---|---|---|
| del(13q) | 387/847 (45.7) | 296/681 (43.5) | 91/166 (54.8) | .009* |
| del(14q) | 93/857 (10.9) | 60/689 (8.7) | 33/168 (19.6) | <.001* |
| del(16q) | 168/861 (19.5) | 0/693 (0) | 168/168 (100) | NA |
| del(17p) | 75/764 (9.8) | 44/614 (7.2) | 31/150 (20.7) | <.001* |
| Any IgHt | 393/857 (45.9) | 325/689 (47.2) | 68/168 (40.5) | .121 |
| t(4;14) | 102/859 (11.9) | 89/691 (12.9) | 13/168 (7.7) | .083 |
| t(6;14) | 14/848 (1.7) | 11/683 (1.6) | 3/165 (1.8) | .742 |
| t(11;14) | 132/861 (15.3) | 106/693 (15.3) | 26/168 (15.5) | 1.000 |
| t(14;16) | 33/861 (3.8) | 30/693 (4.3) | 3/168 (1.8) | .177 |
| t(14;20) | 16/850 (1.9) | 14/685 (2.0) | 2/165 (1.2) | .750 |
| t(8;14) | 22/845 (2.6) | 20/681 (2.9) | 2/164 (1.2) | .282 |
| nonhrd | 329/781 (42.1) | 254/630 (40.3) | 75/151 (47.9) | .043* |
IgHt indicates IgH translocation; nonhrd, non-hyperdiploid.
indicates significant P value.
Clinical correlations and survival analysis
Baseline clinical and laboratory data were available in 501 of 867 cases, of which OS could be assessed in 467. The 366 cases without any baseline laboratory parameters were excluded from further analyses. Univariate and multivariate survival analyses were restricted to the core group of 467 cases to ensure validity of the results. FISH correlations in this group were equivalent to the overall dataset. Del(16q) was negatively associated with the presence of an IgD isotype (P = .04) but there were no other associations between 16q status and other baseline variables (Table 2). Median serum β2-microglobulin (β2m) level was 4.2 mg/L overall, 4.3 mg/L in the del(16q) group, and 3.95 mg/L in the nondeleted group.
Correlation between baseline clinical variables and 16q status
| Clinical data . | All cases, n (%) . | No del(16q), n (%) . | Del(16q), n (%) . | P . |
|---|---|---|---|---|
| Isotype | ||||
| IgA | 112/458 (24.5) | 91/366 (24.9) | 21/92 (22.8) | .786 |
| IgD | 12/458 (2.6) | 12/366 (3.3) | 0/92 (0) | .042* |
| IgG | 271/458 (59.2) | 216/366 (59.0) | 55/92 (59.8) | .906 |
| IgM | 3/458 (0.7) | 2/366 (0.5) | 1/92 (1.1) | .491 |
| LC & NS | 60/458 (13.1) | 45/366 (12.3) | 15/92 (16.3) | .303 |
| International Staging System (ISS) | ||||
| ISS 1 | 84/368 (22.8) | 65/295 (22.0) | 19/73 (26.0) | .533 |
| ISS 2 | 141/368 (38.3) | 116/295 (39.3) | 25/73 (34.2) | .502 |
| ISS 3 | 143/368 (38.9) | 114/295 (38.6) | 29/73 (39.7) | .894 |
| Lytic bone disease present | 338/496 (68.1) | 264/394 (67.0) | 74/102 (72.5) | .340 |
| Hb less than 100 g/L | 198/488 (40.6) | 148/386 (38.3) | 50/102 (49.0) | .055 |
| Plt less than 150 × 109/L | 74/498 (14.9) | 55/396 (13.9) | 19/102 (18.6) | .274 |
| LDH at least 200 U/L | 260/309 (84.1) | 211/252 (83.7) | 49/57 (86.0) | .841 |
| CRP at least 10 mg/L | 159/371 (42.9) | 119/291 (40.9) | 40/80 (50.0) | .161 |
| Calcium at least 2.6 mmol/L | 99/470 (21.1) | 78/373 (20.9) | 21/97 (21.6) | .889 |
| Creatinine at least 140 μmol/L | 109/468 (23.3) | 87/371 (23.5) | 22/97 (22.7) | 1.000 |
| Age at least 70 y | 151/501 (30.1) | 115/399 (28.8) | 36/102 (35.3) | .227 |
| Clinical data . | All cases, n (%) . | No del(16q), n (%) . | Del(16q), n (%) . | P . |
|---|---|---|---|---|
| Isotype | ||||
| IgA | 112/458 (24.5) | 91/366 (24.9) | 21/92 (22.8) | .786 |
| IgD | 12/458 (2.6) | 12/366 (3.3) | 0/92 (0) | .042* |
| IgG | 271/458 (59.2) | 216/366 (59.0) | 55/92 (59.8) | .906 |
| IgM | 3/458 (0.7) | 2/366 (0.5) | 1/92 (1.1) | .491 |
| LC & NS | 60/458 (13.1) | 45/366 (12.3) | 15/92 (16.3) | .303 |
| International Staging System (ISS) | ||||
| ISS 1 | 84/368 (22.8) | 65/295 (22.0) | 19/73 (26.0) | .533 |
| ISS 2 | 141/368 (38.3) | 116/295 (39.3) | 25/73 (34.2) | .502 |
| ISS 3 | 143/368 (38.9) | 114/295 (38.6) | 29/73 (39.7) | .894 |
| Lytic bone disease present | 338/496 (68.1) | 264/394 (67.0) | 74/102 (72.5) | .340 |
| Hb less than 100 g/L | 198/488 (40.6) | 148/386 (38.3) | 50/102 (49.0) | .055 |
| Plt less than 150 × 109/L | 74/498 (14.9) | 55/396 (13.9) | 19/102 (18.6) | .274 |
| LDH at least 200 U/L | 260/309 (84.1) | 211/252 (83.7) | 49/57 (86.0) | .841 |
| CRP at least 10 mg/L | 159/371 (42.9) | 119/291 (40.9) | 40/80 (50.0) | .161 |
| Calcium at least 2.6 mmol/L | 99/470 (21.1) | 78/373 (20.9) | 21/97 (21.6) | .889 |
| Creatinine at least 140 μmol/L | 109/468 (23.3) | 87/371 (23.5) | 22/97 (22.7) | 1.000 |
| Age at least 70 y | 151/501 (30.1) | 115/399 (28.8) | 36/102 (35.3) | .227 |
Abbreviations: LC, light chain; NS, Non-secretory; Hb, hemoglobin; plt, platelet count; LDH, lactate dehydrogenase; CRP, C-reactive protein.
indicates significant P value.
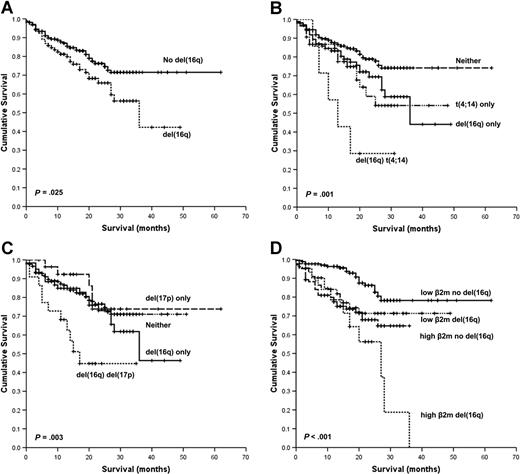
In the univariate analysis, the presence of t(4;14), t(14;16), t(14;20), or del(16q) was associated with an adverse OS. The baseline levels of hemoglobin, platelet count, C-reactive protein (CRP), serum creatinine, age, and International Staging System (ISS) and elevated World Health Organization (WHO) performance status also had a significant survival impact. The presence of del(16q) was associated with a median OS of 36 months versus not reached (P = .025; Figure 1A). OS at 3 years in cases with del(16q) was 42% versus 72% in those without deletion. Moreover, del(16q) was associated with an additional adverse survival impact in combination with the known poor-risk cytogenetic factors t(4;14) and deletion 17p [del(17p)]. The median survival for del(16q) and t(4;14) was 13 months, del(16q) alone 36 months, and t(4;14) alone not reached, P = .001 (Figure 1B). The median survival for del(16q) and del(17p) was 17 months, del(16q) alone 36 months, and del(17p) alone not reached, P = .003 (Figure 1C).
Kaplan-Meier plots of OS. (A) OS according to the presence of del(16q). (B) OS according to the presence of t(4;14) with and without del(16q). (C) OS according to the presence of del(17p) with and without del(16q). (D) OS according to the presence of high (≥ 5.5 mg/L) or low (< 5.5 mg/L) β2m with and without del(16q).
Kaplan-Meier plots of OS. (A) OS according to the presence of del(16q). (B) OS according to the presence of t(4;14) with and without del(16q). (C) OS according to the presence of del(17p) with and without del(16q). (D) OS according to the presence of high (≥ 5.5 mg/L) or low (< 5.5 mg/L) β2m with and without del(16q).
Given that serum β2m is one of the most important variables predicting survival in myeloma, we examined whether del(16q) had prognostic impact in cases with either high (≥ 5.5 mg/L, corresponding to ISS stage III) or low (< 5.5 mg/L, corresponding to ISS stage I and II) β2m.32,33 The presence of del(16q) conferred adverse prognosis in those cases with both low or high β2m: median OS 27 months [del(16q) and high β2m] versus not reached in the remainder (P < .001; Figure 1D). In this series, deletion of 13q [del(13q)] alone or del(13q) with del(16q) had no impact on OS (data not shown).
Multivariate analysis confirmed that del(16q) retained independence as an adverse prognostic marker (P = .003) along with t(14;16) (P = .001), t(4;14) (P < .001), light-chain isotype (P = .021), WHO performance status (P < .001), ISS (P = .002), and age (P < .001).
Global 500K mapping analysis: primary myeloma samples
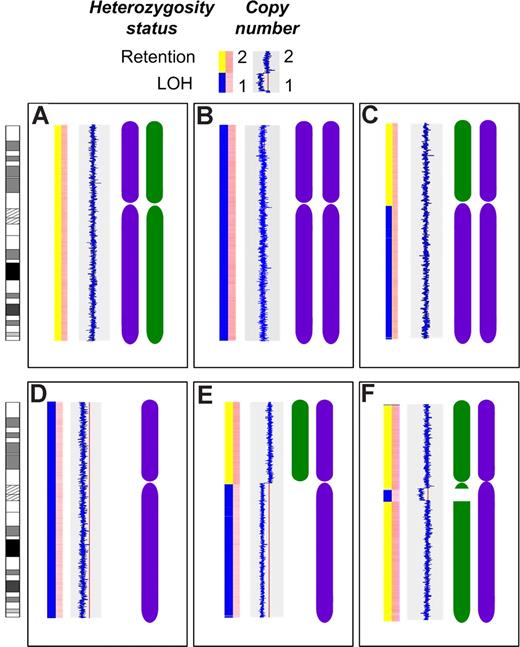
Material was available from 55 cases for gene expression and mapping analysis. These cases did not differ in terms of isotype, ISS stage, and hematologic or biochemical parameters compared with the overall dataset (data not shown). Thirty-one of 55 cases (56%) were hyperdiploid, and 25 of 55 cases (45%) had an IgH translocation, including 10 t(11;14), 4 t(4;14), 3 t(14;16), 2 t(14;20), 1(6;14), and 5 cases with an unidentified translocation partner. The median call rate was 90.22% for the Nsp arrays and 91.84% for the Sty arrays. LOH of all or part of 16q was identified using 500K mapping arrays in 22 of 55 cases (40%). This LOH occurred in 3 distinct patterns: deletion of the entire chromosome 16 or the whole of 16q in 12/55 cases (22%), interstitial deletions of 16q in 7/55 cases (13%), and UPD of the entire chromosome 16 or 16q in 4/55 cases (7%), including one case with both UPD and interstitial deletion (Figure 2). All deletions of 16q were hemizygous.
Patterns of LOH on chromosome 16. The first bar in each panel indicates LOH status with retention of heterozygosity (yellow) and LOH (blue). The second bar in each panel indicates copy number with dark pink indicating normal diploid copy number and pale pink indicating deletion. The blue line indicates copy number relative to the red line (copy number of 2) with a line to the left indicating deletion. The chromosome graphics are representations of each inherited chromosome (purple and green). (A) The normal diploid situation. (B) Acquired UPD of the entire chromosome 16 in 2 cases. (C) Acquired UPD of 16q in 2 cases. (D) Deletion of the entire chromosome 16 in 2 cases. (E) Deletion of 16q in 10 cases. (F) Interstitial deletion of varying regions of 16q in 7 cases.
Patterns of LOH on chromosome 16. The first bar in each panel indicates LOH status with retention of heterozygosity (yellow) and LOH (blue). The second bar in each panel indicates copy number with dark pink indicating normal diploid copy number and pale pink indicating deletion. The blue line indicates copy number relative to the red line (copy number of 2) with a line to the left indicating deletion. The chromosome graphics are representations of each inherited chromosome (purple and green). (A) The normal diploid situation. (B) Acquired UPD of the entire chromosome 16 in 2 cases. (C) Acquired UPD of 16q in 2 cases. (D) Deletion of the entire chromosome 16 in 2 cases. (E) Deletion of 16q in 10 cases. (F) Interstitial deletion of varying regions of 16q in 7 cases.
Integration of gene expression and mapping data
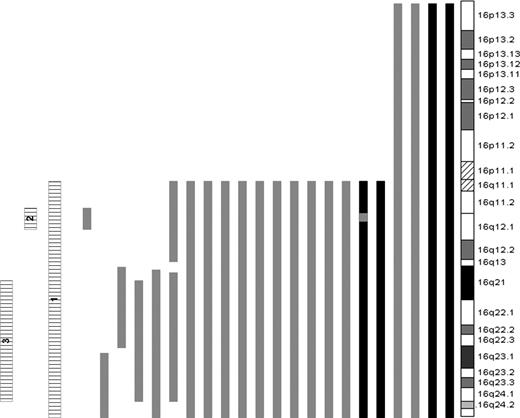
Given the presence of interstitial deletions of 16q, the cases were further subdivided according to the presence or absence of LOH of 3 specific regions of 16q: all of 16q (16 cases, region 1), a 4.2-Mb region located at 16q12.1 (18 cases, region 2), anda 26-Mb region spanning 16q21 to 16q24.1 (21 cases, region 3). There was considerable overlap between the groups. The sizes and positions of all regions of 16q LOH are listed in Table 3 and represented in Figure 3. To determine the genes of likely significance dysregulated as a result of the LOH and the functional consequences of this, we performed supervised hierarchical clustering of cases with each region of LOH versus those without. This revealed differential global gene expression of 185, 165, and 155 genes in cases with LOH of regions 1, 2, and 3, respectively. All such genes located within the regions of interest (38 for region 1 LOH, 4 for region 2, and 17 for region 3) were underexpressed in relation to cases without LOH (Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article). To reduce the complexity of the data, we compared this list of genes with a previous dataset of 60 genes located on 16q that were underexpressed in cases with LOH of that region5 and identified a common set of 16 transcripts. Of these, CYLD was the only gene located in region 2. There were 5 genes located within region 3: TK2, WWP2, DDX19A, GABARAPL2, and WWOX. Of the 5 genes located within region 3, WWOX was identified as being the most likely candidate gene, given its known role as a potential tumor suppressor gene with no such similar function identified for the remainder.14-20
Loss of heterozygosity of 16q identified using 500K mapping arrays
| LOH status . | Cytogenetic band . | Start Position, Mb . | End Position, Mb . | Size, Mb . | Cases, n . |
|---|---|---|---|---|---|
| Deletion | 16 all | All | All | All | 2 |
| UPD | 16 all | All | All | All | 2 |
| Deletion | 16q11.2–16q12.1 | 45.065 | 54.627 | 9.562 | 1 |
| UPD | 16q11.2–16qter | 45.065 | 88.691 | 43.626 | 2 |
| Deletion | 16q11.2–16qter | 45.065 | 88.691 | 43.626 | 10 |
| Deletion | 16q12.1 | 46.718 | 50.872 | 4.154 | 1 |
| Deletion | 16q12.1 | 48.706 | 49.358 | 0.652 | 1 |
| Deletion | 16q13–16q23.1 | 55.398 | 74.985 | 19.587 | 1 |
| Deletion | 16q13–16qter | 55.674 | 88.691 | 33.017 | 1 |
| Deletion | 16q13–16q24.1 | 56.392 | 83.586 | 27.194 | 1 |
| Deletion | 16q21–16q24.1 | 58.469 | 84.511 | 26.042 | 1 |
| Deletion | 16q23.1–16qter | 77.728 | 88.691 | 10.963 | 1 |
| LOH status . | Cytogenetic band . | Start Position, Mb . | End Position, Mb . | Size, Mb . | Cases, n . |
|---|---|---|---|---|---|
| Deletion | 16 all | All | All | All | 2 |
| UPD | 16 all | All | All | All | 2 |
| Deletion | 16q11.2–16q12.1 | 45.065 | 54.627 | 9.562 | 1 |
| UPD | 16q11.2–16qter | 45.065 | 88.691 | 43.626 | 2 |
| Deletion | 16q11.2–16qter | 45.065 | 88.691 | 43.626 | 10 |
| Deletion | 16q12.1 | 46.718 | 50.872 | 4.154 | 1 |
| Deletion | 16q12.1 | 48.706 | 49.358 | 0.652 | 1 |
| Deletion | 16q13–16q23.1 | 55.398 | 74.985 | 19.587 | 1 |
| Deletion | 16q13–16qter | 55.674 | 88.691 | 33.017 | 1 |
| Deletion | 16q13–16q24.1 | 56.392 | 83.586 | 27.194 | 1 |
| Deletion | 16q21–16q24.1 | 58.469 | 84.511 | 26.042 | 1 |
| Deletion | 16q23.1–16qter | 77.728 | 88.691 | 10.963 | 1 |
The size and location of regions of LOH are shown. Note that 2 cases have either two regions of deletion, or UPD and deletion as demonstrated in Figure 3.
LOH of chromosome 16. Bars to the left of the ideogram indicate regions of LOH. LOH caused by deletion (▒); copy neutral LOH, UPD (■); and the 3 regions of LOH used to cluster samples (▤) are shown.
LOH of chromosome 16. Bars to the left of the ideogram indicate regions of LOH. LOH caused by deletion (▒); copy neutral LOH, UPD (■); and the 3 regions of LOH used to cluster samples (▤) are shown.
Hemizygous del(16q) identified CYLD as being an important gene in myeloma and defined as a “low-CYLD signature”
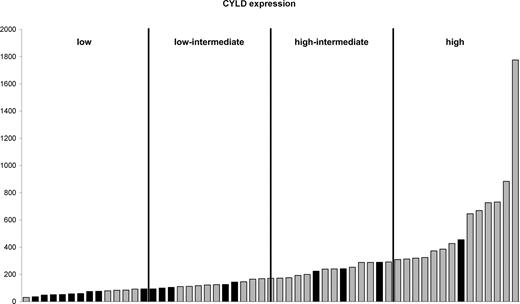
In 18 of 22 cases with LOH of 16q the minimal region included 16q12.1. There were hemizygous interstitial deletions in 2 cases, one of 4.154 Mb and the other of 652 kb, both encompassing the known tumor suppressor gene CYLD. Cases were grouped into quartiles based on CYLD expression. Cases with LOH of region 2 predominantly had CYLD expression in the lowest 2 quartiles (Figure 4). CYLD is a negative regulator of the NF-κB pathway,34-36 raising the potential that this pathway is critical in myeloma. We therefore carried out an analysis of NF-κB and apoptosis-related probe sets in the context of CYLD expression levels. For the low-CYLD vs. high-CYLD sample comparison, 208 of the 640 probe sets were significantly differentially expressed. The empirical P value for finding any 208 of 640 probe sets was less than 10−4; therefore, these 208 probe sets were significant. These data were validated in an independent dataset of 414 newly diagnosed myeloma cases, and we identified a core overlapping set of 73 probe sets with significantly differential expression in both datasets. As such this represents a validated “low-CYLD signature” (Table S2).
CYLD expression. Cases are ordered according to CYLD expression. Cases with LOH at 16q12.1 (■) and those with retention of heterozygosity at 16q12.1 (▒) are shown. The cases are divided into 4 quartiles based on CYLD expression.
CYLD expression. Cases are ordered according to CYLD expression. Cases with LOH at 16q12.1 (■) and those with retention of heterozygosity at 16q12.1 (▒) are shown. The cases are divided into 4 quartiles based on CYLD expression.
Combined mapping and expression analysis identified WWOX as being critically deregulated
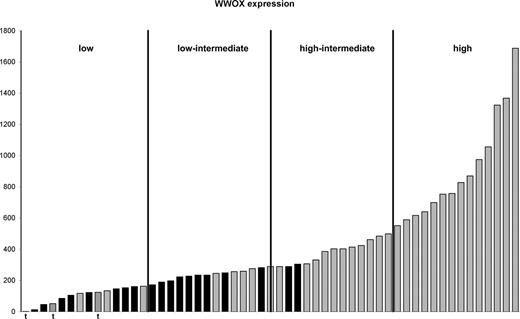
Of the cases with LOH of 16q, 21 of 22 had LOH of region 3, which included 16q23.1, the location of WWOX. This included one t(14;16) case in which the deletion commenced within the gene itself extending to the telomere. In 2 other t(14;16) cases, deletions of 2 Kb and 46 Kb spanning 6 and 27 SNPs, respectively, could be identified within WWOX (Figure 5). The identification of these regions using mapping technology supported the results of previous studies that the IgH translocation breakpoint occurred at common fragile site FRA16D, also the location of WWOX.21,22 WWOX gene expression was examined in those cases either with deletion spanning the gene or translocation and found to be reduced relative to cases without deletion or translocation (Figure 6). Cases with LOH or translocation involving WWOX clustered in quartiles 1 and 2 of WWOX expression.
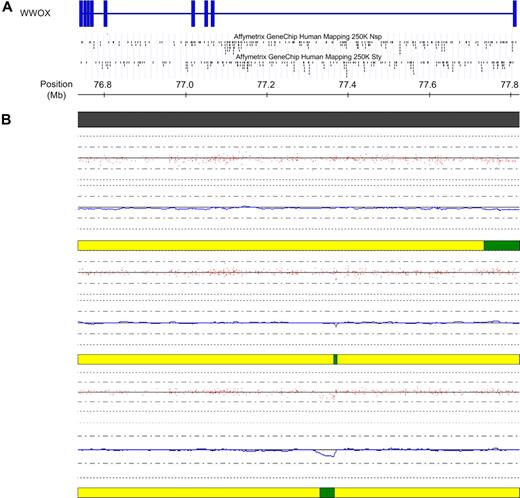
Deletions within WWOX in cases with a t(14;16). (A) Schematic of the WWOX gene located at FRA16D at 16q23. Vertical blue bars indicate exons. The location of the SNPs within the gene on each of the 2 mapping arrays are indicated below the schematic. (B) High-resolution view of the WWOX gene at 16q23. Copy number for each of the 3 t(14;16) samples is indicated below the physical position bar (top). Red dots indicate raw copy number for each SNP. The blue line indicates smoothed copy number. The horizontal bar below each case indicates the presence of normal copy number (yellow) and deletion (green). In all 3 cases, the deletion either commences within WWOX (top case) or lies wholly within WWOX (middle and bottom case), at the presumed translocation breakpoint. The figure was generated using CNAG and the UCSC genome browser.
Deletions within WWOX in cases with a t(14;16). (A) Schematic of the WWOX gene located at FRA16D at 16q23. Vertical blue bars indicate exons. The location of the SNPs within the gene on each of the 2 mapping arrays are indicated below the schematic. (B) High-resolution view of the WWOX gene at 16q23. Copy number for each of the 3 t(14;16) samples is indicated below the physical position bar (top). Red dots indicate raw copy number for each SNP. The blue line indicates smoothed copy number. The horizontal bar below each case indicates the presence of normal copy number (yellow) and deletion (green). In all 3 cases, the deletion either commences within WWOX (top case) or lies wholly within WWOX (middle and bottom case), at the presumed translocation breakpoint. The figure was generated using CNAG and the UCSC genome browser.
Expression of WWOX in cases with LOH of 16q and translocations involving WWOX. Cases are ordered according to WWOX expression. Cases with LOH at 16q23 (■) and those with retention of herozygosity at 16q23 (▒) are shown. Cases with translocation involving WWOX (▨) and “t” below the bar are shown. The cases are divided into 4 quartiles based on WWOX expression.
Expression of WWOX in cases with LOH of 16q and translocations involving WWOX. Cases are ordered according to WWOX expression. Cases with LOH at 16q23 (■) and those with retention of herozygosity at 16q23 (▒) are shown. Cases with translocation involving WWOX (▨) and “t” below the bar are shown. The cases are divided into 4 quartiles based on WWOX expression.
WWOX is known to have a proapoptotic effect by participating in the tumor necrosis factor (TNF) apoptotic pathway and via direct physical interaction with p53 and its homolog p73.37-39 We therefore examined the apoptotic response in cases classified according to WWOX expression. A total of 175 of 640 probe sets were significantly differentially expressed (2-tailed t test, P < .05) in cases with low versus high WWOX expression. The empirical P value for finding any 175 of 640 probe sets was less than 10−4. A core overlapping set of 71 probe sets was identified when the same analysis was performed on the independent set of 414 cases. These 71 probe sets represent a validated “low-WWOX signature” (Table S3).
HMCLs
Gene expression and mapping on 9 HMCLs were performed. Deletion (JIM1) or UPD (U266) of the whole of 16q was present in 2 cell lines, and deletion of 16q11-q12.1 and 16q13-q23.1 was present in a third (KMS26), spanning the loci of CYLD and WWOX in all 3. In 3 cell lines, hemizygous deletions were identified located entirely within WWOX (KMS11, RPMI-8266, and U266), including a 149-kb homozygous deletion located entirely within WWOX (KMS11). Gene expression analysis confirmed that all HMCLs with LOH had low or low-normal expression levels of these genes. These data support the importance of 16q abnormalities in myeloma cell lines derived from advanced clinical samples as well as in presenting myeloma cases.
Discussion
Clinical impact of 16q deletion
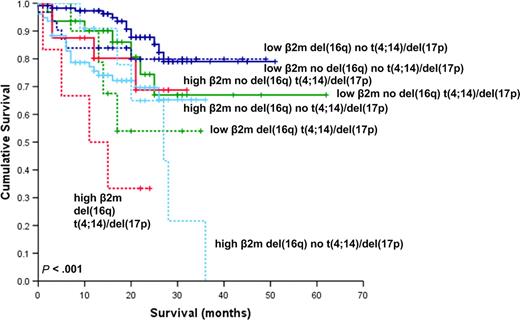
In this study we have carried out the first large-scale analysis of the effects of del(16q) on prognosis in newly diagnosed multiple myeloma. In this series, the presence of del(16q) was associated with worse OS. Given the association we found between del(16q) with deletions of 13 and 17p, we examined the effect of combinations of del(16q) with these and other generally recognized adverse prognostic abnormalities. The addition of del(16q) did not impact OS in cases with and without del(13q) but was associated with a significantly worse outcome in combination with either t(4;14) or del(17p). We also determined that the presence of del(16q) impacts adversely on prognosis in cases with both a low and high β2m. Multivariate analysis confirmed del(16q) to be an independent prognostic marker when other cytogenetic and clinical variables were taken into consideration. However, despite the apparent independence of del(16q) in determining clinical outcome, Figure 7 shows that this difference is not apparent in those cases with neither high β2m, nor the presence of t(4;14) or del(17p). These data mirror that of the Intergroupe Francophone Myélome (IFM), which has shown that t(4;14) and del(17p) have a lesser impact in cases with a low β2m.40 It therefore appears that β2m still needs to be considered alongside del(16q) and other cytogenetic factors in determining prognosis.
Kaplan-Meier plot of OS considering the influence of del(16q), β2m, t(4;14), and/or del(17p). The continuous and dashed lines indicate the absence and presence of del(16q), respectively, in combination with high (≥ 5.5 mg/L) or low (< 5.5 mg/L) β2m and the presence of t(4;14) and/or del(17p).
Kaplan-Meier plot of OS considering the influence of del(16q), β2m, t(4;14), and/or del(17p). The continuous and dashed lines indicate the absence and presence of del(16q), respectively, in combination with high (≥ 5.5 mg/L) or low (< 5.5 mg/L) β2m and the presence of t(4;14) and/or del(17p).
Gene mapping provides mechanistic insights into myeloma initiation and progression
The prognostic importance of del(16q) led us to examine the genes of potential importance dysregulated by the deletion. We previously examined 30 myeloma cases using 50K mapping arrays and identified the presence of del(16q) in 5 cases, all of which involved the entire of 16q,5 making it difficult to determine which gene or genes were of greatest importance. In this study we have performed copy number and LOH analysis using 500K mapping arrays on a subset of 55 of the 861 cases in which FISH had been performed. In addition to 10-fold greater resolution compared with the 50K arrays, we also had the advantage in this study of paired peripheral blood DNA and therefore the ability to identify allele-specific copy number. This has enabled identification of smaller deletions with greater accuracy and confident determination of true LOH and UPD. We have shown that 16q LOH is not restricted solely to deletions of the whole of 16q but ranges from deletions of the entire chromosome to interstitial deletions of less than 1 Mb. The presence of small regions of LOH outside the area covered by the MAF FISH probe on 16q and the identification of UPD of 16q in 4 cases resulted in the identification of 16q LOH in 22 of 55 cases (40%), considerably higher than the frequency identified by FISH for 16q23 deletion alone (19%). The impact of this on prognosis is unclear from this dataset because of sample size, but warrants further investigation.
Combining our current observations with what is known about the molecular biology of myeloma, it is possible to extrapolate further about the initiation and progression of myeloma. It is accepted that there are 2 broad groups of myeloma, those with reciprocal translocations and those with hyperdiploidy. Translocations are thought to involve aberrant physiologic class switch rearrangements, which deregulate oncogenes on partner chromosomes. It seems unlikely that these events, in isolation, are sufficient to give rise to myeloma as they are often evenly distributed between MGUS, SMM, and MM. The negative impact on survival of del(16q) in cases with t(4;14) suggests that genomic changes leading to functional loss of tumor suppressor genes is an important mechanism of tumor progression in these cases. The presence of 16q deletions and UPD in HMCLs supports this concept but warrants further investigation in both MGUS and relapsed myeloma cases. This type of observation can help to explain clinical heterogeneity within an apparently homogeneous group defined by a translocation. While deletion is a key mechanism in this respect, we also identify copy neutral LOH as being important (Figures 2,3).
Definition of deregulated genes by integrating gene mapping with expression analysis
We have previously identified 60 genes located on 16q that were underexpressed in cases with LOH of the whole region.5 However, in this study, we have used more stringent comparison criteria and identified a shorter list of 39 transcripts located on 16q that were underexpressed in cases with 16q LOH (Table S1). Of these, the 16 genes present on both lists include CYLD and WWOX, both of which show underexpression in the cases with LOH of 16q12 and 16q23, respectively, emphasizing their importance.
WWOX
WWOX is located at common fragile site FRA16D, which is relevant to the pathogenesis of myeloma as it is the location of t(14;16) breakpoints that juxtapose the IgH enhancer centromeric to MAF, resulting in MAF overexpression.21,22 Subsequently, the WWOX gene itself was identified within FRA16D, consisting of 9 exons and spanning in excess of 1 Mb.14 There is a high incidence of LOH of WWOX, including homozygous deletions, in a wide range of human cancers.18 The tumor suppressor function of WWOX has been demonstrated in a number of studies in which ectopic WWOX protein expression has been shown to result in inhibition of tumor growth in breast, lung, and prostate cancer cell lines and suppressed tumorgenicity of xenografts in nude mice.17,41-43 In a recent study, not only did Wwox−/− mice develop spontaneous tumors, but there was also a higher incidence of ethyl nitrosurea-induced tumors, including lymphoma, in Wwox+/− mice. This not only confirms the tumor suppressor function of Wwox, but it also confirms that haploinsufficiency is sufficient to induce tumorigenesis.44 WWOX has been shown to act via a number of pathways to induce apoptosis. Murine Wwox-transfected L929 cells were shown to have an enhanced sensitivity to TNF cytotoxicity, which was associated with an up-regulation of p53 and down-regulation of antiapoptotic BCL2 and BCL-xL.38 WWOX protein also directly interacts with tumor suppressor proteins p53 and its homolog p73, enhancing the proapoptotic effects of these proteins.37,39 Using WWOX expression as a marker, we were able to determine a “low-WWOX signature” composed of 71 probe sets that characterized these cases. This signature needs further evaluation, particularly with regard to clinical response to chemotherapy agents acting via differing apoptosis pathways.
The interaction of WWOX protein with p53 and p73 is particularly interesting given the positive association between the presence of del(16q) and del(17p) by FISH (Table 1). In other studies, del(17p) has been reported as being a prognostic factor in myeloma.40,45,46 In our series, del(17p) showed a trend toward adverse survival, but when the effects of del(16q) were taken into consideration, the presence of del(17p) alone resulted in a survival curve equivalent to that of cases with neither del(16q) nor del(17p) (Figure 1C). Del(16q) alone conferred moderately adverse prognosis (median OS 36 months), whereas the combination of del(16q) with del(17p) was associated with considerably worse outlook (median OS 17 months). Given that p73 is able to activate p53 target genes, this observation suggests that with loss of p53 alone the intact WWOX/p73 pathway is able to regulate apoptosis. However, disruption of p73-dependent apoptosis caused by deletion of WWOX in combination with loss of p53 heralds rapid clinical deterioration. Further studies are warranted to investigate this and the association between t(4;14) and del(16q) further.
The high-resolution mapping arrays used in this study have allowed us to analyze in some detail the genomic structure of the t(14;16). In 2 of 3 cases with a t(14;16), we have been able to identify intragenic deletions within WWOX at the presumed translocation breakpoint. In the third t(14;16) case, a deletion of 16q23 commenced within WWOX at this site. In all 3 cases, the translocation had the same functional consequences as deletion of the entire gene by diminishing the expression of WWOX to a level equivalent to cases with deletion of the entire gene (Figure 5). Therefore, the poor prognosis of the t(14;16) may not solely be caused by the overexpression of MAF resulting from the translocation but also by the simultaneous inactivation of WWOX during the molecular events leading to the translocation. In total, WWOX was deregulated in 22 of 55 cases (40%) as a result of LOH. Methylation of WWOX has been identified as being a mechanism of gene inactivation in breast and prostate cancer47 and probably explains the low expression of WWOX in the cases without LOH or translocation of this gene. It is likely that other mechanisms of WWOX inactivation may actually have diminished the prognostic impact demonstrated by FISH of deletion of 16q23.1 in our series, given that cases with WWOX inactivation without deletion would have been included in the nondeleted group.
CYLD.
CYLD, located at 16q12.1, was first identified as the relevant tumor suppressor gene in cylindroma tumors, a form of hereditary skin tumor.48 Mutation of CYLD has been detected in cylindroma cases,49 and mutations have been noted in myeloma.50 CYLD is known to function as a negative regulator of the NF-κB pathway,34-36 and recent studies suggest that CYLD acts via the noncanonical NF-κB pathway.50 LOH of CYLD provides further evidence of the central role of deregulation of this pathway in myeloma. In this study, we found that LOH cases clustered in the lowest quartile of CYLD expression levels. We used CYLD expression levels to define a validated list of 73 apoptosis-related probe sets. Further investigation is warranted to determine the clinical significance of alterations of genes contained within this signature.
Combined loss of tumor suppressor genes.
Using high-resolution gene mapping has enabled us to target specific regions of 16q as the location of key genes responsible for the poor prognosis associated with del(16q). However, it remains the case that in the majority of cases with 16q abnormalities, with the exception of cases with t(14;16), there is LOH of the whole of 16q rather than being solely the regions of interest outlined above. Given that loss of CYLD acts via the NF-κB pathway to promote proliferation, and loss of WWOX expression results in loss of the proapoptotic effects of p53 and p73, it seems highly likely that it is the combined loss of these 2 genes that is the crucial factor in determining prognosis in this group of cases. As a result, it is difficult to determine the precise impact of each of the 2 genes in isolation; therefore, further studies are required to fully determine the relative importance of different genetic events in the generation of specific gene signatures and their clinical impact.
Summary
In this study, we provide significant additional information contributing to our understanding of the pathogenesis and clinical outcome of myeloma. Using FISH, we have shown for the first time the adverse prognostic impact associated with 16q deletion in a large series of newly diagnosed myeloma cases. In particular, del(16q) confers additional adverse prognosis in cases with established poor risk abnormalities such as the presence of t(4;14), del(17p), and high β2m. Using 500K mapping and expression arrays, we have identified the 2 most likely genes responsible for the outcome of patients with del(16q), WWOX and CYLD. These genes and their corresponding pathways also provide a focus for the identification of other aberrations that may be important in myeloma progression.
The online version of the article contains a data supplement
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Research grants and financial support were received from the Leukaemia Research Fund, Cancer Research UK, the Bud Flanagan Research Fund, the David Adams Leukaemia Fund, the Kay Kendall Leukaemia Fund, and the United Kingdom Department of Health.
Authorship
Contribution: M.W.J. performed research, collected and analyzed data, and wrote the paper; P.E.L. and B.A.W. performed research and analyzed data; D.C.J. performed research; L.C., E.D.C., G.P.D., M.N., R.K.M.P., and D.S. performed research and collected data; N.J.D. and M.E. analyzed data; F.M.R. performed research and collected and analyzed data; D.G., N.C.P.C., and F.E.D. designed research; G.J.M. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gareth Morgan, Section of Haemato-Oncology, Institute of Cancer Research, 15 Cotswold Road, Sutton, Surrey SM2 5NG, United Kingdom; e-mail: gareth.morgan@icr.ac.uk.