Abstract
The PU.1 gene contains a 237–base pair distal enhancer located 14 kilobases upstream of its promoter. We have identified 2 sites within the PU.1 enhancer that strongly bind C/EBPα in a gel shift assay, and interaction with endogenous C/EBPα was confirmed by chromatin immunoprecipitation. Mutation of these DNA elements reduced activity of a distal enhancer-promoter construct 2- or 5-fold in a myeloid cell line, while mutation of a weaker C/EBPα-binding site located in the promoter minimally reduced activity in this context. These findings strengthen the link between C/EBPα and PU.1 expression. Reduction of C/EBPα activity in cases of acute myeloid leukemia may therefore contribute to transformation by reducing PU.1 levels. In addition, induction of PU.1 by C/EBPα during normal hematopoiesis may contribute to stem cell commitment to the myeloid lineages and further commitment to monopoiesis. Consistent with a requirement for C/EBPα induction of PU.1 during myeloid development, we demonstrate that C/EBPα induces monocytic development when expressed in PU.1+/+, PU.1+/−, or PU.1+/kd marrow myeloid progenitors but induces granulocyte lineage commitment in PU.1kd/kd cells lacking the PU.1 distal enhancer and does not induce either lineage in PU.1−/− cells.
Introduction
C/EBPα and PU.1 are both key regulators of granulocytic and monocytic myeloid development.1 Within hematopoiesis, C/EBPα is found predominantly in immature myeloid cells, whereas both lymphoid and myeloid cells express PU.1.2,3 In keeping with these expression profiles, C/EBPα−/− mice have greatly reduced granulocyte colony-forming units (CFU-Gs) and macrophage colony-forming units (CFU-Ms) due to a block in the transition from the common myeloid progenitor (CMP) to the granulocyte-monocyte progenitor (GMP), whereas PU.1−/− mice lack B cells and monocytes and have markedly reduced neutrophils.4-8 Hematopoietic stem cells lacking PU.1 do not generate lymphoid or myeloid progenitors.9
These findings suggest that PU.1 acts at an earlier stage than C/EBPα to regulate hematopoietic lineage determination. However, the situation is more complicated as several lines of evidence indicate that increased PU.1 favors monocyte over granulocyte or B-lineage commitment. Lineage-negative fetal liver progenitors develop into B cells when exogenous PU.1 is introduced at low levels and into monocytes when increased PU.1 is expressed, and siRNA knockdown of PU.1 in embryonic stem cell (ESC)–derived CD34+ cells favors B-cell formation.10,11 Marrow from PU.1+/−;G-CSF−/− mice generates increased CFU-Gs and reduced CFU-Ms compared with PU.1+/+;G-CSF−/− mice, as do PU.1+/− compared with control ESCs.12 PU.1 knockdown (PU.1kd/kd) mice, having homozygous deletion of the PU.1 distal enhancer located at −14 kb, demonstrate 20% of normal PU.1 expression and loss of monopoiesis with preservation of granulopoiesis, and CRE-mediated deletion of PU.1 in adult mice preserves granulocytes at the expense of monocytes.13,14
Transcriptional induction of PU.1 by C/EBPα may play a role in myeloid lineage specification. C/EBPα or PU.1 induces monocytic development when expressed via retroviral transduction in B-lineage cells.15 Similarly, exogenous C/EBPα induces PU.1 mRNA and increases the formation of monocytic cells when expressed in lineage-negative myeloid progenitors.16 Activation of C/EBPα-ER with estradiol in the Ba/F3 pro-B-cell or 32Dcl3 myeloid cell lines rapidly induces PU.1 mRNA even in the presence of cycloheximide, and C/EBPα binds and activates the PU.1 promoter.17,18 However, interaction of C/EBPα with the PU.1 promoter site in a gel shift assay was several-fold weaker than its interaction with the neutrophil elastase (NE) promoter, and mutation of the PU.1 promoter site reduced reporter activity less than 2-fold in myeloid cells.
To further establish that C/EBPα regulates PU.1 gene transcription, we have now investigated its ability to bind and activate the proximal region of the recently identified PU.1 distal enhancer.19 In addition to identifying 2 strong, functional C/EBP-binding sites within the enhancer, we demonstrate that C/EBPα cannot induce granulocyte or monocyte differentiation in the absence of PU.1 and preferentially induces granulopoiesis in the absence of the PU.1 distal enhancer. These results suggest that induction of PU.1 by C/EBPα plays a central role in the CMP to GMP transition and in subsequent commitment to the monocyte lineage. In addition, mutations in acute myeloid leukemia (AML) that diminish C/EBPα activity may contribute to transformation by reducing PU.1 expression.
Materials and methods
Plasmids
A BamHI fragment spanning from −334 to +151 of the murine PU.1 promoter was ligated into the BglII site of pXP2 (ATCC, Manassas, VA) to generate PUProm-Luc. The 237-bp proximal region of the PU.1 distal enhancer (DE) was amplified by polymerase chain reaction (PCR) and ligated as a HindIII/XhoI fragment to generate DE-PUProm-Luc. Three C/EBP-binding sites in the DE or the C/EBP-binding site in the promoter were individually mutated by PCR-based mutagenesis, leading to the following changes, with mutant bases italicized: C1 (5′-TGTGGTAAT to 5′-TGTGGTTCC), C2 (5′-TTTTGCAAT to 5′-TTACGCGTT), C5 (5′-TGTGGAAAC to 5′-TGTGCATCC), or Pr (5′-TAGCGCAAG to 5′-TAGCCCTGG). pBabePuro-C/EBPα-ER and pBabePuro-PU.1-ER(T) were previously described.17
Cell culture and transfection
32DPKCδ or 32Dcl3 cells18,20 were maintained in Iscoves modified Dulbecco medium (IMDM) with 10% heat-inactivated fetal bovine serum (HI-FBS) and 1 ng/mL murine IL-3 (Peprotech, Rocky Hill, NJ). 32Dcl3 cells were washed twice with phosphate-buffered saline before transfer to IMDM/HI-FBS with 20 ng/mL granulocyte colony-stimulating factor (G-CSF; Amgen, Thousand Oaks, CA). HF1 myeloid cells21 were maintained in IMDM medium with 10% HI-FBS and 2.5 ng/mL murine granulocyte-macrophage colony-stimulating factor (GM-CSF; Peprotech), 503 PU.1−/− cells22 were maintained in IMDM with 20% HI-FBS and 2.5 ng/mL murine IL-3, and U937 cells were cultured in RPMI 1640 with 10% HI-FBS. 293T cells were cultured in Dulbecco modified Eagle medium with 10% HI-FBS. 32DPKCδ cells were transfected with luciferase reporter plasmids using a DEAE-dextran procedure, followed by luciferase assay of cell extracts 2 days later, as described.23 CMV-βGal was included in each transfection as an internal control, and extracts were subjected to β-galactosidase assay using the Galacto-Light Plus kit per the manufacturer's instructions (Tropix, Bedford, MA). 293T cells on 100-mm dishes were transfected with 8 μg pBabePuro or its derivatives and 2 μg pkat2ecopac24 using 16 μL Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Supernatants collected 2 and 3 days later were adsorbed to retronectin-coated dishes (Takara, Shiga, Japan), after which 503 cells were added with 4 μg/mL polybrene for 2 days. Pools of transductants were subsequently obtained by selection in 2 μg/mL puromycin. Subclones were isolated by limiting dilution in 96-well dishes. 503 lines were cultured with 1 μM estradiol (E2) or 100 nM 4-hydroxytamoxifen (4HT). Transduction of murine mononuclear marrow cells followed by lineage depletion and subsequent liquid culture with or without E2 were as described.16 Marrow cells were obtained from PU.1+/+, PU.1+/−, PU.1+/kd, and PU.1kd/kd mice.8,13,25 For fluorescence-activated cell sorting (FACS) analysis, cells were stained with PE–anti–Mac-1 and FITC–anti–Gr-1 (BD Pharmingen, San Diego, CA) or with PE–anti-F4/80 (Caltag, Burlingame, CA) and FITC–anti–Gr-1. For colony-forming unit (CFU) assay, cells were plated at 2500 cells/mL in IL-3/IL-6/SCF as described.16 Morphology was assessed by Wright-Giemsa staining. Photomicrographs of cells were taken using a Zeiss Axiophot microscope (Carl Zeiss, Thornwood, NY), a Kontron Electronik Progress 3012 camera (Kontron, Munich, Germany), and a 63×/1.40 NA oil objective.
Chromatin immunoprecipitation, gel shift assay, and Western blotting
Chromatin immunoprecipitation (ChIP) analysis for interaction with the PU.1 DE was carried out as described18 using rabbit–anti-C/EBPα (14AA), rabbit–anti-C/EBPβ (C19), or rabbit Ig (Santa Cruz Biotechnology, Santa Cruz, CA). Oligonucleotides for PCR amplification after ChIP were as follows: PU.1 DE, 5′-CTGGTGGCAAGAGCGTTTC-3′ and 5′-CAGCAAGGCCGGTGCCTG-3′; actin, 5′-GCGAGGCCGGTGAGTGAG-3′ and 5′-GTGGACGCGACTCGACAG-3′; and PU.1 (−20 kb), 5′-CTTGTGCCTGCATGAGTTTAC-3′ and 5′-AAAGCCAAGTCAAGTCACATC-3′.
Nuclear extracts were obtained from control or C/EBPα-transfected 293T cells or from U937 cells and subjected to gel shift assay as described.18 Double-stranded oligonucleotides used as probes were radiolabeled by Klenow reaction of 4 bp TCGA or GATC overhangs with 10 mM TTP, dATP, and dGTP with 32P-labeled dCTP. The sense strands of the wild-type probes, with C/EBP sites italicized, were as follows: NE, 5′-TCGAGGCCAGGATGGGGCAATACAACCCG-3′; C1, 5′-GATCTGTGGTGCCTGTGGTAATGGGGCTGTT-3′; C2, 5′-GATCCTGTTGGC-GTTTTGCAATGGGCAGGGG-3′; C3, 5′-GATCGCGGGCCTGGTGGCAAGAGCGTTTC-3′; C4, 5′-GATCGCACCGGCCTTGCTGCTGCC-GATGT-3′; and C5, 5′-GATCGCTGCCGATGTGGAAACAGGGGCAG-3′.
The mutant mC1, mC2, and mC5 probes carry the same changes described for the luciferase reporters. Western blotting was carried out using ERα (HC20) or ERα (MC20) antiserum as described.17
Results
C/EBPα binds the PU.1 distal enhancer
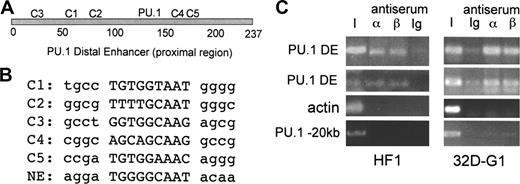
The 237-bp proximal region of the PU.1 distal enhancer contains 5 sites that match the C/EBP consensus binding site, T(T/G)NNGNAA(T/G), or differ by one base pair. The locations of these sites, designated C1 to C5, and the previously identified, functional PU.1-binding site are diagrammed (Figure 1A). The sequences of the C1 to C5 sites and the active C/EBP-binding site in the NE promoter are also shown (Figure 1B).
C/EBP consensus binding sites in the PU.1 distal enhancer and interaction of endogenous C/EBPs with the enhancer. (A) Positions of the 5 potential C/EBP-binding sites, C1 to C5, and the PU.1 site19 in the distal enhancer. (B) Sequences of the C1 to C5 and NE C/EBP sites, with flanking sequences in lower case. (C) HF1 myeloid cells or 32Dcl3 myeloid cells cultured in G-CSF for one day (32D-G1) were subjected to the ChIP assay using C/EBPα or C/EBPβ antiserum or rabbit Ig followed by PCR for a 154–base pair PU.1 −14-kb distal enhancer fragment (PU.1 DE), a 138-bp β-actin promoter fragment, or a 329-bp fragment located at −20 kb in the PU.1 gene. Bands were visualized after agarose gel electrophoresis by ethidium bromide staining. I indicates input DNA.
C/EBP consensus binding sites in the PU.1 distal enhancer and interaction of endogenous C/EBPs with the enhancer. (A) Positions of the 5 potential C/EBP-binding sites, C1 to C5, and the PU.1 site19 in the distal enhancer. (B) Sequences of the C1 to C5 and NE C/EBP sites, with flanking sequences in lower case. (C) HF1 myeloid cells or 32Dcl3 myeloid cells cultured in G-CSF for one day (32D-G1) were subjected to the ChIP assay using C/EBPα or C/EBPβ antiserum or rabbit Ig followed by PCR for a 154–base pair PU.1 −14-kb distal enhancer fragment (PU.1 DE), a 138-bp β-actin promoter fragment, or a 329-bp fragment located at −20 kb in the PU.1 gene. Bands were visualized after agarose gel electrophoresis by ethidium bromide staining. I indicates input DNA.
To demonstrate interaction of endogenous C/EBPα with the −14-kb PU.1 distal enhancer within chromatin in myeloid cells, we carried out the ChIP assay (Figure 1C). Specific interaction of both C/EBPα and C/EBPβ with the enhancer was evident in HF1 cells and in 32Dcl3 cells exposed to G-CSF for one day, compared with the actin promoter or to a DNA segment located −20-kb upstream of the PU.1 start site.
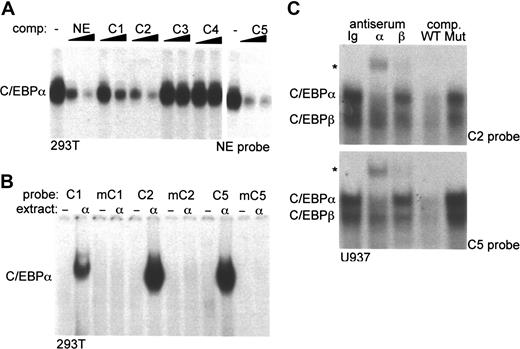
To evaluate the relative affinities of the potential binding sites C1 to C5 for C/EBPα, a gel shift competition assay was performed using a radiolabeled NE-C/EBP probe (Figure 2A). Unlabeled C2 or C5 sites competed as well as the NE-C/EBP oligonucleotide for binding. C1 competed 5-fold less well, as 25 ng C1 was required to obtain the same reduction in binding seen with 5 ng of the NE-C/EBP, C2, or C5 oligonucleotides. Similar relative competition with C1, C2, and C5 was seen in an independent experiment (not shown). Neither C3 nor C4 competed for C/EBPα binding to the NE probe.
Identification of C/EBPα-binding sites in the PU.1 distal enhancer. (A) Gel shift assay was carried out using 10 μg nuclear extract from 293T cells transfected with CMV-C/EBPα and 1 ng radiolabeled NE-C/EBP probe. Where indicated, 5 ng or 25 ng unlabeled NE or C1 to C5 double-stranded oligonucleotides were added as competitors (comp.) prior to addition of radiolabeled probe. Data with C5 are from a separate gel. (B) Gel shift assay was carried out using 10 μg nuclear extract from 293T cells transfected with CMV (−) or CMV-C/EBPα (α) and 1 ng of the indicated wild-type and mutant radiolabeled C/EBP site probes. See “Materials and methods” for a description of the point mutations in mC1, mC2, and mC5. (C) Gel shift assay was carried out using 12 μg nuclear extract from U937 cells and 1 ng radiolabeled C2 or C5 probes. Rabbit Ig (Ig), C/EBPα (α) or C/EBPβ (β) antiserum, or 25 ng unlabeled wild-type (WT) C2 or C5 or mutant (Mut) mC2 or mC5 competitor oligonucleotides were included in the gel shift reactions. The C/EBPα and C/EBPβ gel shift species and the supershifted species (*) are indicated.
Identification of C/EBPα-binding sites in the PU.1 distal enhancer. (A) Gel shift assay was carried out using 10 μg nuclear extract from 293T cells transfected with CMV-C/EBPα and 1 ng radiolabeled NE-C/EBP probe. Where indicated, 5 ng or 25 ng unlabeled NE or C1 to C5 double-stranded oligonucleotides were added as competitors (comp.) prior to addition of radiolabeled probe. Data with C5 are from a separate gel. (B) Gel shift assay was carried out using 10 μg nuclear extract from 293T cells transfected with CMV (−) or CMV-C/EBPα (α) and 1 ng of the indicated wild-type and mutant radiolabeled C/EBP site probes. See “Materials and methods” for a description of the point mutations in mC1, mC2, and mC5. (C) Gel shift assay was carried out using 12 μg nuclear extract from U937 cells and 1 ng radiolabeled C2 or C5 probes. Rabbit Ig (Ig), C/EBPα (α) or C/EBPβ (β) antiserum, or 25 ng unlabeled wild-type (WT) C2 or C5 or mutant (Mut) mC2 or mC5 competitor oligonucleotides were included in the gel shift reactions. The C/EBPα and C/EBPβ gel shift species and the supershifted species (*) are indicated.
To confirm interaction of C/EBPα with the C1, C2, or C5 sites, each of these was radiolabeled and subjected to gel shift analysis using either a control 293T nuclear extract or an extract expressing exogenous C/EBPα. Radiolabeled probes carrying mutations in the C1, C2, or C5 sites were studied similarly (Figure 2B). Consistent with the gel shift competition assay, C1, C2, and C5 each interacted with C/EBPα, with C1 having approximately 5-fold reduced affinity in this single assessment. Mutation of the C/EBP consensus sites prevented binding to each of these probes, confirming interaction at the predicted consensus sequences.
Gel shift assay was also carried out with nuclear extracts derived from the U937 myeloid cell line and the C2 or C5 probes (Figure 2C). C/EBPα antiserum specifically shifted the upper gel shift species and C/EBPβ antiserum, the lower gel shift species. A similar supershift pattern was obtained previously using myeloid nuclear extracts and either the PU.1 promoter or NE promoter C/EBP site.18 Competition with 25-fold excess of unlabeled wild-type C2 or C5 competed for both C/EBP species, whereas the mutant oligonucleotides carrying point mutations in their C/EBP consensus sites did not. Similar results were obtained using 32Dcl3 nuclear extracts (not shown). These data indicate that C/EBPα and C/EBPβ are the predominant endogenous myeloid nuclear proteins that interact with the C2 and C5 probes.
The C/EBP-binding sites are functional in myeloid cells
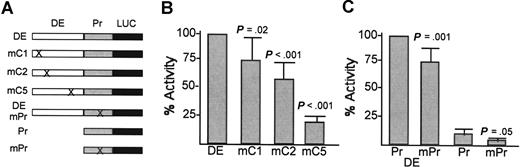
The PU.1 distal enhancer was linked to a 500-bp PU.1 promoter region and the luciferase cDNA. Point mutations were then introduced into the C1, C2, C5, or promoter C/EBP sites. The promoter site was also mutated in the absence of the enhancer (Figure 3A). The activities of these constructs were compared in the readily transfectable 32DPKCδ myeloid cell line that we used previously to determine the effect of mutating the PU.1 promoter C/EBP-binding site (Figure 3B, C).18 Mutation of C1 only mildly reduced enhancer activity, mutation of C2 reduced activity 2-fold, and mutation of C5 reduced activity 5-fold. Disruption of the promoter C/EBPα-binding site only minimally reduced activity when the enhancer was present but reduced activity approximately 2-fold in its absence. Together with the ChIP and gel shift results, these data indicate that C/EBPα strongly binds and activates the PU.1 distal enhancer.
Activity of the PU.1 distal enhancer C/EBP-binding sites in myeloid cells. (A) Diagram of reporter constructs containing point mutations in the C1, C2, C5, or promoter C/EBP-binding sites. DE indicates 237-bp PU.1 distal enhancer; Pr, PU.1 485-bp PU.1 promoter; and LUC, luciferase cDNA. (B) 32DPKCδ cells were transfected with 15 μg of the indicated reporters and 0.5 μg CMV-βGal. Two days later, cell extracts were analyzed for luciferase and β-galactosidase activities. The ratio of these activities for DE was set to 100% in each experiment. Results shown are mean and SE from 6 determinations. (C) The indicated constructs were assayed similarly, with DE linked to wild-type promoter (Pr) set to 100% activity in each experiment. Results are mean and SE from 6 determinations. P values versus DE-Luc (or versus Pr-Luc for mPr) are from the Student t test.
Activity of the PU.1 distal enhancer C/EBP-binding sites in myeloid cells. (A) Diagram of reporter constructs containing point mutations in the C1, C2, C5, or promoter C/EBP-binding sites. DE indicates 237-bp PU.1 distal enhancer; Pr, PU.1 485-bp PU.1 promoter; and LUC, luciferase cDNA. (B) 32DPKCδ cells were transfected with 15 μg of the indicated reporters and 0.5 μg CMV-βGal. Two days later, cell extracts were analyzed for luciferase and β-galactosidase activities. The ratio of these activities for DE was set to 100% in each experiment. Results shown are mean and SE from 6 determinations. (C) The indicated constructs were assayed similarly, with DE linked to wild-type promoter (Pr) set to 100% activity in each experiment. Results are mean and SE from 6 determinations. P values versus DE-Luc (or versus Pr-Luc for mPr) are from the Student t test.
C/EBPα requires the PU.1 gene and its distal enhancer to induce monopoiesis
We evaluated the ability of C/EBPα to induce monocytic development in PU.1+/+ versus PU.1+/− murine marrow cells using a strategy we recently optimized that allows the effects of C/EBPα on early myeloid differentiation events to be evaluated while minimizing bias due to lineage-specific cell cycle inhibition.16 C/EBPα inhibits G1 to S cell cycle progression in myeloid cells via interaction with E2F1.26,27 Key features of this strategy are transduction with C/EBPα-ER, a fusion protein inactive prior to addition of E2, and lineage depletion only after transduction and drug selection. Cells are then split and cultured with or without E2 for 2 days in liquid culture prior to FACS analysis for Mac-1 and Gr-1. Representative results of this assay for PU.1+/+ compared with PU.1+/− cells from littermate mice are shown (Figure 4A). Mac-1+Gr-1− cells are monocytes and Mac-1+Gr-1+ cells are granulocytes.16 In the absence of E2, PU.1+/− cells developed 26% monocytes, while PU.1+/+ cells developed 55% monocytes. In a second experiment, PU.1+/− cells developed 38% and PU.1+/+ cells 49% monocytes prior to E2 addition (not shown). Despite reduced basal monopoiesis in PU.1+/− marrow cells, activation of C/EBPα-ER with E2 markedly increased the production of monocytes from both PU.1+/+ and PU.1+/− cells, shifting the majority of the cells to the upper left quadrant in the experiment shown and in a second experiment in which 78% of PU.1+/− and 87% of PU.1+/+ cells were Mac-1+Gr-1− after activation of C/EBPα-ER for 2 days (not shown). E2 does not increase monocyte formation from pBabePuro vector–transduced cells.16 Thus, loss of one copy of the PU.1 gene reduces basal monopoiesis but still allows a monocytic response to exogenous C/EBPα.
Induction of monocyte development by C/EBPα requires the distal enhancer. (A) Marrow mononuclear cells from PU.1+/+ or PU.1+/− mice exposed 5 days earlier to 5-fluorouracil were transduced with pBabePuro-C/EBPαER for 3 days and then exposed to 2 μg/mL puromycin for one day. Viable cells were then lineage-depleted and finally cultured with or without E2 for 2 days before FACS analysis. Percentage monocyte (% M) was calculated as (Mac-1+Gr-1−)/(Mac-1+Gr-1− + Mac-1+Gr-1+) cells × 100%. (B) PU.1+/kd or PU.1kd/kd mice were analyzed similarly. (C) Morphology of PU.1+/kd or PU.1kd/kd cells transduced with C/EBPα-ER and cultured with or without E2 for 2 days. (D) Percentage of CFU-Gs or CFU-Ms compared with CFU-Gs + CFU-Ms obtained in methylcellulose culture with IL-3/IL-6/SCF without E2 after transduction of C/EBPα-ER into PU.1+/+, PU.1+/kd, or PU.1kd/kd cells and cultured for 24 hours ± E2 prior to plating. Results shown are from 2 independent transduction experiments (top and bottom panels). The graphs represent the mean and SE from 4 determinations within each experiment (P values are from the Student t test).
Induction of monocyte development by C/EBPα requires the distal enhancer. (A) Marrow mononuclear cells from PU.1+/+ or PU.1+/− mice exposed 5 days earlier to 5-fluorouracil were transduced with pBabePuro-C/EBPαER for 3 days and then exposed to 2 μg/mL puromycin for one day. Viable cells were then lineage-depleted and finally cultured with or without E2 for 2 days before FACS analysis. Percentage monocyte (% M) was calculated as (Mac-1+Gr-1−)/(Mac-1+Gr-1− + Mac-1+Gr-1+) cells × 100%. (B) PU.1+/kd or PU.1kd/kd mice were analyzed similarly. (C) Morphology of PU.1+/kd or PU.1kd/kd cells transduced with C/EBPα-ER and cultured with or without E2 for 2 days. (D) Percentage of CFU-Gs or CFU-Ms compared with CFU-Gs + CFU-Ms obtained in methylcellulose culture with IL-3/IL-6/SCF without E2 after transduction of C/EBPα-ER into PU.1+/+, PU.1+/kd, or PU.1kd/kd cells and cultured for 24 hours ± E2 prior to plating. Results shown are from 2 independent transduction experiments (top and bottom panels). The graphs represent the mean and SE from 4 determinations within each experiment (P values are from the Student t test).
We carried out a similar experiment with marrow cells from PU.1+/kd and PU.1kd/kd mice, the latter lacking both copies of the distal enhancer (Figure 4B). Activation of C/EBPα-ER in PU.1+/kd cells again increased monocytic compared with granulocytic development. Consistent with the FACS profile, cytospins of PU.1+/kd cells cultured without E2 consisted mainly of small, granulated cells, whereas addition of E2 led to a predominance of larger monocytic cells (Figure 4C left panels). For PU.1kd/kd cells transduced with C/EBPα-ER, in the absence of E2 only a small proportion developed into Mac-1+Gr-1− monocytes or Mac1+Gr-1+ granulocytes. Seventy-two percent of PU.1kd/kd cells that demonstrated maturation in the absence of E2 on FACS expressed Mac-1 but not Gr-1, although their level of Mac-1 was lower than that achieved by the monocytes obtained with PU.1+/kd cells. In addition, the large majority of PU.1kd/kd cells transduced with C/EBPα-ER but not exposed to E2 had an immature appearance, further indicating that these are not mature monocytes (Figure 4C upper right panel). Activation of C/EBPα-ER in PU.1kd/kd cells increased the overall numbers of both lineages, with granulocytes predominating on FACS (Figure 4B) and becoming evident morphologically (Figure 4C lower right panel).
To assess the ability of C/EBPα to induce monocyte versus granulocyte lineage commitment of immature progenitors in the absence of the PU.1 distal enhancer, transduced PU.1+/+, PU.1+/kd, or PU.1kd/kd cells were cultured with or without E2 for 24 hours and then plated in the absence of E2 in methylcellulose with IL-3/IL-6/SCF (Figure 4D). Removal of E2 at the time of plating prevents CFU suppression by C/EBPα-ER via cell cycle inhibition.16 As seen previously, activation of C/EBPα-ER in either PU.1+/+ or PU.1+/kd cells increased the proportion of CFU-Ms compared with CFU-Gs. In contrast, and consistent with results obtained in liquid culture, C/EBPα-ER significantly increased the proportion of CFU-Gs when expressed in lin− marrow cells isolated from PU.1kd/kd mice, in 2 independent experiments.
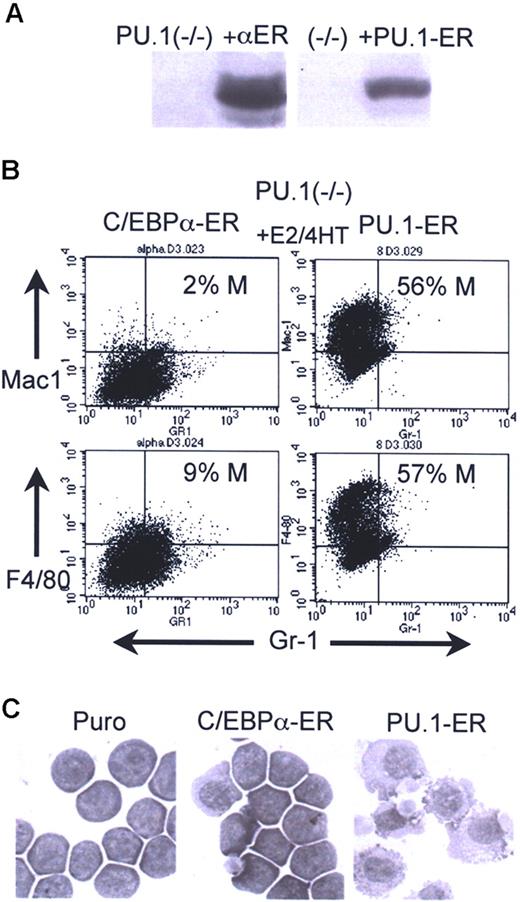
Finally, we introduced C/EBPα-ER into 503 PU.1−/− cells (Figure 5A left). PU.1-ER(T), a fusion between PU.1 and the 4HT-responsive variant of the murine ER ligand-binding domain, was also expressed in these cells as a positive control (Figure 5A right). Activation of C/EBPα-ER in pooled transductants did not induce monocyte or granulocyte formation, as judged by FACS analysis for Mac-1+/Gr-1− or F4/80+/Gr-1− monocytes, and Mac-1+/Gr-1+ granulocytes on day 2 (Figure 5B) and by morphologic analysis on day 3 (Figure 5C). In contrast, activation of PU.1-ER(T) with 4HT induced a marked induction of monocytic differentiation, demonstrating the capacity of these cells to differentiate along this pathway. Data representative of 2 independent assessments of monocytic induction using 503:C/EBPα-ER and 503:PU.1-ER(T) cells are shown. 503 PU.1−/− cells transduced with empty pBabePuro had less than 1% Mac-1+Gr-1− and 5% to 9% F4/80+Gr-1− cells, with or without E2 or 4HT, and 3 C/EBPα-ER–expressing subclones did not differentiate in response to E2 based on FACS or morphologic analysis (not shown). Thus, complete absence of the PU.1 gene prevents C/EBPα from inducing either monocytic or granulocytic maturation.
Inability of C/EBPα to induce myeloid maturation of PU.1−/− cells. (A) Western blots showing expression of C/EBPα-ER or PU.1-ER(T) in 503 PU.1−/− cells. Lane 1 in each blot contains extracts from the parental cell line. The filters were probed with human (left) or murine (right) ERα antisera. (B) C/EBPα-ER–expressing cells exposed to E2 and PU.1-ER(T) cells exposed to 4HT for 2 days were subjected to FACS analysis. Percentage monocytes is indicated (% M). (C) C/EBPα-ER–expressing cells exposed to E2 and PU.1-ER(T) cells exposed to 4HT for 3 days were cytospun and visualized by Wright-Giemsa staining. See “Cell culture and transfection” for image acquisition information.
Inability of C/EBPα to induce myeloid maturation of PU.1−/− cells. (A) Western blots showing expression of C/EBPα-ER or PU.1-ER(T) in 503 PU.1−/− cells. Lane 1 in each blot contains extracts from the parental cell line. The filters were probed with human (left) or murine (right) ERα antisera. (B) C/EBPα-ER–expressing cells exposed to E2 and PU.1-ER(T) cells exposed to 4HT for 2 days were subjected to FACS analysis. Percentage monocytes is indicated (% M). (C) C/EBPα-ER–expressing cells exposed to E2 and PU.1-ER(T) cells exposed to 4HT for 3 days were cytospun and visualized by Wright-Giemsa staining. See “Cell culture and transfection” for image acquisition information.
Discussion
Development of the mature blood lineages from pluripotent hematopoietic stem cells (HSCs) requires a series of commitment decisions regulated by transcription factors in response to environmental cues. Gene regulatory and protein-protein interaction mechanisms ensure the fidelity of this process. For example, GATA-1 is required for erythroid and megakaryocyte development, while PU.1 is required for lymphoid and myeloid development.7,8,28,29 PU.1, GATA-1, and C/EBPα activate their own promoters and enhancers as a positive-feedback mechanism to fix commitment decisions,19,30-32 and cross-inhibition between PU.1 and GATA-1 via direct interaction also ensures that cells continue to mature along a committed pathway once initiated.33-35 Similarly, cross-inhibition between Pax5 and C/EBPα may be essential for maintaining commitment of HSCs to the GMP versus the B-lymphoid progenitor stages.15,36 Our finding that C/EBPα strongly binds and activates the PU.1 distal enhancer solidifies a link between C/EBPα and PU.1 expression important to our understanding of both normal and malignant hematopoiesis. Of note, we previously demonstrated that C/EBPα-ER induces PU.1 mRNA when activated in transduced, lineage-depleted myeloid progenitors and that PU.1-ER(T) also induces monocytic maturation of these cells.16
The greater affinity of C/EBPα for the C2 or C5 enhancer sites compared with the promoter site and the larger impact of C5 mutation compared with promoter mutation on reporter activity in myeloid cells indicate that the distal enhancer is a key mediator of PU.1 gene regulation by C/EBPα. Of the 5 DE sites evaluated, C3 and C4 showed no interaction with C/EBPα. Of note, these 2 sites have a G or A instead of a T at the first position of the 9-bp core sequence. Mutation of C5 had greater effect than mutation of C2 on DE activity, although each bound C/EBPα with similar affinity in gel shift assays. The reason for this difference is unclear but may reflect synergy with other factors that bind nearby. Alternatively, mutation of C2 may allow cryptic binding of an activator whose binding is prevented by interaction of the enhancer with C/EBPα. In support of this latter possibility, a C2/C5 double mutant was consistently more active than the C5 mutant (not shown).
The inability of C/EBPα to induce myeloid differentiation of the 503 PU.1−/− line indicates that C/EBPα cannot induce another protein to bypass dependence on PU.1 for monopoiesis or granulopoiesis. PU.1+/− cells have 2 copies of the PU.1 distal enhancer, but only one is linked to a functional PU.1 gene. C/EBPα-ER increased monocyte formation from lineage-negative myeloid progenitors derived from either PU.1+/− or PU.1+/kd mice, although not as effectively as from PU.1+/+ cells. In contrast, C/EBPα-ER induced PU.1kd/kd cells to develop as granulocytes rather than monocytes in liquid culture and increased CFU-Gs relative to CFU-Ms in methylcellulose.
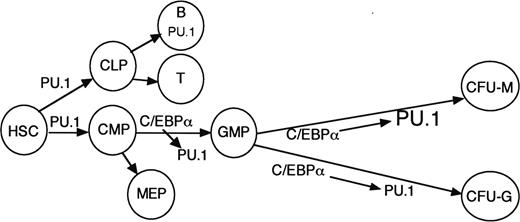
Overall, our findings indicate that C/EBPα directly regulates PU.1 transcription via interaction with its promoter and −14-kb distal enhancer and that availability of the distal enhancer for interaction with C/EBPα guides monocyte versus granulocyte lineage commitment in our in vitro transduction system. These results and the phenotype of C/EBPα−/− mice suggest that C/EBPα induces PU.1 to commit CMP to the GMP stage and that further induction of PU.1 contributes to specification of CFU-Ms and monopoiesis as opposed to CFU-Gs and granulopoiesis (Figure 6). Cytokine signals and cooperating factors might allow C/EBPα to induce PU.1 to a greater extent in CFU-Ms compared with CFU-Gs. PU.1 and Runx1 also bind and transactivate the PU.1 enhancer.19,37 These factors may provide the level of PU.1 expression necessary for lymphoid development while priming the enhancer for further activation by C/EBPα. The finding that C/EBPα induces monocytic development more potently than PU.1 in immature B cells suggests that C/EBPα may activate additional genes besides PU.1 required for monocytic development.15 Adult mice lacking PU.1 do not develop CMPs, whereas adult mice lacking C/EBPα retain CMPs but lack GMPs.5,9,14 Therefore, although C/EBPα inhibits HSC proliferation,5 obtaining the level of PU.1 required to specify the CMPs apparently does not require C/EBPα (Figure 6).
Model for the role of C/EBPα induction of PU.1 during hematopoiesis. PU.1 achieves sufficient levels in the absence of C/EBPα to help specify the CMPs. C/EBPα increases PU.1 to allow commitment of the CMP to the GMP stage. C/EBPα helps maintain PU.1 expression in the granulocyte lineage but further induces PU.1 in cooperation with cytokine signals and additional transcription factors to allow monocyte lineage specification.
Model for the role of C/EBPα induction of PU.1 during hematopoiesis. PU.1 achieves sufficient levels in the absence of C/EBPα to help specify the CMPs. C/EBPα increases PU.1 to allow commitment of the CMP to the GMP stage. C/EBPα helps maintain PU.1 expression in the granulocyte lineage but further induces PU.1 in cooperation with cytokine signals and additional transcription factors to allow monocyte lineage specification.
Mice lacking the PU.1 distal enhancer express 20% of normal PU.1 levels and develop acute myeloid leukemia (AML) with high penetrance,13 and yet mutation of PU.1 is rare in AML cases.38 In contrast, although C/EBPα−/− mice do not develop acute leukemia,4,5 several mechanisms reduce C/EBPα levels or activity in more than 50% of human AMLs: Mutations occur within the C/EBPα protein-coding region, AML1-ETO or flt3-ITD repress CEBPA transcription, bcr-abl inhibits CEBPA translation, and flt3-ITD and potentially other kinases that activate ERK perturb C/EBPα activity via phosphorylation of serine 21.39-43 Our findings suggest that these alterations contribute to transformation, at least in part, by reducing PU.1 expression. Therapies that increase PU.1 levels may be useful in reversing their transformed phenotype. Analogously, PML-RARα reduces PU.1 expression in the FAB M3 subset of AML, and exogenous PU.1 induced differentiation of myeloblasts expressing this oncoprotein.44
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health (grant HL082948) and by the Children's Cancer Foundation (A.D.F.).
National Institutes of Health
Authorship
Contribution: C.Y., D.W., and I.P.-P. performed the research and analyzed the results; A.D.F., D.G.T., and B.E.T. designed the study and analyzed the results; C.Y. and A.D.F. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alan D. Friedman, Johns Hopkins University, CRB I, Rm 253, 1650 Orleans St, Baltimore, MD 21231; e-mail: afriedm2@jhmi.edu.