Abstract
Cited2 (cAMP-responsive elementbinding protein [CBP]/p300-interacting transactivators with glutamic acid [E] and aspartic acid [D]–rich tail 2) is a newly identified transcriptional modulator. Knockout of the Cited2 gene results in embryonic lethality with embryos manifesting heart and neural tube defects. Cited2−/− fetal liver displayed significant reduction in the numbers of Lin−c-Kit+Sca-1+ cells, Lin−c-Kit+ cells, and progenitor cells of different lineages. Fetal liver cells from Cited2−/− embryos gave rise to markedly reduced number of colonies in the colony-forming unit assay. Primary and secondary transplantation studies showed significantly compromised reconstitution of T-lymphoid, B-lymphoid, and myeloid lineages in mice that received a transplant of Cited2−/− fetal liver cells. Competitive reconstitution experiments further showed that fetal liver hematopoietic stem cell (HSC) function is severely impaired due to Cited2 deficiency. Microarray analysis showed decreased expression of Wnt5a and a panel of myeloid molecular markers such as PRTN3, MPO, Neutrophil elastase, Cathepsin G, and Eosinophil peroxidase in Cited2−/− fetal livers. Decreased expression of Bmi-1, Notch1, LEF-1, Mcl-1, and GATA2 was also observed in Cited2−/− Lin−c-Kit+ cells. The present study uncovers for the first time a novel role of Cited2 in the maintenance of hematopoietic homeostasis during embryogenesis and thus provides new insights into the molecular regulation of hematopoietic development.
Introduction
The hematopoietic system is composed of a well-organized hierarchy of cells at different stages of differentiation. Hematopoietic stem cells (HSCs) are the most primitive component of this hierarchy and are responsible for life-long regeneration of blood cells. HSCs have 2 major functional features: one is the ability to produce new stem cells, a function normally referred to as self-renewal and the other is the commitment to differentiation. Progenitors or colony-forming cells (CFCs) are primitive hematopoietic cells capable of producing mature cells of one or more lineages. During murine ontogeny, HSCs and CFCs migrate from their respective tissue origins to fetal liver at approximately day 10 after coitus (10 dpc), and at or near birth, migrate from fetal liver to bone marrow, where they remain throughout the adult life.1
Cited2 (cAMP-responsive element–binding protein [CBP]/p300-interacting transactivators with glutamic acid [E] and aspartic acid [D]–rich tail 2) is one of the founding members of a new family of transcriptional modulators.2-5 As a CBP/p300-dependent transcription factor, Cited2 binds directly with high affinity to the first cysteine-histidine–rich (CH1) region of p300 and CBP.6 Cited2 is induced by many biologic stimuli, such as cytokines, serum, and lipopolysaccharide, in different cell types. Cited2 transforms cells when overexpressed by inducing loss of cell contact inhibition in Rat1 cells and tumor formation in nude mice.2 These initial in vitro studies underscore the potential roles of Cited2 in different biologic processes. Deletion of Cited2 gene results in embryonic lethality in the mid to late gestation with embryos displaying cardiac malformations, neural tube defects,7 adrenal agenesis,8-10 left-right patterning defects,9,11 and placental defects.12 Further mechanistic studies have provided evidence that Cited2 plays pivotal roles in these processes through its transcriptional modulator functions for HIF-18,13 or AP-2α signaling.9-11 Accumulated evidence has implicated the role of Cited2 in hematopoiesis because Bmi-1, which is essential for adult hematopoietic stem cell self-renewal,14 is induced by Cited2 in mouse embryonic fibroblast (MEF) cells.15 CBP and p300 are fate decision factors for HSCs, responsible for HSC self-renewal and hematopoietic differentiation, respectively.16 A recent gene expression profiling study to identify specific genes with long-term reconstitution (LTR) stem cell activity showed that expression of Cited2 correlates positively with LTR HSC activity.17 Cited2 expression during development is detected at multiple sites that form mesodermal structures5,18 from which HSCs are derived.19,20 All of these findings are suggestive of a potential role of Cited2 in hematopoiesis.
In this report, a series of studies were carried out to characterize the potential hematopoietic defects in Cited2−/− fetal liver. We demonstrate for the first time the gross aberrations in Cited2−/− fetal liver hematopoiesis, indicating that Cited2 is required for hematopoietic development.
Materials and methods
Mice
Cited2-deficient mouse line8 was maintained on C57BL/6 (CD45.2+) background. B6.SJL/BoyJ (CD45.1+) mice were purchased from Jackson Laboratory (Bar Harbor, ME). Mice were maintained in microisolator cages in pathogen-free facility. All husbandry and experiments were conducted in accordance with institutional guidelines of Case Western Reserve University.
Clonogenic assay
Fetal liver cells (2 × 104) were plated in triplicate in 35-mm cell culture dishes with 2-mm grid (Nalge Nunc International, Rochester, NY). Methylcellulose-based medium supplemented with 3 units/mL Epo, 10 ng/mL mouse recombinant IL-3, 10 ng/mL human recombinant IL-6, and 50 ng/mL mouse recombinant stem-cell factor (M3434; StemCell Technologies, Vancouver, BC) was used in the clonogenic assay. Colony formation of burst-forming unit–erythroid (BFU-Es), colony-forming unit–granulocyte/macrophage (CFU-GMs), and CFU–granulocyte/erythrocyte/monocyte/macrophage (CFU-GEMMs) was analyzed after 7 to 12 days.
Immunophenotypic analysis
For the analysis of Lin−c-Kit+Sca-1+ cells, 5 × 105 fetal liver cells were blocked with HBSS/10% rabbit serum and then stained with an antibody cocktail containing phycoerythrin-conjugated antibodies against lineage markers (BD Pharmingen, San Diego, CA). Antibodies included PE-conjugated CD3 (clone 500A2), CD4 (clone RM4–5), B220 (clone RA3–6B2), Gr-1 (clone RB6–8C5), and Ter119 (TER-119), an APC-conjugated antibody against c-Kit (clone 2B8), and an FITC-conjugated antibody against Sca-1 (D7). Fetal liver cells were also stained with antibodies against FITC-conjugated CD45 (clone 104), PE-conjugated Ter119, or Gr-1 separately. Fluorescence activated cell sorting (FACS) analysis was performed on the flow cytometer and the data were analyzed using CellQuest software (Becton Dickinson, Mountain View, CA). For each assay, 30 000 cells were counted.
Fetal liver cell reconstitution and bone marrow cell transplantation
For fetal liver cell transplantation, 8- to 12-week-old male B6.SJL/BoyJ mice were lethally irradiated with 9.5 Gy using γ 137Cs Shepherd Mark I irradiator (JL Shepherd, San Fernando, CA). For each recipient mouse, 106 14.5 dpc fetal liver cells were transplanted by intravenous injection through the tail lateral vein. Reconstitution of donor-derived lymphoid and myeloid cells was monitored by staining the peripheral blood using CD3, B220, Gr-1, and Mac-1 10 and 15 weeks after transplantation. After 8 months of reconstitution, the peripheral blood was analyzed with CD45.2 and lineage markers again, and bone marrow cells of mice that underwent primary transplantation were harvested for secondary transplantation. Bone marrow cells were obtained by flushing with complete IMDM media plus 10 U/mL heparin (Sigma, St Louis, MO) through the femur and tibia using a 26-gauge needle. Bone marrow cells were then filtered through a 100-μm cell strainer (BD Biosciences, San Jose, CA). After centrifugation of cell suspension at 300g (Allegra 6R; Beckman Coulter, Hialeah, FL) for 5 minutes, cell pellets were resuspended in IMDM supplemented with 2% FBS (StemCell Technologies). Cell number was counted using a hemocytometer (Sigma) after diluting cells in 3% acetic acid. Cells (107) were injected into 9 Gy–irradiated B6.SJL/BoyJ mice, and hematopoietic reconstitution was analyzed at 7 weeks after transplantation. Recipient mice were provided with sterile water supplemented with neomycin and bacitracin (Sigma) and sterile food after transplantation.
Fetal liver cell competitive reconstitution and chimerism analysis
For competitive reconstitution, lethally irradiated B6.SJL/BoyJ mice received a transplant of 106 14.5 dpc fetal liver cells from CD45.2+ Cited2+/+, Cited2+/−, or Cited2−/− donors and the same number of fetal liver cells from CD45.1+ mice. Peripheral blood of mice that underwent transplantation was analyzed 7 weeks after transplantation for donor chimerism by FITC-conjugated antibody against CD45.2 and PE-conjugated antibody against CD45.1. For each assay, 30 000 cells were counted. Donor chimerism was determined as: [%CD45.2+/(%CD45.1+ + %CD45.2+)] × 100.
CRU assay
The limiting dilution assay for competitive repopulating cells (CRUs) with long-term, lymphomyeloid repopulation function was performed as previously described.21 Lin−c-Kit+Scal-1+ cells from Cited2−/− (n = 4) and Cited2+/+ (n = 4) fetal livers were sorted and intravenously injected, along with 105 competitor B6.SJL/BoyJ bone marrow cells, into lethally irradiated B6.SJL/BoyJ recipients. Cell doses were 20, 50, 100, and 300, and 5 recipients from each group were injected at each cell dose in a dilution series. Repopulation of the hematopoietic system in the recipients was evaluated by taking aliquots of peripheral blood at 15 weeks after transplantation, and analyzing samples for the presence of CD45.2+ myeloid and B-lymphoid cells using the following antibodies: FITC-conjugated anti-CD45.2 antibody, PE-conjugated anti-B220 antibody, PE-conjugated anti–Gr-1 antibody, and PE-conjugated anti–Mac-1 antibody. Mice that had more than 1% test sample–derived (CD45.2+) cells in both lymphoid (B220+) and myeloid (Gr-1+ and Mac-1+) subpopulations were considered to be repopulated by test donor cells. The CRU frequency in the test fetal liver Lin−c-Kit+Scal-1+ cells was calculated by applying Poisson statistics to the proportion of negative recipients at different dilutions using limiting dilution analysis software (L-Calc; StemCell Technologies).
Retrovirus transduction of fetal liver cells
GP+E86 retrovirus producer cells were established by infection of GP+E86 cells with retrovirus supernatant after transfection of Phoenix 293 cell with MSCV-Cited2-IRES-GFP plasmid or MSCV-IRES-GFP control plasmid. Cited2−/− and Cited2+/+ 13.5 dpc fetal liver cells were cocultured with Cited2-expressing GP+E86 producer cells and control cells for 2 days. After coculture, GFP+Lin−c-Kit+ cells were sorted and 500 cells were plated in triplicate in methylcellulose-based medium (M3434; StemCell Technologies) for colony-forming unit (CFU) assay. Colonies were counted 12 days after plating. The rest of the GFP+c-Kit+Lin− cells were prepared for RNA extraction and real-time polymerase chain reaction (PCR) analysis for Cited2 expression (see Table 1 for primers).
Real-time PCR primers
| Primer . | Forward . | Reverse . |
|---|---|---|
| Cited2 | 5′-cgcatcatcaccagcagcag-3′ | 5′-cgctcgtggcattcatgttg-3′ |
| PRTN3 | 5′-agattgtaggtgggcacgag-3′ | 5′-agcaccactgtcacaagctg-3′ |
| Neutrophil elastase | 5′-actctggctgccatgctact-3′ | 5′-ccggaaatttagaccgttca-3′ |
| MPO | 5′-caccctctttgttcgagagc-3′ | 5′-caacaccaagggcaggtagt-3′ |
| Cathepsin G | 5′-tctcctgctcctgttgacct-3′ | 5′-cctttctcgcatttggatgt-3′ |
| Eosinophil peroxidase | 5′-ccgacaacattgacatctgg-3′ | 5′-tgaaaactccccatttctgc-3′ |
| Wnt5a | 5′-caaataggcagccgagagac-3′ | 5′-tctagcgtccacgaactcct-3′ |
| LEF-1 | 5′-tcactgtcaggcgacacttc-3′ | 5′-tgaggcttcacgtgcattag-3′ |
| GATA1 | 5′-gatggaatccagacgaggaa-3′ | 5′-gccctgacagtaccacaggt-3′ |
| GATA2 | 5′-ccagcaaatccaagaagagc-3′ | 5′-agactggaggaagggtggat-3′ |
| GATA3 | 5′-ctggaggaggaacgctaatg-3′ | 5′-cagggatgacatgtgtctgg-3′ |
| Mcl-1 | 5′-agagcgctggagaccctg-3′ | 5′-ctatcttattagatatgccagacc-3′ |
| Notch1 | 5′-attgacgagtgtgaccctga-3′ | 5′-ggcactcattgatgttgggtc-3′ |
| Bmi-1 | 5′-gctctccagcattcgtcagtc-3′ | 5′-agcagcaatgactgtgatgca-3′ |
| BMP4 | 5′-tgatacctgagaccgggaag-3′ | 5′-ctgctcttcctcctcctcct-3′ |
| HPRT | 5′-gttggatacaggccagactttgttg-3′ | 5′-gagggtaggctggcctataggct-3′ |
| 18S | 5′-cgtctgccctatcaactttcg-3′ | 5′-ccttggatgtggtagccgtt-3′ |
| Primer . | Forward . | Reverse . |
|---|---|---|
| Cited2 | 5′-cgcatcatcaccagcagcag-3′ | 5′-cgctcgtggcattcatgttg-3′ |
| PRTN3 | 5′-agattgtaggtgggcacgag-3′ | 5′-agcaccactgtcacaagctg-3′ |
| Neutrophil elastase | 5′-actctggctgccatgctact-3′ | 5′-ccggaaatttagaccgttca-3′ |
| MPO | 5′-caccctctttgttcgagagc-3′ | 5′-caacaccaagggcaggtagt-3′ |
| Cathepsin G | 5′-tctcctgctcctgttgacct-3′ | 5′-cctttctcgcatttggatgt-3′ |
| Eosinophil peroxidase | 5′-ccgacaacattgacatctgg-3′ | 5′-tgaaaactccccatttctgc-3′ |
| Wnt5a | 5′-caaataggcagccgagagac-3′ | 5′-tctagcgtccacgaactcct-3′ |
| LEF-1 | 5′-tcactgtcaggcgacacttc-3′ | 5′-tgaggcttcacgtgcattag-3′ |
| GATA1 | 5′-gatggaatccagacgaggaa-3′ | 5′-gccctgacagtaccacaggt-3′ |
| GATA2 | 5′-ccagcaaatccaagaagagc-3′ | 5′-agactggaggaagggtggat-3′ |
| GATA3 | 5′-ctggaggaggaacgctaatg-3′ | 5′-cagggatgacatgtgtctgg-3′ |
| Mcl-1 | 5′-agagcgctggagaccctg-3′ | 5′-ctatcttattagatatgccagacc-3′ |
| Notch1 | 5′-attgacgagtgtgaccctga-3′ | 5′-ggcactcattgatgttgggtc-3′ |
| Bmi-1 | 5′-gctctccagcattcgtcagtc-3′ | 5′-agcagcaatgactgtgatgca-3′ |
| BMP4 | 5′-tgatacctgagaccgggaag-3′ | 5′-ctgctcttcctcctcctcct-3′ |
| HPRT | 5′-gttggatacaggccagactttgttg-3′ | 5′-gagggtaggctggcctataggct-3′ |
| 18S | 5′-cgtctgccctatcaactttcg-3′ | 5′-ccttggatgtggtagccgtt-3′ |
Statistical analysis
Comparison between 2 cell types was assessed by Student t test. P less than .05 was considered statistically significant.
Results
Cited2 is expressed in Lin−c-Kit+Scal-1+ and Lin−c-Kit+ fetal liver cells
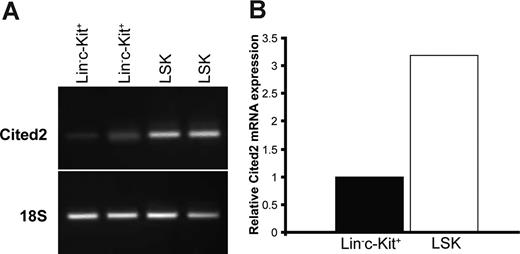
Cited2 expression has been detected in mouse bone marrow HSCs and shown to be positively related to LTR stem cell activity.17 We have detected Cited2 expression in fetal liver–derived hepatoblasts and hepatocytes (data not shown). Detailed analysis of Cited2 expression in Lin−c-Kit+Scal-1+ (LSK) and Lin−c-Kit+ fetal liver cells was further performed in sorted cells by reverse-transcription (RT)–PCR. As shown in Figure 1A, Cited2 expression was detected in LSK and Lin−c-Kit+ fetal liver cells at 14.5 dpc. Real-time PCR analysis detected higher expression of Cited2 in LSK cells than in Lin−c-Kit+ cells from fetal liver (Figure 1B), which is consistent with the finding that Cited2 is highly expressed in LTR stem cells17 and suggests that Cited2 might be associated with the HSC function in mouse fetal liver.
Cited2 is expressed in Lin−c-Kit+, Lin−c-Kit+Sca-1+ (LSK) fetal liver cells. Fetal liver cells from 14.5 dpc were sorted for Lin−c-Kit+ and LSK subpopulations. Sorted cells were processed for mRNA expression of Cited2 by RT-PCR, and the expression was further quantified by real-time PCR with 18S as the internal control for verifying the presence of cDNA after reverse transcription and for quantification purposes. (A) Cited2 expression was detected in Lin−c-Kit+ and LSK cells. (B) Cited2 expression in LSK cells was approximately 3-fold higher than that in Lin−c-Kit+ cells. Two sets of independently sorted cells were performed in parallel for Cited2 mRNA expression analysis.
Cited2 is expressed in Lin−c-Kit+, Lin−c-Kit+Sca-1+ (LSK) fetal liver cells. Fetal liver cells from 14.5 dpc were sorted for Lin−c-Kit+ and LSK subpopulations. Sorted cells were processed for mRNA expression of Cited2 by RT-PCR, and the expression was further quantified by real-time PCR with 18S as the internal control for verifying the presence of cDNA after reverse transcription and for quantification purposes. (A) Cited2 expression was detected in Lin−c-Kit+ and LSK cells. (B) Cited2 expression in LSK cells was approximately 3-fold higher than that in Lin−c-Kit+ cells. Two sets of independently sorted cells were performed in parallel for Cited2 mRNA expression analysis.
Decreased numbers of differentiated erythroid and myeloid cells in Cited2−/− fetal liver
Fetal liver cellularity at 14.5 dpc and 15.5 dpc was quantified based on the observation of smaller sized fetal liver in Cited2−/− embryos. Cited2−/− fetal liver showed a significant reduction in cellularity (P < .01) compared with Cited2+/+ littermate controls at 14.5 dpc and 15.5 dpc (Figure S1A,B, available on the Blood website; see the Supplemental Materials link at the top of the online article). Because fetal liver is the major hematopoietic organ during murine embryonic development, hypoplasia of Cited2−/− fetal liver suggests that hematopoiesis might be affected accordingly.
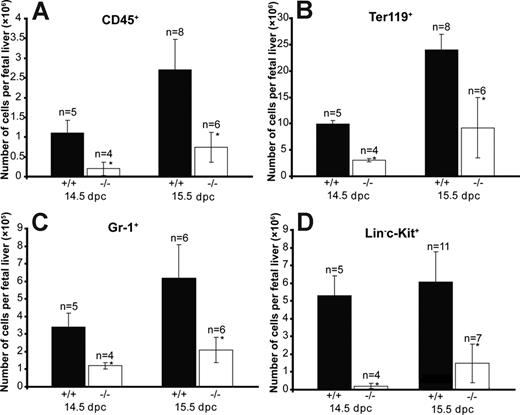
To characterize the potential hematopoietic defects due to Cited2 deficiency, we first analyzed the expression of a panel of hematopoietic lineage markers, such as Ter119 for erythrocytes, CD45 for leukocytes,22 and Gr-1 for granulocytes. There was an overall reduction of cells from multiple hematopoietic lineages. At 14.5 and 15.5 dpc, the number of Cited2−/− CD45+ cells was reduced to approximately 18% and 27% of that of Cited2+/+ control, respectively (P < .01) (Figure 2A). The number of Cited2−/− Ter119+ cells was 32% of the number of Cited2+/+ control (P < .01) at 14.5 dpc and 38.5% of the number of cells from Cited2+/+ control (P < .01) at 15.5 dpc (Figure 2B). The number of Cited2−/− Gr-1+ cells was 36% of the number of Cited2+/+ control (P < .01) at 14.5 dpc and 33.9% of the number of cells from Cited2+/+ control (P < .01) at 15.5 dpc (Figure 2C). The reduction in the numbers of CD45+ hematopoietic cells, Ter119+ erythroid cells, and Gr-1+ myeloid cells suggests impaired hematopoietic homeostasis as a result of Cited2 deficiency.
Reduction of hematopoietic cells in Cited2−/− fetal liver at 14.5 dpc and 15.5 dpc. Fetal livers from 14.5 dpc and 15.5 dpc were harvested and single cell suspensions were prepared by passing them through a 1-mL pipette and filtering through a 100-μm cell strainer. Cell number was counted using a hemocytometer. Fetal liver cells (5 × 105) were used for the FACS analysis. The absolute number of CD45+ (A), Ter119+ (B), Gr-1+ (C), and Lin−c-Kit+ (D) cells from Cited2−/− and Cited2+/+ controls at 14.5 dpc and 15.5 dpc is presented (*P < .05; average ± SEM).
Reduction of hematopoietic cells in Cited2−/− fetal liver at 14.5 dpc and 15.5 dpc. Fetal livers from 14.5 dpc and 15.5 dpc were harvested and single cell suspensions were prepared by passing them through a 1-mL pipette and filtering through a 100-μm cell strainer. Cell number was counted using a hemocytometer. Fetal liver cells (5 × 105) were used for the FACS analysis. The absolute number of CD45+ (A), Ter119+ (B), Gr-1+ (C), and Lin−c-Kit+ (D) cells from Cited2−/− and Cited2+/+ controls at 14.5 dpc and 15.5 dpc is presented (*P < .05; average ± SEM).
Significantly reduced numbers of Lin−c-Kit+ cells and LSK cells in Cited2−/− fetal liver
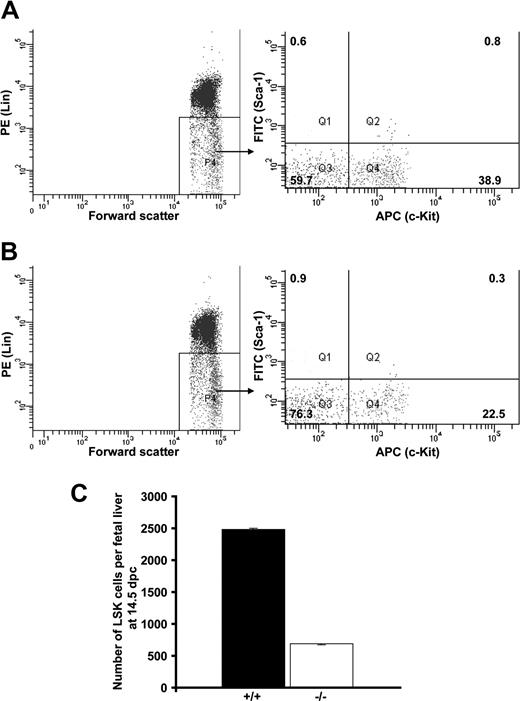
To test whether Cited2 deficiency affects hematopoiesis at the level of immature hematopoietic progenitors, which might result in quantitative reduction of the differentiated hematopoietic cells, we quantified Lin−c-Kit+ cells from Cited2-deficient fetal liver and wild-type littermate control. c-Kit is a widely used marker for the detection of immature hematopoietic progenitor cells.23 The number of Cited2−/− Lin−c-Kit+ phenotypically defined immature hematopoietic cells was significantly reduced compared with Cited2+/+ control at 14.5 dpc and 15.5 dpc (P < .01) (Figure 2D). The decreased numbers of definitive hematopoietic cells and phenotypically defined immature progenitor cells might result from an insufficient HSC pool. The number of LSK cells from Cited2−/− fetal liver was further analyzed by sorting Lin−c-Kit+Sca-1+ cells to obtain a precise enumeration of the LSK cells in individual fetal liver. The result revealed decreased frequency of LSK cells in Cited2−/− fetal liver (Figure 3B) compared with Cited2+/+ littermates at 14.5 dpc (Figure 3A). The absolute number of LSK cells obtained after sorting also was reduced in Cited2−/− fetal liver compared with Cited2+/+ littermate control (P < .01) (Figure 3C). Similar observation was made when we compared the frequency of Lin−CD45+c-Kit+Sca-1+ cells in Cited2−/− fetal liver and Cited2+/+ littermate controls at the same stage (Figure S2A,B). These results indicate that Cited2 deficiency significantly affects the number of phenotypically defined hematopoietic stem cells in fetal liver. The impaired HSC pool could be one of the major causes leading to the reduced number of hematopoietic progeny in Cited2-deficient embryos.
Reduced number of LSK cells in Cited2−/− fetal liver. Fetal liver cells from (A) Cited2+/+ littermate control (n = 5) and (B) Cited2−/− (n = 4) at 14.5 dpc were sorted as shown by the representative lineage and sorting gates (LSK cells were collected from Q2). Ten thousand counts were presented in panels A,B. The numbers within the quadrants represent the percentage of cells. The yield of LSK cells sorted from Q2 was too low to be reanalyzed. (C) The cumulative counts of LSK cells obtained by flow cytometer during the sorting procedure, which was compared between Cited2−/− and Cited2+/+ littermate control and revealed significant reduction in Cited2−/− fetal liver (P < .01; average ± SEM).
Reduced number of LSK cells in Cited2−/− fetal liver. Fetal liver cells from (A) Cited2+/+ littermate control (n = 5) and (B) Cited2−/− (n = 4) at 14.5 dpc were sorted as shown by the representative lineage and sorting gates (LSK cells were collected from Q2). Ten thousand counts were presented in panels A,B. The numbers within the quadrants represent the percentage of cells. The yield of LSK cells sorted from Q2 was too low to be reanalyzed. (C) The cumulative counts of LSK cells obtained by flow cytometer during the sorting procedure, which was compared between Cited2−/− and Cited2+/+ littermate control and revealed significant reduction in Cited2−/− fetal liver (P < .01; average ± SEM).
Cited2 is required for hematopoietic progenitor proliferation and differentiation
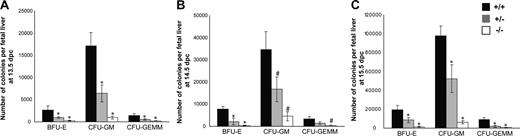
In vitro colony-forming unit (CFU) assay was performed to study the influence of Cited2 deficiency on the proliferation and differentiation of hematopoietic progenitors of various lineages. We compared the clonogenic cells from Cited2+/+, Cited2+/−, as well as Cited2−/− fetal livers from 13.5 dpc to 15.5 dpc. We observed reduced frequency of erythroid (BFU-E), granulocyte/macrophage (CFU-GM), and multipotential progenitors (CFU-GEMM) from Cited2−/− fetal liver compared with Cited2+/+ littermate control at different developmental stages (Figure S3A-C). CFU-GEMM was undetectable from some of Cited2−/− fetal liver CFU culture at 13.5 dpc and 15.5 dpc, in contrast to an average of 3 to 4 CFU-GEMMs that were readily detected from Cited2+/+ littermate control culture. The total number of BFU-Es, CFU-GMs, and CFU-GEMMs per fetal liver was calculated, which reflected a significant reduction (P < .01) in Cited2−/− embryos at 13.5 dpc (Figure 4A), 14.5 dpc (Figure 4B), and 15.5 dpc (Figure 4C). Thus, Cited2 deficiency significantly decreased the number and impaired the proliferation of erythroid, granulocyte/macrophage, as well as multipotential progenitors. Reduced number of BFU-Es and CFU-GMs per fetal liver from 13.5 dpc to 15.5 dpc was also noted in Cited2+/− embryos (P < .01-.05) (Figure 4A-C). Decreased number of CFU-GEMMs was observed in Cited2+/− fetal liver at 13.5 dpc and 15.5 dpc (P < .01) (Figure 4A,C). These results indicate that generation of hematopoietic progenitor cells is sensitive to Cited2 gene dosage.
Number and proliferation of hematopoietic progenitor cells were compromised due to Cited2 deficiency. Fetal liver cells were prepared and 2 × 104 fetal liver cells from each sample were plated in triplicate cultures of methylcellulose-based medium supplemented with 3 units/mL Epo, 10 ng/mL mouse recombinant IL-3, 10 ng/mL human recombinant IL-6, and 50 ng/mL mouse recombinant stem cell factor. The frequency of BFU-Es, CFU-GMs, and CFU-GEMMs was determined after 7 to 12 days of culture. Total number of BFU-Es, CFU-GMs, and CFU-GEMMs per fetal liver was obtained by multiplying the frequency of BFU-Es, CFU-GMs, and CFU-GEMMs per 2 × 104 cells by the total fetal liver cell number. The data were expressed as average (± SD). (A) At 13.5 dpc, the number of BFU-Es, CFU-GMs, and CFU-GEMMs was reduced in Cited2−/− fetal liver (n = 5) compared with the wild-type littermate control (n = 5). Cited2+/− fetal liver (n = 4) had fewer numbers of BFU-Es, CFU-GMs, and CFU-GEMMs as well. (B) At 14.5 dpc, decreased number of BFU-Es, CFU-GMs, and CFU-GEMMs was observed in Cited2−/− fetal liver (n = 4) compared with the wild-type littermate control (n = 3). Cited2+/− fetal liver (n = 5) showed decreased number of BFU-Es and CFU-GMs. (C) Decreased number of BFU-Es, CFU-GMs, and CFU-GEMMs was observed in Cited2−/− fetal liver (n = 4) compared with the wild-type littermate control (n = 6) at 15.5 dpc. Cited2+/− fetal liver (n = 4) showed decreased number of BFU-Es, CFU-GMs, and CFU-GEMMs. All comparisons were relative to wild-type controls (# indicates P < .05, *P < .01).
Number and proliferation of hematopoietic progenitor cells were compromised due to Cited2 deficiency. Fetal liver cells were prepared and 2 × 104 fetal liver cells from each sample were plated in triplicate cultures of methylcellulose-based medium supplemented with 3 units/mL Epo, 10 ng/mL mouse recombinant IL-3, 10 ng/mL human recombinant IL-6, and 50 ng/mL mouse recombinant stem cell factor. The frequency of BFU-Es, CFU-GMs, and CFU-GEMMs was determined after 7 to 12 days of culture. Total number of BFU-Es, CFU-GMs, and CFU-GEMMs per fetal liver was obtained by multiplying the frequency of BFU-Es, CFU-GMs, and CFU-GEMMs per 2 × 104 cells by the total fetal liver cell number. The data were expressed as average (± SD). (A) At 13.5 dpc, the number of BFU-Es, CFU-GMs, and CFU-GEMMs was reduced in Cited2−/− fetal liver (n = 5) compared with the wild-type littermate control (n = 5). Cited2+/− fetal liver (n = 4) had fewer numbers of BFU-Es, CFU-GMs, and CFU-GEMMs as well. (B) At 14.5 dpc, decreased number of BFU-Es, CFU-GMs, and CFU-GEMMs was observed in Cited2−/− fetal liver (n = 4) compared with the wild-type littermate control (n = 3). Cited2+/− fetal liver (n = 5) showed decreased number of BFU-Es and CFU-GMs. (C) Decreased number of BFU-Es, CFU-GMs, and CFU-GEMMs was observed in Cited2−/− fetal liver (n = 4) compared with the wild-type littermate control (n = 6) at 15.5 dpc. Cited2+/− fetal liver (n = 4) showed decreased number of BFU-Es, CFU-GMs, and CFU-GEMMs. All comparisons were relative to wild-type controls (# indicates P < .05, *P < .01).
To demonstrate that the hematopoietic defects are due to Cited2 deficiency in hematopoietic progenitor cells, 13.5-dpc Cited2−/− and Cited2+/+ fetal liver cells were transduced with Cited2-expressing retrovirus as well as control retrovirus and analyzed by the CFU assay. GFP+Lin−c-Kit+ cells were sorted after retrovirus transduction, and 500 cells were plated in triplicate culture for the CFU analysis. In parallel, mRNA expression of Cited2 after retrovirus transduction was analyzed by real-time PCR. An approximately 8-fold increase of Cited2 expression in Cited2−/− GFP+Lin−c-Kit+ cells transduced with Cited2-expressing virus was observed compared with Cited2−/− GFP+Lin−c-Kit+ cells transduced with control virus (Figure 5A). The CFU assay revealed that compared with vector control, the frequency of BFU-Es, CFU-GMs, and CFU-GEMMs was significantly enhanced by Cited2 expression in Cited2−/− Lin−c-Kit+ fetal liver cells (P < .01), and there was no difference compared with vector control–transduced Cited2+/+ fetal liver cells (P > .05) (Figure 5B). These results indicate that Cited2 is required for hematopoietic activity at the progenitor level during development.
Retrovirus-mediated Cited2 expression rescues the defective hematopoietic colony-forming activity in Lin− c-Kit+ Cited2−/− fetal liver cells. MSCV-Cited2-IRES-GFP plasmid– or MSCV-IRES-GFP control plasmid–mediated retrovirus transduction was performed on Cited2−/− and Cited2+/+ 13.5 dpc fetal liver cells. Briefly, after coculture with retrovirus producer cells, GFP+Lin−c-Kit+ fetal liver cells were sorted for GFP expression followed by analysis of Cited2 mRNA expression via real-time PCR. Meanwhile, 500 cells of the analyzed cell population were plated in triplicate in methylcellulose-based medium (M3434; StemCell Technologies) for colony-forming unit (CFU) assay. Colonies were counted 12 days after plating. (A) Cited2 was expressed in GFP+Lin−c-Kit+ Cited2−/− fetal liver after transduction with Cited2-expressing retrovirus. (B) Retrovirus-mediated Cited2 expression in GFP+Lin−c-Kit+ Cited2−/− fetal liver cells at 13.5 dpc significantly increased the frequency of BFU-Es, CFU-GMs, and CFU-GEMMs (n = 3) compared with the vector control (n = 3; *P < .01; average ± SD). (Cited2+/+ fetal liver cells transduced with control virus [■]; Cited2−/− fetal liver cells transduced with control virus [■]; Cited2−/− fetal liver cells transduced with Cited2-expressing virus [□].)
Retrovirus-mediated Cited2 expression rescues the defective hematopoietic colony-forming activity in Lin− c-Kit+ Cited2−/− fetal liver cells. MSCV-Cited2-IRES-GFP plasmid– or MSCV-IRES-GFP control plasmid–mediated retrovirus transduction was performed on Cited2−/− and Cited2+/+ 13.5 dpc fetal liver cells. Briefly, after coculture with retrovirus producer cells, GFP+Lin−c-Kit+ fetal liver cells were sorted for GFP expression followed by analysis of Cited2 mRNA expression via real-time PCR. Meanwhile, 500 cells of the analyzed cell population were plated in triplicate in methylcellulose-based medium (M3434; StemCell Technologies) for colony-forming unit (CFU) assay. Colonies were counted 12 days after plating. (A) Cited2 was expressed in GFP+Lin−c-Kit+ Cited2−/− fetal liver after transduction with Cited2-expressing retrovirus. (B) Retrovirus-mediated Cited2 expression in GFP+Lin−c-Kit+ Cited2−/− fetal liver cells at 13.5 dpc significantly increased the frequency of BFU-Es, CFU-GMs, and CFU-GEMMs (n = 3) compared with the vector control (n = 3; *P < .01; average ± SD). (Cited2+/+ fetal liver cells transduced with control virus [■]; Cited2−/− fetal liver cells transduced with control virus [■]; Cited2−/− fetal liver cells transduced with Cited2-expressing virus [□].)
Impaired reconstitution activity of Cited2−/− fetal liver HSCs
To test whether the function of the Cited2−/− HSCs is impaired, we examined the ability of Cited2+/+ and Cited2−/− fetal liver cells in multilineage hematopoietic reconstitution. Lethally irradiated congenic recipient mice received a transplant of 106 of 14.5 dpc Cited2+/+ and Cited2−/− fetal liver cells. Peripheral blood was harvested and long-term reconstitution of lymphoid and myeloid lineages was evaluated 8 months after transplantation. Donor chimerism was very high for both Cited2+/+ and Cited2−/− transplantations (Figure S4A); however, there was a 5- to 8-fold reduction in the absolute numbers of donor-derived T-lymphoid, B-lymphoid, and myeloid cells in mice that received a transplant of Cited2−/− versus Cited2+/+ fetal liver cells (Figure 6A). This indicates impaired reconstitution of multiple hematopoietic lineages due to Cited2 deficiency.
Cited2 deficiency results in impaired reconstitution of multiple lineages in primary and secondary transplantations. (A) Fetal liver cells (106) from each of Cited2−/− (n = 3) and Cited2+/+ (n = 3) embryos were injected via tail vein into lethally irradiated congenic recipient mice, and the peripheral blood reconstitution of T-lymphoid, B-lymphoid, and myeloid cells was analyzed 8 months after transplantation. The long-term reconstitution of T-lymphoid, B-lymphoid, and myeloid lineages was impaired as shown by significantly decreased absolute number of donor-derived cells of each lineage (P < .01) in Cited2−/− mice that underwent transplantation. Donor chimerism was determined as: [%CD45.2+CD3+/%CD3+] × 100, [%CD45.2+B220+/%B220+] × 100, [%CD45.2+Mac-1+/%Mac-1+] × 100, [%CD45.2+Gr-1+/%Gr-1+] × 100. (B) Secondary transplantation was performed 8 months after the primary transplantation. Bone marrow cells (107) were harvested from primary transplants and transplanted into recipient mice after irradiation (9 Gy). Significantly decreased numbers of donor-derived T-lymphoid, B-lymphoid, and myeloid cells were observed in Cited2−/− mice that underwent transplantation (P < .01).
Cited2 deficiency results in impaired reconstitution of multiple lineages in primary and secondary transplantations. (A) Fetal liver cells (106) from each of Cited2−/− (n = 3) and Cited2+/+ (n = 3) embryos were injected via tail vein into lethally irradiated congenic recipient mice, and the peripheral blood reconstitution of T-lymphoid, B-lymphoid, and myeloid cells was analyzed 8 months after transplantation. The long-term reconstitution of T-lymphoid, B-lymphoid, and myeloid lineages was impaired as shown by significantly decreased absolute number of donor-derived cells of each lineage (P < .01) in Cited2−/− mice that underwent transplantation. Donor chimerism was determined as: [%CD45.2+CD3+/%CD3+] × 100, [%CD45.2+B220+/%B220+] × 100, [%CD45.2+Mac-1+/%Mac-1+] × 100, [%CD45.2+Gr-1+/%Gr-1+] × 100. (B) Secondary transplantation was performed 8 months after the primary transplantation. Bone marrow cells (107) were harvested from primary transplants and transplanted into recipient mice after irradiation (9 Gy). Significantly decreased numbers of donor-derived T-lymphoid, B-lymphoid, and myeloid cells were observed in Cited2−/− mice that underwent transplantation (P < .01).
Secondary transplantation was subsequently performed. Bone marrow cells (107) harvested from primary mice that underwent primary transplantation transplanted into recipient mice after irradiation with a dose of 9 Gy, and the reconstitution of the hematopoietic system was monitored 7 weeks later. The chimerism of donor-derived cells of multiple lineage was analyzed and revealed a universally reduced reconstituting ability of HSCs due to Cited2 deficiency (P < .01) (Figure S4B), consistent with a self-renewal defect. The absolute number of donor-derived cells of each lineage was greatly reduced (P < .01) (Figure 6B). Partial instead of full reconstitution of T lymphoid cells was observed in Cited2+/+ secondary transplantations (Figure S4B), and this might be due to the irradiation dose applied in this specific experiment, which did not fully ablate recipient HSCs.
To increase the stringency of HSC activity assay, competitive reconstitution was performed with the same number of CD45.2+ and CD45.1+ fetal liver cells. The donor chimerism was analyzed in the peripheral blood. In this setting, Cited2−/− fetal liver cells showed severely impaired reconstitution ability, reflected by 0.011% donor chimerism of blood cells compared with 51.247% for Cited2+/+ fetal liver cells (P < .01) (Figure 7B). Cited2+/− fetal liver HSCs also displayed impaired reconstituting ability in comparison with the wild-type control (Figure 7B). The reconstitution assay indicates that Cited2 deficiency significantly impairs the activity of fetal liver HSCs, and fetal liver HSC activity is sensitive to Cited2 gene dosage.
Impaired HSC activity due to Cited2 deficiency. Competitive reconstitution was performed by transplanting 106 fetal liver cells from the recipient strain as competitor cells (CD45.1+) and 106 fetal liver cells from Cited2−/− (n = 3), Cited2+/− (n = 3), and Cited2+/+ littermate control (n = 3) at 14.5 dpc (CD45.2+). Percentage of CD45.2+ and CD45.1+ cells in the peripheral blood was analyzed and donor chimerism was determined as [%CD45.2+/(%CD45.1+ + %CD45.2+)] × 100. The chimerism data were expressed as average plus or minus SD (below “Donor chimerism”). (A) The gating was performed in 2 steps to exclude the CD45.1 and CD45.2 double-positive cells and artifacts in the analysis. First, viable nucleated cells (R1) according to forward and side scatter characteristics were gated to gain CD45.1 and CD45.2 positivity. Then, CD45.1 and CD45.2 double-positive cells (ranging from 0.95% to 4.5%) were gated out and the R2 was retained for further analysis, which includes UL, LL, and LR quadrants. (B) Representative histograms plotted after gating on R1 and R2. Compared with Cited2+/+ littermate controls, Cited2−/− fetal liver cells exhibited severely impaired reconstitution ability reflected by significantly decreased donor chimerism (*P < .01). Cited2+/− fetal liver cells showed significant reduced reconstitution as well (*P < .01). The percentages above the brackets represent the CD45.2+ cells.
Impaired HSC activity due to Cited2 deficiency. Competitive reconstitution was performed by transplanting 106 fetal liver cells from the recipient strain as competitor cells (CD45.1+) and 106 fetal liver cells from Cited2−/− (n = 3), Cited2+/− (n = 3), and Cited2+/+ littermate control (n = 3) at 14.5 dpc (CD45.2+). Percentage of CD45.2+ and CD45.1+ cells in the peripheral blood was analyzed and donor chimerism was determined as [%CD45.2+/(%CD45.1+ + %CD45.2+)] × 100. The chimerism data were expressed as average plus or minus SD (below “Donor chimerism”). (A) The gating was performed in 2 steps to exclude the CD45.1 and CD45.2 double-positive cells and artifacts in the analysis. First, viable nucleated cells (R1) according to forward and side scatter characteristics were gated to gain CD45.1 and CD45.2 positivity. Then, CD45.1 and CD45.2 double-positive cells (ranging from 0.95% to 4.5%) were gated out and the R2 was retained for further analysis, which includes UL, LL, and LR quadrants. (B) Representative histograms plotted after gating on R1 and R2. Compared with Cited2+/+ littermate controls, Cited2−/− fetal liver cells exhibited severely impaired reconstitution ability reflected by significantly decreased donor chimerism (*P < .01). Cited2+/− fetal liver cells showed significant reduced reconstitution as well (*P < .01). The percentages above the brackets represent the CD45.2+ cells.
Cited2 is required for HSC function in a cell-autonomous manner
Cited2 is expressed in phenotypically defined hematopoietic progenitor cells, HSCs, hepatoblasts, and hepatocytes in the fetal liver. To test whether Cited2 plays a cell-autonomous role in HSC functions, we performed the CRU assay with sorted LSK cells from Cited2−/− and Cited2+/+ fetal livers at 14.5 dpc. The same number of Cited2−/− and Cited2+/+ fetal liver LSK cells at different cell dosage along with competitor cells were transplanted, and the reconstitution for both lymphoid and myeloid was assayed 15 weeks after transplantation. As shown in Table 2, Cited2−/− fetal liver LSK cells displayed significantly impaired reconstitution ability compared with LSK cells from Cited2+/+ fetal liver (P = .002), indicating that loss of Cited2 results in an HSC intrinsic defect.
CRU frequency in Cited2+/+ and Cited2−/− fetal liver LSK cells
| Cell dose injected . | No. of mice with donor-derived reconstitution . | No. of mice analyzed . | CRUfrequency . | 95% Cl . |
|---|---|---|---|---|
| +/+ | ||||
| 20 | 0 | 2 | — | — |
| 50 | 2 | 5 | — | — |
| 100 | 3 | 4 | — | — |
| 300 | 5 | 5 | — | — |
| 1/83 | 1/39-1/175 | |||
| −/− | ||||
| 20 | 0 | 5 | — | — |
| 50 | 0 | 4 | — | — |
| 100 | 0 | 5 | — | — |
| 300 | 1 | 5 | — | — |
| 1/2147 | 1/307-1/14 988* | |||
| Cell dose injected . | No. of mice with donor-derived reconstitution . | No. of mice analyzed . | CRUfrequency . | 95% Cl . |
|---|---|---|---|---|
| +/+ | ||||
| 20 | 0 | 2 | — | — |
| 50 | 2 | 5 | — | — |
| 100 | 3 | 4 | — | — |
| 300 | 5 | 5 | — | — |
| 1/83 | 1/39-1/175 | |||
| −/− | ||||
| 20 | 0 | 5 | — | — |
| 50 | 0 | 4 | — | — |
| 100 | 0 | 5 | — | — |
| 300 | 1 | 5 | — | — |
| 1/2147 | 1/307-1/14 988* | |||
Lethally irradiated B6.SJL/BoyJ recipient mice (5 recipient mice for each cell dose) received a transplant of sorted LSK cells (CD45.2+) from Cited2+/+ (n = 5) and Cited2−/− (n = 4) fetal liver at 14.5 dpc at different cell doses as indicated. Donor-derived reconstitution was assessed by monitoring the percentage for CD45.2+ lymphoid (B220+) and myeloid (Gr-1+ and Mac-1+) cells in the peripheral blood at 15 weeks after transplantation. Mice that had more than 1% test sample–derived (CD45.2+) cells in both lymphoid and myeloid subpopulations were considered to be repopulated by test donor cells. The CRU frequency in the test fetal liver LSK cells was calculated by L-Calc software.
CI indicates confidence interval; —, not applicable.
P = .002.
Decreased expression of myeloid-specific genes in Cited2−/− fetal liver
To elucidate mechanisms underlying hematopoietic deficiency in Cited2−/− fetal liver, gene expression profiles for Cited2+/+ (n = 3) and Cited2−/− (n = 3) fetal livers at 14.5 dpc were compared by microarray analysis. Nine comparisons were performed among data collected from Cited2−/− and Cited2+/+ fetal livers. Genes with altered expression from more than 3 of 9 comparisons were further confirmed by real-time PCR (see Table 1 for primers). Decreased expression of several myeloid molecular markers, such as Neutrophil elastase (Ela2), Proteinase 3 (Prtn3), Myeloperoxidase (Mpo), Cathepsin G, and Eosinophil peroxidase, was detected in unsorted and sorted Lin−c-Kit+ Cited2−/− fetal liver cells (Table 3). Expression of ELA2 and PRTN3 is restricted to the promyelocytic stage of granulocytic differentiation, and they are coordinately expressed in a granulocyte-specific fashion.24-26 MPO represents an early-appearing and highly reliable intracellular myeloid lineage marker,27-29 which is detected in a subset of human hematopoietic bone marrow progenitor cells and in granulomonocytic cells.30 Cathepsin G is highly expressed at the promyelocytic stage of myeloid development and also functions to stimulate the proliferation of lymphocytes.31,32 Eosinophil peroxidase is a molecular marker for eosinophils and eosinophil lineage-committed progenitors.33 The down-regulated expression of these genes provides molecular evidence of impaired myelopoiesis in Cited2-deficient fetal liver, which is consistent with the findings from the in vivo studies.
Decreased expression of genes restricted to myeloid lineage in Cited2−/− fetal liver and Lin−c-Kit+ cells at 13.5 dpc
| Gene name . | Microarray . | Real-time PCR . | |
|---|---|---|---|
| Fetal liver . | Fetal liver . | Lin−c-Kit+ cells . | |
| Pre-pro proteinase3 | −2.99 | −2.78 ± 0.11 | −2.9 ± 0.35 |
| Myeloperoxidase | −2.53 | −2.14 ± 0.02 | −4.28 ± 0.16 |
| Neutrophil elastase | −3.276 | −3.275 ± 0.46 | −5.77 ± 0.47 |
| Cathepsin G | −2.738 | −2.015 ± 0.12 | −3.47 ± 0.07 |
| Eosinophil peroxidase | −12.931 | −6 ± 1.47 | −6.12 ± 0.22 |
| Wnt5a | −9.413 | −4.68 ± 1.39 | ND |
| Gene name . | Microarray . | Real-time PCR . | |
|---|---|---|---|
| Fetal liver . | Fetal liver . | Lin−c-Kit+ cells . | |
| Pre-pro proteinase3 | −2.99 | −2.78 ± 0.11 | −2.9 ± 0.35 |
| Myeloperoxidase | −2.53 | −2.14 ± 0.02 | −4.28 ± 0.16 |
| Neutrophil elastase | −3.276 | −3.275 ± 0.46 | −5.77 ± 0.47 |
| Cathepsin G | −2.738 | −2.015 ± 0.12 | −3.47 ± 0.07 |
| Eosinophil peroxidase | −12.931 | −6 ± 1.47 | −6.12 ± 0.22 |
| Wnt5a | −9.413 | −4.68 ± 1.39 | ND |
Microarray data collected from Cited2−/− fetal liver (n=3) and Cited2+/+ littermate controls (n=3) were compared, and real-time PCR was performed to verify the expression of differentially expressed genes. HPRT was used as an internal control for real-time PCR, and the expression of each gene was normalized to HPRT and compared between Cited2−/− and Cited2+/+ littermate controls. The table shows mean (± SD) fold changes in Cited2−/− (n = 3-5) fetal liver at 14.5 dpc and Lin−c-Kit+ cells at 13.5 dpc after comparison with wild-type littermate controls (n = 3-5).
ND indicates not detected.
Decreased expression of genes involved in hematopoietic stem cell self-renewal and survival in Cited2−/− hematopoietic progenitors
Bmi-1, a polycomb transcription factor essential for adult HSC self-renewal, has been reported to be a Cited2 target gene.14,15 Significantly decreased Bmi-1 expression in Cited2−/− fetal liver was revealed by Northern blot analysis (data not shown) and further quantified by real-time PCR in sorted Lin−c-Kit+ fetal liver cells (Table 4), thus supporting the involvement of Bmi-1 in Cited2−/− hematopoietic deficiency. Microarray analysis also revealed a 9-fold decrease in Wnt5a expression, and this down-regulation was confirmed by real-time PCR, which showed a 5.6-fold decrease in Cited2-deficient fetal liver (Table 3). The expression of Wnt5a was not detected in sorted Lin−c-Kit+ fetal liver cells, which is consistent with the report that Wnt family members are expressed in the microenvironment of fetal liver.34,35 Because LEF-1 is one of the important signaling molecules in Wnt signaling pathway and is involved in B lymphogenesis,35 we further analyzed the expression of LEF-1 by real-time PCR and found a 6-fold reduction in Cited2−/− fetal liver (data not shown) and 15.9-fold reduction in sorted Lin−c-Kit+ cells (Table 4). Wnt signaling is important for hematopoietic stem cell self-renewal, and this function is mediated by genes including Notch1.36 Notch signaling is capable of enhancing the in vitro generation of hematopoietic progenitor cells and is critical for lymphoid specification.37 In Lin−c-Kit+ Cited2−/− fetal liver cells, Notch1 expression was down-regulated approximately 2.9-fold (Table 4). Altered expression of GATA2 was found by screening a number of genes that have been shown to be involved in hematopoiesis. A 4.4-fold decreased expression of GATA2 in sorted Lin−c-Kit+ Cited2−/− fetal liver cells suggests that altered expression of GATA2 might contribute to hematopoietic defects in Cited2−/− fetal liver (Table 4). Mcl-1 is one of the members of the Bcl-2 family and is required for the HSC survival.38 A 4.2-fold decrease in the expression of Mcl-1 was also observed in Lin−c-Kit+ Cited2−/− fetal liver cells (Table 4). The down-regulated expression of these genes provides molecular evidence of impaired HSCs in Cited2-deficient fetal liver, which is consistent with hematopoietic reconstitution studies.
Decreased expression of genes essential for HSC function in Cited2−/− Lin−c-Kit+ cells at 13.5 dpc
| Gene name . | Real-time PCR Lin−c-Kit+ cells . |
|---|---|
| Bmi-1 | -6.08 ± 0.81 |
| GATA-1 | ND |
| GATA2 | -4.42 ± 0.36 |
| GATA3 | ND |
| LEF-1 | -15.5 ± 0.67 |
| Mcl-1 | -4.13 ± 0.1 |
| Notch1 | -2.9 ± 0.25 |
| BMP4 | NC |
| Gene name . | Real-time PCR Lin−c-Kit+ cells . |
|---|---|
| Bmi-1 | -6.08 ± 0.81 |
| GATA-1 | ND |
| GATA2 | -4.42 ± 0.36 |
| GATA3 | ND |
| LEF-1 | -15.5 ± 0.67 |
| Mcl-1 | -4.13 ± 0.1 |
| Notch1 | -2.9 ± 0.25 |
| BMP4 | NC |
Real-time PCR was performed to analyze an array of genes essential for HSC function. HPRT was used as an internal control for real-time PCR, and the expression of each gene was normalized to HPRT and compared between Cited2−/− and wild-type littermate controls. The table shows mean (± SD) fold changes in Cited2−/− (n = 3) Lin−c-Kit+ cells at 13.5 dpc after comparison with wild-type littermate controls (n = 3).
ND indicates not detected; NC, no change.
Discussion
The present study demonstrates for the first time that fetal liver hematopoiesis is disturbed due to Cited2 deficiency. A broad and quantitative developmental aberration of HSCs, hematopoietic progenitors, and differentiated hematopoietic cells is evident in Cited2-deficient fetal liver, indicating an essential role for Cited2 in hematopoietic maintenance during development.
Cited2 competes with HIF-1α in binding to the CH1 domain of CBP/p300, thus interfering with hypoxia-driven transcription.39,40 Hypoxic condition is important for many physiological processes including hematopoiesis.41 Up-regulated HIF-1 signaling is observed in Cited2-deficient embryonic hearts and has been shown to be partially responsible for the defective heart morphogenesis due to Cited2 deficiency.8,13 However, Cited2-deficient fetal liver manifested multiple lineage hematopoietic defects during development, which could not be explained by dysregulated HIF-1 signaling alone based on the known functions of HIF family members in hematopoiesis and the negative regulatory role of Cited2 in HIF-1–mediated responses revealed by other studies.8,39,40 Hypoxia also promotes the undifferentiated cell state in various stem and precursor cell types, and Notch signaling has been shown to be in part responsible for hypoxia-mediated processes in myogenic and neuronal precursor cells.42 Notch signaling is active in HSCs and is important during the earliest stage of hematopoietic development.43 Overexpression of the intracellular domain of Notch leads to enhanced self-renewal of HSCs.37,44 Conversely, HSC development and functions are impaired in vitro and in vivo due to the loss of Notch signaling and Notch deficiency results in defective establishment and maintenance of HSCs.45,46 We detected decreased expression of Notch1 in Cited2-deficient hematopoietic progenitors. An 18-fold decrease in Notch1 expression was also detected by microarray in Cited2-deficient embryonic hearts (unpublished data, July 2001). Thus, it is possible that hypoxic responses might be impaired in Cited2-deficient fetal liver due to decreased Notch1 expression, which partially contributes to the hematopoietic defects in Cited2-deficient embryos. Involvement of Cited2 in TGFβ signaling47 also cannot exclusively explain the hematopoietic defects due to Cited2 deficiency, because TGFβ signaling is thought to be a negative regulator for hematopoiesis by in vitro studies and deficiency of TGFβ signaling in vivo does not affect proliferation of HSCs and lineage determination.48
MEF cells lacking Cited2 ceased proliferation prematurely and had reduced expression of Bmi-1. Complementation with Cited2-expressing retrovirus induced Bmi-1 expression, and Bmi-1–expressing retrovirus enhanced the proliferation of Cited2-deficient MEF cells, indicating Bmi-1 may work downstream of Cited2.15 Bmi-1 is essential for adult HSC self-renewal because the number of HSCs is markedly reduced in postnatal Bmi-1−/− mice and there is no detectable self-renewal of adult HSCs, indicating a cell-autonomous defect in Bmi-1−/− mice. Competitive reconstitution with Bmi-1−/− fetal liver cells showed severe defects of Bmi-1−/− fetal liver HSCs in maintaining hematopoiesis, with impaired B-lymphoid reconstitution being the earliest event detected.14 This is consistent with the hematopoietic phenotypes we observed in Cited2−/− fetal liver. Significantly decreased expression of Bmi-1 in hematopoietic progenitor cells in Cited2-deficient fetal liver supports the involvement of Bmi-1 in hematopoietic defects due to Cited2 deficiency and indicates that impaired function of Cited2-deficient fetal liver HSCs might partially result from the decreased expression of Bmi-1.
In addition to Bmi-1, several other genes of critical importance to HSC functions showed decreased expression in Cited2-deficient fetal liver, including Wnt5a, LEF-1, GATA2, Notch1, and Mcl-1. Wnt proteins have been shown to act as potent growth factors in a variety of cell types and tissues. During embryogenesis, Wnt proteins, such as Wnt5a and Wnt10b, are expressed in murine yolk sac and fetal liver microenvironment and Wnt5a is expressed in fetal liver stromal cells. Accumulated evidence has shown that Wnt signaling pathway regulates stem cell fate in many organs, including the hematopoietic system. Wnt proteins function as hematopoietic regulatory factors that can directly stimulate the proliferation and survival of hematopoietic stem cells and the expansion of multipotential colony-forming cells and progenitor cells.49 Furthermore, Wnt proteins activate a LEF-1/TCF reporter in HSCs residing in their normal microenvironment, indicating that HSCs respond to Wnt signaling in vivo and Wnt proteins play critical roles in HSC homeostasis and HSC self-renewal.34-36 In our study, the decreased expression of Wnt5a and LEF-1 was detected in Cited2-deficient fetal liver and enriched hematopoietic progenitor cells, respectively. These results correlate with the disturbed hematopoietic homeostasis and impaired HSC function in the absence of Cited2, thus strongly indicating the potential connection between Cited2 and Wnt signaling during hematopoietic development. As discussed above, Notch signaling is involved in enhancing HSC self-renewal and regulating hematopoietic differentiation.37 Interestingly, Notch1 expression is up-regulated following the activation of Wnt signaling in HSCs,36 and Notch signaling is required for Wnt-mediated maintenance of undifferentiated HSCs.43 Thus, one possible interpretation of our findings is that down-regulated Wnt signaling is in part responsible for the decreased Notch1 expression in Cited2−/− hematopoietic progenitor cells, thus contributing to Cited2−/− hematopoietic defects synergistically.
GATA2 expression level is high in hematopoietic progenitor cells and is required for the proliferation of progenitor cells. GATA2 is also a critical regulator for the maintenance and expansion of hematopoietic stem cells and is essential for definitive hematopoiesis.50,51 GATA2 is stimulated by CBP,52 a Cited2 interacting protein that integrates hematopoietic transcription. GATA2 expression is significantly decreased in hematopoietic progenitor cells in Cited2-deficient fetal liver and thus might contribute to the impaired function of Cited2-deficient HSCs.
Apoptosis is one of the mechanisms that regulates the size of the hematopoietic stem cell pool.53 The Bcl-2 family members are critical regulators of apoptosis. Mcl-1 is one of the antiapoptotic members of the Bcl-2 family. Inducible deletion of Mcl-1 results in ablation of bone marrow leading to the early loss of bone marrow hematopoietic progenitors including HSCs. Mcl-1 is regulated by growth factors54,55 to augment the survival of early hematopoietic progenitors and HSCs.38 In our study, decreased expression of Mcl-1 is associated with decreased number of hematopoietic progenitors and reduced size of the hematopoietic stem cell pool in Cited2-deficient fetal liver, which may explain the compromised survival of Cited2−/− fetal liver HSCs.
A potential role of Cited2 as a molecule with stem cell–specific activity has been predicted by a study comparing gene expression profile of murine long-term reconstituting versus short-term reconstituting hematopoietic stem cells.17 Cited2 is ubiquitously expressed, and its expression has been detected in the hematopoietic and nonhematopoietic lineages in the fetal liver. We have shown by the limiting dilution/CRU assay that Cited2 plays a cell-autonomous role in fetal liver hematopoiesis; however, our study cannot rule out the possible extrinsic effect of Cited2 on HSC functions. Additional studies will be required to explore the possible involvement of Cited2 in the microenvironment to affect fetal liver hematopoiesis.
The ability of pluripotent hematopoietic stem cells to self-renew, proliferate, and differentiate into mature blood cells is coordinated by a complex series of transcriptional events. So far, numerous transcriptional factors have been identified to be important for hematopoiesis at different stages and lineages. However, the proper onset and development of hematopoiesis are not achieved by a single transcription factor. Unique combinations of cell type–specific and ubiquitously expressed nuclear factors account for the specificity and diversity in gene expression profiles and the resulting biologic outcomes in hematopoiesis. As a transcriptional modulator, Cited2 might be recruited by complexes of transcriptional factors and is very likely to play important roles in stem and/or progenitor cell function in an integrative manner. Because the majority of Cited2 molecules bind directly with high affinity to the first cysteine-histidine–rich (CH1) region of CBP and p300,6,39 which play different roles in hematopoietic self-renewal and differentiation,16 it is possible that some of Cited2's functions in hematopoiesis are partially mediated by CBP and p300. It is also not surprising that Cited2 deficiency has a broad impact in hematopoiesis. It is reasonable to hypothesize that CBP and p300 may exert their differential effects in HSC self-renewal and differentiation through the recruitment of certain chromatin remodeling proteins allowing the regulation of expression of stage- and lineage-specific target genes. In this regard, the observation that Cited2 controls the expression of Bmi-1, a polycomb-group chromatin remodeling gene indispensable for self-renewal or normal and leukemic stem cells, may also in part explain some of the phenotypes in Cited2−/− fetal liver.
Our study is the first demonstration uncovering a novel role of Cited2 in hematopoietic maintenance and HSC function during development. As a transcriptional modulator, Cited2 is involved in a variety of signaling pathways. The crosstalk and synergistic effects among these signaling pathways may complicate the interpretation of the hematopoietic phenotypes observed in Cited2−/− fetal liver. A detailed connection of Cited2 with possible regulatory mechanisms could be further explored by overexpression of genes identified from the current study in Cited2−/− hematopoietic stem/progenitor cells or generating conditional knockouts of Cited2 at specific hematopoietic lineages at specific developmental stages. Nevertheless, our current study reveals a novel function of Cited2 in hematopoiesis and thus provides a new insight into the molecular regulation of hematopoietic development.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants R01HL075436–02 and R01HL076919–07 (Y.-C.Y.), and R01HL073738 and R01DK059380 (K.D.B).
We thank Dr Lili Liu, Dr Youngji Park, Dr Zhengqi Wang, Eleonora Haviernikova, Tami Stefan, Alex Rodriguez, and members of Dr Yang's laboratory for technical support.
National Institutes of Health
Authorship
Contribution: Y.C. designed and performed the experiments, analyzed the data, and wrote the paper; P.H. performed the experiments and analyzed the data; K.D.B. designed the experiments, analyzed the data, and helped write the paper; Y.-C.Y. designed the experiments, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yu-Chung Yang, Department of Pharmacology and Cancer Center, Case Western Reserve University School of Medicine, 10900 Euclid Ave, W319, Cleveland, OH 44106; e-mail: yu-chung.yang@case.edu.

![Figure 5. Retrovirus-mediated Cited2 expression rescues the defective hematopoietic colony-forming activity in Lin− c-Kit+ Cited2−/− fetal liver cells. MSCV-Cited2-IRES-GFP plasmid– or MSCV-IRES-GFP control plasmid–mediated retrovirus transduction was performed on Cited2−/− and Cited2+/+ 13.5 dpc fetal liver cells. Briefly, after coculture with retrovirus producer cells, GFP+Lin−c-Kit+ fetal liver cells were sorted for GFP expression followed by analysis of Cited2 mRNA expression via real-time PCR. Meanwhile, 500 cells of the analyzed cell population were plated in triplicate in methylcellulose-based medium (M3434; StemCell Technologies) for colony-forming unit (CFU) assay. Colonies were counted 12 days after plating. (A) Cited2 was expressed in GFP+Lin−c-Kit+ Cited2−/− fetal liver after transduction with Cited2-expressing retrovirus. (B) Retrovirus-mediated Cited2 expression in GFP+Lin−c-Kit+ Cited2−/− fetal liver cells at 13.5 dpc significantly increased the frequency of BFU-Es, CFU-GMs, and CFU-GEMMs (n = 3) compared with the vector control (n = 3; *P < .01; average ± SD). (Cited2+/+ fetal liver cells transduced with control virus [■]; Cited2−/− fetal liver cells transduced with control virus [■]; Cited2−/− fetal liver cells transduced with Cited2-expressing virus [□].)](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/8/10.1182_blood-2007-01-066316/4/m_zh80220708950005.jpeg?Expires=1765902027&Signature=ALfBrfnbSc6qamrr2iUyIUm903EpbrZ0wPPKG64ZvT8q6g6IHd0-~FtTqkZ9RtqtiTN8otDzMCPE2dgFy1sWFx~OdeZQOBkuNOlgNKBvXY-Njq-xn4jRT16GG0dDVYsCtOtuQmnWGMGbCSZrT-p2SikAWAC6DhCSxZYyJrWaan5xDIsRI05oi~-9~gv8B1eJikmiUwKPt5i9uk7FDOQCxCKiL-P87pw5PfY1BUu2jSEpnNPg~GumOpj5jrJP6pE9jDS7WLJIrEKX5QnmxGVkdYqLN2v5K4-v4VthI2INQyv7KFuYFCqIXrJ~1tXuzRBRYheo2lhBvVrcZ85mqWgZ7A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Cited2 deficiency results in impaired reconstitution of multiple lineages in primary and secondary transplantations. (A) Fetal liver cells (106) from each of Cited2−/− (n = 3) and Cited2+/+ (n = 3) embryos were injected via tail vein into lethally irradiated congenic recipient mice, and the peripheral blood reconstitution of T-lymphoid, B-lymphoid, and myeloid cells was analyzed 8 months after transplantation. The long-term reconstitution of T-lymphoid, B-lymphoid, and myeloid lineages was impaired as shown by significantly decreased absolute number of donor-derived cells of each lineage (P < .01) in Cited2−/− mice that underwent transplantation. Donor chimerism was determined as: [%CD45.2+CD3+/%CD3+] × 100, [%CD45.2+B220+/%B220+] × 100, [%CD45.2+Mac-1+/%Mac-1+] × 100, [%CD45.2+Gr-1+/%Gr-1+] × 100. (B) Secondary transplantation was performed 8 months after the primary transplantation. Bone marrow cells (107) were harvested from primary transplants and transplanted into recipient mice after irradiation (9 Gy). Significantly decreased numbers of donor-derived T-lymphoid, B-lymphoid, and myeloid cells were observed in Cited2−/− mice that underwent transplantation (P < .01).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/8/10.1182_blood-2007-01-066316/4/m_zh80220708950006.jpeg?Expires=1765902027&Signature=FLlhSTIGd9HUfPFHNbKwYbK8Cd2iPCZv-3WYJT3vQeJ4-qydEd~X5LznIEDoF6fWmoBtFUZ27NhhQANuuGqKfCRPkM2kagFSWiLaZG9GzPAfTHkkC6uabW2MQUrJOfuW8-1YtJUvasg5kO6IrKysmrwp~kdJwrQ4CovwwN1eXfdUwsS2uQUL1J3J7Y9PyY3nWbKKQQKoG~qqU5ZPBwyP6ykMuTriUFlBQdubdYNuRUkXiRHJkPmBIn9A52GDkMs4pvNdwIXy7yDQgK8rX793bqFX6gKryjYnDyvwxFc7tvFRdn0SFlGVsk7oFo0TtLQfBs7kfdIuq2aXmEd~zV7JvQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. Impaired HSC activity due to Cited2 deficiency. Competitive reconstitution was performed by transplanting 106 fetal liver cells from the recipient strain as competitor cells (CD45.1+) and 106 fetal liver cells from Cited2−/− (n = 3), Cited2+/− (n = 3), and Cited2+/+ littermate control (n = 3) at 14.5 dpc (CD45.2+). Percentage of CD45.2+ and CD45.1+ cells in the peripheral blood was analyzed and donor chimerism was determined as [%CD45.2+/(%CD45.1+ + %CD45.2+)] × 100. The chimerism data were expressed as average plus or minus SD (below “Donor chimerism”). (A) The gating was performed in 2 steps to exclude the CD45.1 and CD45.2 double-positive cells and artifacts in the analysis. First, viable nucleated cells (R1) according to forward and side scatter characteristics were gated to gain CD45.1 and CD45.2 positivity. Then, CD45.1 and CD45.2 double-positive cells (ranging from 0.95% to 4.5%) were gated out and the R2 was retained for further analysis, which includes UL, LL, and LR quadrants. (B) Representative histograms plotted after gating on R1 and R2. Compared with Cited2+/+ littermate controls, Cited2−/− fetal liver cells exhibited severely impaired reconstitution ability reflected by significantly decreased donor chimerism (*P < .01). Cited2+/− fetal liver cells showed significant reduced reconstitution as well (*P < .01). The percentages above the brackets represent the CD45.2+ cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/8/10.1182_blood-2007-01-066316/4/m_zh80220708950007.jpeg?Expires=1765902027&Signature=AAPKfYTzp86wz7EiK5vEbPeShdoWPUDcfMFHaflJ1TzyyRlZk6BqwdOmWmZVUsWEbZcxSan2z9oAKpUFK4t4wgicrFX7sOroEx98iDcFTKybHnekPAlqSli7q1TO0wmgl13obh6c9nWgOxT-GCnFWx8fXHZuOONFnGEJm9EZanq~vYOiNjtKUxCGUr2DiJxH3eXJqF9D5wlcoOQzwglnS3w8RilkUBGbAcr9hoMTgs6DeS4l6lBk1Dor7b-2N4qTwvLQx0V4pAZFQnO52Sru4bmiTsltacLX-7Hp1IWY9F9C81DveOEga-JJOyb7QIwGvktvPoJ4iUjd8ShZSSbzEA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal