Myeloid leukemia arises from leukemia stem cells (LSCs), which are resistant to standard chemotherapy agents and likely to be a major cause of drug-resistant disease and relapse. To investigate the in vivo properties of LSCs, we developed a mouse model in which the biologic features of human LSCs are closely mimicked. Primitive normal hematopoietic cells were modified to express the BCR/ABL and Nup98/HoxA9 translocation products, and a distinct LSC population, with the aberrant immunophenotype of lineage−, Kit+/−, Flt3+, Sca+, CD34+, and CD150−, was identified. In vivo studies were then performed to assess the response of LSCs to therapeutic insult. Treatment of animals with the ABL kinase inhibitor imatinib mesylate induced specific modulation of blasts and progenitor cells but not stem- cell populations, thereby recapitulating events inferred to occur in human chronic myelogenous leukemia (CML) patients. In addition, challenge of leukemic mice with total body irradiation was selectively toxic to normal hematopoietic stem cells (HSCs), suggesting that LSCs are resistant to apoptosis and/or senescence in vivo. Taken together, the system provides a powerful means by which the in vivo behavior of LSCs versus HSCs can be characterized and candidate treatment regimens can be optimized for maximal specificity toward primitive leukemia cells.
Introduction
A broad range of studies indicate that chronic and acute forms of human myelogenous leukemia arise from mutations occurring at the hematopoietic stem or progenitor cell level. These reports show that leukemia stem cells (LSCs) share many properties of normal hematopoietic stem cells (HSCs), including a distinct immunophenotype, relatively quiescent cell-cycle status, and low frequency.1,,,,–6 In addition, LSCs possess the canonical stem-cell properties of self-renewal, multipotentiality, and strong proliferative capacity.7,–9 Thus, the organization of leukemic populations appears to mirror the hierarchical structure of normal hematopoietic tissues, with a central stem cell driving the overall system. Consequently, from a biologic and therapeutic perspective, characterizing the leukemia stem cell is key to understanding leukemia pathogenesis and for developing more effective therapeutic regimens.10,–12
In order to study the in vivo biology of LSCs, several recent studies have begun to characterize the genesis of malignant stem cells in different murine models.13,,–16 These studies have shown that HSCs as well as various progenitors can serve as targets for leukemic transformation. Further, while details vary depending on the specific mutations and experimental procedures employed, initial reports have also shown that phenotypically identifiable and functionally distinct LSCs can be found in murine models,15,17,–19 thereby validating the mouse as a useful system in which to study LSCs. Moreover, recent therapeutic modeling of drug effects in mouse chronic myelogenous leukemia (CML) has provided intriguing potential insights on new strategies for targeting primitive populations.20 To date, murine studies of LSCs have employed expression of a single mutation, which induces subsequent evolution of acute disease with varying kinetics. To generate a more defined genetic system, we have employed previously described manipulations involving well-known leukemia mutations. Beginning with the studies of Daley et al21 in 1990, it has been known that expression of the BCR/ABL translocation in murine marrow cells is sufficient to induce a model of chronic-phase CML.22 More recently, studies have demonstrated that simultaneous expression of an appropriate second mutation leads to an acute form of disease that resembles blast-crisis CML (bcCML).23,–25 Importantly, such “2 hit” systems appear to be genetically sufficient to induce disease and thus provide a defined background in which biologic studies may be performed. For the studies presented herein, we have pursued a model that employs retroviral vectors to coexpress the BCR/ABL and Nup98/HoxA9 translocation products.24,25 These 2 mutations have previously been documented in leukemia patients,26,27 thereby corroborating the relevance of these particular genetic aberrancies to human disease. Further, the nature of the translocations is consistent with proposed theories on the molecular genetics of acute leukemia in which the combination of a strong mitogenic signal (ie, constitutive kinase activation such as seen with BCR/ABL, Flt3, Jak2, etc) along with mutations that inhibit differentiation (eg, transcription factor–based anomalies such as Nup98/HoxA9, AML1-ETO, etc) are key components of transformation.28 Indeed, based on the biology of the system, interaction between BCR/ABL and Nup98/HoxA9 appears to fulfill the criteria for true oncogene cooperativity.
Our findings in the BCR/ABL and Nup98/HoxA9 system indicate that LSCs are relatively rare, possess a distinct cell-surface immunophenotype, and retain physiologic and homeostatic mechanisms reminiscent of normal stem cell–based hematopoietic populations. Moreover, the consequences of drug or radiation challenge can be readily ascertained and appear to recapitulate physiologic responses one might expect from human LSCs in vivo. Therefore, we propose that the model provides a platform from which the properties of normal versus malignant stem cells can be examined and that the relative effects of therapeutic regimens can be evaluated in vivo.
Materials and methods
Plasmids and virus production
The MSCV-BCR/ABL-IRES-GFP vector was kindly provided by Dr Richard Van Etten. The MSCV-Nup98/HoxA9-NEO plasmid was kindly provided by Dr Guy Sauvageau. The neo gene was removed from this vector by digestion with NcoI and ClaI, and the YFP gene was inserted by standard cloning procedure to yield the MSCV-Nup98/HoxA9-YFP vector used in the present study. Retroviral vector plasmids were transfected into phoenix-eco cells (ATCC, Manassas, VA) using lipofectamine 2000 per manufacturer's instructions (10 μg DNA/100 000 cells in a 6-well tissue culture dish). At 36 hours after transfection, viral supernatants were collected, filtered, and stored at −80°C.
Generation of animal models
Six- to 8-week-old female C57BL6/J mice were purchased from Jackson Laboratories, Bar Harbor, ME, or the National Cancer Institute (NCI). Donor animals were killed and marrow was flushed from femora and tibiae. Single-cell suspensions were depleted of lin+ cells using BD IMag (San Jose, CA) immunoaffinity system per manufacturer's instructions. Purified populations of HSCs, common myeloid progenitors (CMPs), and granulocyte macrophage progenitors (GMPs) were isolated as previously described.29 Donor cell populations were plated in 24-well dishes coated with retronectin (Takara, Shiga, Japan; day 1) and cultured overnight in IMDM containing SCF (25 ng/mL), Flt3 (25 ng/mL), IL6 (10 ng/mL), and IL3 (10 ng/mL). The following day (day 2), 50% of culture media was replaced with viral supernatant, once in the morning and again in the evening. The viral infection procedure was repeated again on day 3. On the morning of day 4, cells were harvested, resuspended in cold PBS, and injected intravenously into recipient C57BL6/J mice. For the CML model (ie, BCR/ABL alone), recipient animals were sublethally irradiated (6 Gy) to facilitate engraftment. This dose was chosen based on the work of Mardiney and Malech,30 who previously demonstrated high levels of engraftment using a nonlethal dose of approximately 6 Gy. We note that to successfully establish the CML model using 6 Gy of conditioning for recipient mice, it is important to follow the donor cell infection protocol outlined earlier in this section rather than more conventional methods that employ 5-fluorouracil treatment of donor animals and nonpurified populations. For the blast-crisis model (BCR/ABL + Nup/98), no irradiation was required. Transplantations were performed to correspond with naturally occurring frequencies of each population. For example, since CMPs and GMPs are approximately 10-fold more abundant that HSCs, we typically infected and transplanted 10-fold more of the myeloid progenitor cell types than LSKs (Lin−Sca+Kit+cells). Infected LSKs were transplanted at approximately 20 000 cells per recipient for all experiments shown. Infection efficiencies were typically 5% to 10%.
Flow-cytometry and LSC phenotyping
All flow-cytometry studies were performed using either a Becton Dickinson FACSAria or LSRII flow cytometer. To discriminate between GFP and YFP, a 525-nm LP mirror was used in front of the YFP detector (with a 530/30 band pass filter), and a 502-nm LP mirror was used in front of the GFP detector (with a 510/20 band pass filter). This configuration provides excellent resolution of the GFP versus YFP signals, as shown in Figure 1A. For phenotyping of major lineages (Table 1), animals were killed at advanced stages of disease and marrow cells were labeled with antibodies to red cell precursors (Ter119), B cells (B220), T cells (CD3), myeloid cells (Gr-1), or a cocktail of all lineage-specific markers (lin; all antibodies from Becton Dickinson). For sorting experiments, lineage-depleted (lin−) bone marrow cells were labeled with antibodies to Sca-1, CD34, Flt3, CD150, c-kit, and the Fc-gamma receptor (2.4G2; all antibodies from Becton Dickinson) and sorted to isolate the desired subpopulations using a FACSAria flow cytometer and standard procedures. Chronic-phase LSCs were defined as cells capable of generating disease in animals within 6 weeks of transplantation. Chronic disease was defined using the following criteria: white blood cell count in excess of 50 × 109/L (50 000/μL), spleen weight in excess of 4-fold normal controls. In all cases of BCR/ABL-mediated disease, this profile represented animals destined to succumb to lethal myeloproliferative disease within 2 to 3 days. Blast-crisis LSCs were defined as cells capable of inducing acute leukemia within 21 days of transplantation into a naive recipient animal. Acute disease was defined using the following criteria: white blood cell count in excess of 30 × 109/L (30 000/μL), spleen weight in excess of 4-fold normal controls, and marrow blasts in excess of 25%. In all cases studied, animals displaying these features succumbed to disease within 24 to 36 hours. Occasionally, experimental animals died at considerably later stages after transplantation, however, those deaths were from atypical forms of disease or secondary infection and not interpreted as a direct result of primary LSC outgrowth. For studies employing a limiting dilution approach, calculations were performed using the L-Calc software (version 1.1, Stemsoft, Vancouver, BC). Cell doses were tested over at least a 3-log range (cohorts of 4 mice per cell dose). Typical doses were 50, 500, and 5000 cells per animal. For cell-cycle studies, antibody-labeled populations were fixed in 1% ultrapure formaldehyde (Polysciences, Warrington, PA) for 20 minutes and permeabilized with 0.1% Triton X-100 for 20 minutes. Fixed cells were then labeled with DAPI (5 μg/mL) and analyzed for DNA cycle profiles using FlowJo 8.2 software (Tree Star, Ashland, OR). For BrdU incorporation experiments, animals were administered 2.0 mg BrdU via intraperitoneal injection at 90 minutes prior to being killed. Bone marrow cells were collected, subjected to cell-surface labeling, then fixed in 1.0% ultrapure formaldehyde overnight at 4°C. Cells were then permeabilized with 0.1% Triton X-100 for 20 minutes and processed by DNase digestion and labeling using the Becton Dickinson BrdU Flow kit (catalog no. 552598) per manufacturer's instructions.
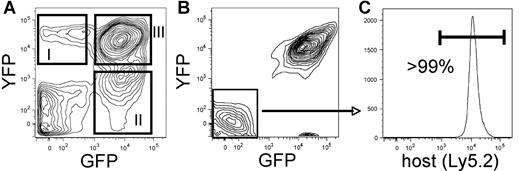
Characterization of leukemia models. (A) Example of bone marrow derived from a primary recipient of coinfected cells. The BCR/ABL-transduced cells are detected by GFP expression and the Nup98/HoxA9-infected cells are detected by YFP expression. Panels I and II indicate singly infected populations, whereas panel III shows cells that are successfully transduced with both the BCR/ABL and Nup98/HoxA9 vectors. (B) Example of bone marrow derived from a secondary recipient of coinfected cells. The doubly transduced cells typically outgrow the singly infected populations shown in panel A. (C) GFP−/YFP− cells from the secondary recipient in panel B were analyzed with respect to Ly5.1 versus Ly5.2 expression as a means to verify host origin and to establish that potential donor leukemic cells have not lost GFP/YFP expression (host cells in this experiment were of Ly5.2 origin). Fidelity of the system is demonstrated by greater than 99% purity of the host marker.
Characterization of leukemia models. (A) Example of bone marrow derived from a primary recipient of coinfected cells. The BCR/ABL-transduced cells are detected by GFP expression and the Nup98/HoxA9-infected cells are detected by YFP expression. Panels I and II indicate singly infected populations, whereas panel III shows cells that are successfully transduced with both the BCR/ABL and Nup98/HoxA9 vectors. (B) Example of bone marrow derived from a secondary recipient of coinfected cells. The doubly transduced cells typically outgrow the singly infected populations shown in panel A. (C) GFP−/YFP− cells from the secondary recipient in panel B were analyzed with respect to Ly5.1 versus Ly5.2 expression as a means to verify host origin and to establish that potential donor leukemic cells have not lost GFP/YFP expression (host cells in this experiment were of Ly5.2 origin). Fidelity of the system is demonstrated by greater than 99% purity of the host marker.
Lineage analysis of normal versus leukemic populations
| Population . | Naive ctrl, % . | GFP ctrl, % . | BCR, % . | BCR + Nup, % . | |||
|---|---|---|---|---|---|---|---|
| GFP+ . | GFP− . | GFP+ . | GFP− . | GFP+/YFP+ . | GFP−/YFP− . | ||
| B cells | 30 | 47 | 21 | 37 | 16 | 15 | 13 |
| T cells | 14 | 8 | 21 | 10 | 14 | 6 | 18 |
| RBCs | 9 | 7 | 20 | 6 | 12 | 1 | 15 |
| Myeloid | 36 | 28 | 31 | 34 | 46 | 46 | 43 |
| Lin− | 11 | 10 | 6 | 12 | 12 | 32 | 11 |
| Population . | Naive ctrl, % . | GFP ctrl, % . | BCR, % . | BCR + Nup, % . | |||
|---|---|---|---|---|---|---|---|
| GFP+ . | GFP− . | GFP+ . | GFP− . | GFP+/YFP+ . | GFP−/YFP− . | ||
| B cells | 30 | 47 | 21 | 37 | 16 | 15 | 13 |
| T cells | 14 | 8 | 21 | 10 | 14 | 6 | 18 |
| RBCs | 9 | 7 | 20 | 6 | 12 | 1 | 15 |
| Myeloid | 36 | 28 | 31 | 34 | 46 | 46 | 43 |
| Lin− | 11 | 10 | 6 | 12 | 12 | 32 | 11 |
GFP ctrl indicates animals that received cells infected with the GFP control retrovirus; GFP+, percentage of indicated lineage within GFP+ (ie, infected) bone marrow cells; GFP−, percentage of indicated lineage within GFP− (ie, noninfected) bone marrow cells; BCR, animals that received cells infected with the BCR/ABL retrovirus; BCR + Nup, animals that received cells infected with the BCR/ABL and Nup98/HoxA9 retroviruses; and RBCs, red blood cells.
Southern blots
Genomic DNA from bone marrow was digested with EcoRI, separated by electrophoresis on 0.8% agarose Tris-acetate-EDTA gels, and transferred onto GeneScreen Plus membranes (Perkin Elmer, Shelton, CT). Blots were sequentially hybridized with [32P]-labeled probes for EGFP, using the entire EGFP coding sequence from a pEGFP-C2 plasmid (Clontech Laboratories, Palo Alto, CA). The blots were washed at high stringency, and hybridization was detected after exposure onto Molecular Dynamics Phosphor Screens using a Storm 860 Imaging System scanner and ImageQuant (version 5.0) software (Molecular Dynamics, Piscataway, NJ).
In vitro colony-forming assays
The indicated cell populations were plated in methylcellulose medium (Stem Cell Technologies, Vancouver, BC, Canada; M3134) supplemented with 15% FBS, 20% BIT (BSA, insulin, transferrin; Stem Cell Technologies, no. 09500), SCF (50 ng/mL), IL3 (10 ng/mL), Flt-3 (50 ng/mL), and IL6 (10 ng/mL). Cultures were incubated at 37°C for 10 to 14 days before scoring colonies (100 or more cells/colony).
In vivo studies
For drug challenge, cohorts of leukemic mice were established as described in “Generation of animal models.” At advanced stages of disease, 100 mg/kg of ara-C was administered by intraperitoneal injection. Animals were killed 4 or 20 hours later and subjected to immunophenotyping and cell-cycle analysis. Alternatively, imatinib mesylate (100 mg/kg in water) was administered by oral gavage twice daily for 3 consecutive days before mice were killed and analyzed. For studies requiring pretransplantation conditioning, animals were exposed to 5.5 Gy from a Rad Source 2000 X-ray irradiator (Alpharetta, GA). At 7 days after treatment, animals were killed and analyzed as described.
Results
Model systems for chronic and blast-crisis CML
Initial studies were performed to validate systems using both BCR/ABL alone (CML model) and in combination with Nup98/HoxA9 (bcCML model) to induce disease. For gene transfer, retroviral vectors encoding each translocation product and a distinct fluorescent protein marker (GFP and YFP) were employed to allow delineation of cells transduced with one or both vectors. Enriched marrow stem-cell populations from donor C57BL/6-Ly5.1 mice were infected with one or both vectors and transplanted into C57BL/6-Ly5.2 congenic recipients. Figure 1A shows a typical example of the resulting disease where expression of the GFP and YFP markers can be used to identify singly versus doubly transduced populations. As noted by comparing boxes I-III (Figure 1A), in primary recipients we detect cells transduced with either vector alone or in combination. The relative proportions of cells expressing each vector vary from experiment to experiment but are typically in the range of 5% to 20% for each of the singly transduced populations and 20% to 40% for the double-positive cells. These numbers are also a function of the relative stage of disease progression, with increased double-positive events at later stages. Notably, upon transplantation of cells to secondary recipients (Figure 1B), the more aggressive acute disease typically predominates. Data shown in Figure 1C verify that expression of GFP and YFP accurately reflects the donor-host origin of cells in the model. Table 1 provides a representative example experiment comparing naive, GFP control, CML, and bcCML models with respect to the frequencies of various lineages and populations. As expected, with disease progression an increase in both primitive lin− and myeloid cell types was observed, with a concomitant reduction in lymphoid cell types. Table 1 also shows comparison of transduced (GFP+) versus normal (GFP−) cells coresident within in each model.
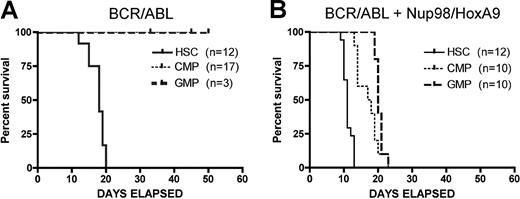
Using par system, we first examined the developmental origin of LSCs by transducing various primitive populations with BCR/ABL. For these studies, we used cells that were phenotypically negative for a standard panel of lineage markers (Lin−) and positive for expression of Sca-1 and c-kit (termed LSK cells) as our candidate stem-cell population. For convenience, we refer to these cells as “HSCs,” however, it should be noted that LSK cells are relatively heterogeneous, containing long-term and short-term repopulating HSCs as well as multipotent progenitors (MPPs). Nonetheless, they do represent a commonly used primitive population that is distinct from later-stage myeloid progenitors. As shown in Figure 2A, evolution of the CML-like disease typically requires 16 to 18 days to cause death and is manifest only when LSK cells are used as the originating population. In good agreement with the previous studies of Huntly et al,14 transduction of myeloid progenitors CMPs or GMPs with BCR/ABL is not sufficient to induce disease. In contrast, the bcCML disease develops more quickly, with advanced disease evident in as little as 10 days. Moreover, simultaneous expression of both translocations permits myeloid progenitors (CMPs and GMPs) to serve as targets for transformation, albeit with somewhat slower kinetics (Figure 2B). Notably, while transplantation of the CML disease requires sublethal irradiation of host animals (6 Gy), bcCML was successfully transferred without any preconditioning regimen. Thus, for the experiments in this study, no irradiation of host animals was required for the bcCML model. This finding indicates that LSCs arising from the combined translocations are both quantitatively and qualitatively more aggressive than stem cells transduced with BCR/ABL alone.
Kaplan-Meier analysis of survival for CML and bcCML models. (A) Donor populations of purified HSCs, CMPs, or GMPs were infected with the BCR/ABL retrovirus and transplanted into primary mice. (B) Donor populations of purified HSCs, CMPs, or GMPs were infected with the BCR/ABL + Nup98/HoxA9 retroviruses and transplanted into primary mice. Numbers of recipients indicated in parentheses for each donor cell type. Days of survival after transplantation are indicated for each cell type on the x-axis.
Kaplan-Meier analysis of survival for CML and bcCML models. (A) Donor populations of purified HSCs, CMPs, or GMPs were infected with the BCR/ABL retrovirus and transplanted into primary mice. (B) Donor populations of purified HSCs, CMPs, or GMPs were infected with the BCR/ABL + Nup98/HoxA9 retroviruses and transplanted into primary mice. Numbers of recipients indicated in parentheses for each donor cell type. Days of survival after transplantation are indicated for each cell type on the x-axis.
Previous studies have shown that expression of BCR/ABL alone is sufficient to induce CML-like myeloproliferative disease and that no additional mutations are required. This finding is based on the relatively rapid kinetics of disease formation and the polyclonal nature of malignant cells arising in vivo.31 Similarly, the bcCML form of disease also appears to arise directly from expression of BCR/ABL + Nup98/HoxA9, without requiring additional mutations. The pathogenesis is extremely rapid and leukemic animals display a highly polyclonal disease profile (Figure 3 left panel). Notably, animals within the same cohort display distinct retroviral integration patterns, indicating the presence of multiple independent clones sufficient to mediate disease. In addition, secondary recipients of bcCML marrow display largely stable polyclonal integration patterns (Figure 3 right panel), further suggesting that the 2 mutations are sufficient and that no further genetic changes need occur. Thus, combination of BCR/ABL + Nup98/HoxA9 provides a genetically defined background in which to explore the features of leukemia stem cells. We note that during pathogenesis of human disease, acquisition of the 2 mutations would typically occur in a sequential rather than simultaneous fashion. Therefore, primary animals were established expressing BCR/ABL-GFP alone. Lin− marrow derived from these animals was then harvested and used for transduction with the Nup98/HoxA9 virus. Upon transplantation into secondary recipients, the sequentially infected cells generated a bcCML disease that was indistinguishable from the model generated by simultaneous infection (data not shown).
Southern-blot analysis of bcCML. Bone marrow DNA from 5 primary recipient mice (left panel) was digested with EcoRI and hybridized with a GFP probe, which detects bands of unique size for each independent viral integrant. For primary animals nos. 2, 3, and 4, 3 secondary recipients (a, b, and c) were established. The right panel shows EcoRI digestion and GFP probe analysis of the secondary recipients in comparison to each primary donor.
Southern-blot analysis of bcCML. Bone marrow DNA from 5 primary recipient mice (left panel) was digested with EcoRI and hybridized with a GFP probe, which detects bands of unique size for each independent viral integrant. For primary animals nos. 2, 3, and 4, 3 secondary recipients (a, b, and c) were established. The right panel shows EcoRI digestion and GFP probe analysis of the secondary recipients in comparison to each primary donor.
Leukemia stem-cell phenotype
To identify the leukemia-initiating cell for the chronic and blast-crisis models, we infected purified LSK cells with one or both vectors, transplanted these cells into naive recipients, and isolated leukemic cells from primary mice in advanced stages of disease. Bone marrow from such animals was labeled with a range of antibodies (myeloid, T cell, B cell, and erythroid; see “Flow cytometry and LSC phenotyping”), sorted to purify specific subpopulations, and transplanted into secondary recipient mice to assess leukemic potential. For the chronic-phase model, we first separated lin+ versus lin− cells and determined that all detectable leukemia-initiating activity was in the lin− cells (2 independent experiments; n = 15 mice). Subsequent fractionation of the lin− population into lin−/kit+ versus lin−/kit− showed all leukemic potential in the Lin−/kit+ cells (3 independent experiments; n = 18 mice). Finally, comparison of Lin−/Kit+/Sca+ versus Lin−/kit+/Sca− indicated that 100% of transplantable LSC activity was in the Lin−/Kit+/Sca+ cells (3 independent experiments; n = 18 mice). Thus, the CML LSC displays a phenotype similar to normal HSCs (Figure 4), and only cells bearing the Lin−/Sca+/kit+ phenotype were able to generate disease in secondary recipients. This finding is in good agreement with the studies of Hu et al,19 who previously reported that CML LSCs are identified by the LSK phenotype. Parallel studies of in vitro colony-forming ability were also performed and indicated that colony-forming unit (CFU) potential is highly enriched in the Lin−/Sca+/kit+ population, as is observed for normal cells (Table 2). Therefore, the developmental and hierarchical structure of cells arising in chronic-phase disease appears to be very similar to normal populations, both with regard to in vitro CFU and transplantation potential.
Phenotypic analysis of normal versus leukemic stem-cell populations. Examples of stem-cell phenotyping are shown for GFP control (top left), CML (top right), and bcCML (bottom panels) populations. In each case, cells were first gated on the Lin− population. GFP and CML populations show typical labeling for stem-cell markers c-kit and sca-1. Labeling of bcCML shows reduced c-kit expression (bottom left). The CD150− and Flt3+ subpopulation of bcCML cells (bottom right) is highly enriched for LSCs (1 in 7 cells by limiting dilution). Percentages shown in each panel represent the overall frequency in the marrow population.
Phenotypic analysis of normal versus leukemic stem-cell populations. Examples of stem-cell phenotyping are shown for GFP control (top left), CML (top right), and bcCML (bottom panels) populations. In each case, cells were first gated on the Lin− population. GFP and CML populations show typical labeling for stem-cell markers c-kit and sca-1. Labeling of bcCML shows reduced c-kit expression (bottom left). The CD150− and Flt3+ subpopulation of bcCML cells (bottom right) is highly enriched for LSCs (1 in 7 cells by limiting dilution). Percentages shown in each panel represent the overall frequency in the marrow population.
CFU frequency for sorted subpopulations
| Cells and fraction . | CFU/5000 cells . |
|---|---|
| Naive BM | |
| Total MNCs | 10 ± 2 |
| Lin− | 101 ± 30 |
| Lin−/Kit+/Sca− | 256 ± 60 |
| Lin−/Kit+/Sca+ | 1800 ± 372 |
| CML BM, GFP+ | |
| Total MNCs | 9 ± 3 |
| Lin− | 69 ± 19 |
| Lin−/Kit+/Sca− | 104 ± 37 |
| Lin−/Kit+/Sca+ | 1743 ± 314 |
| bcCML BM, GFP+/YFP+ | |
| Lin+ | 20 ± 5 |
| Lin−/Sca−/CD150+ | 1575 ± 525 |
| Lin−/Sca−/CD150− | 3300 ± 1100 |
| Lin−/Flt3−/Fc−low/CD150+ | 1750 ± 585 |
| Lin−/Flt3−/Fc−low/CD150− | 2210 ± 735 |
| Lin−/Sca+/Flt+/CD150− | 2395 ± 800 |
| Cells and fraction . | CFU/5000 cells . |
|---|---|
| Naive BM | |
| Total MNCs | 10 ± 2 |
| Lin− | 101 ± 30 |
| Lin−/Kit+/Sca− | 256 ± 60 |
| Lin−/Kit+/Sca+ | 1800 ± 372 |
| CML BM, GFP+ | |
| Total MNCs | 9 ± 3 |
| Lin− | 69 ± 19 |
| Lin−/Kit+/Sca− | 104 ± 37 |
| Lin−/Kit+/Sca+ | 1743 ± 314 |
| bcCML BM, GFP+/YFP+ | |
| Lin+ | 20 ± 5 |
| Lin−/Sca−/CD150+ | 1575 ± 525 |
| Lin−/Sca−/CD150− | 3300 ± 1100 |
| Lin−/Flt3−/Fc−low/CD150+ | 1750 ± 585 |
| Lin−/Flt3−/Fc−low/CD150− | 2210 ± 735 |
| Lin−/Sca+/Flt+/CD150− | 2395 ± 800 |
CFU/5000 indicates no. of colony-forming units (± standard deviation) normalized to 5000 cells; naive BM, bone marrow from naive control animals; MNCs, mononuclear cells; CML BM, bone marrow from animals bearing BCR/ABL-transduced cells; and bcCML BM, bone marrow from animals bearing BCR/ABL + Nup98/HoxA9–transduced cells.
LSCs for the bcCML model were defined using a series of sorting and limiting dilution experiments that included analysis of multiple markers associated with primitive stem and progenitor populations. We found that Lin+ cells contained only 1 in 1959 cells capable of initiating disease, a level consistent with the purity of transplanted populations. Thus, as observed for the chronic-phase model, essentially all LSCs reside in the Lin− compartment. Further fractionation of Lin− cells was undertaken using antibodies for several stem cell–associated markers: Sca-1, c-kit, Flt3, CD150, CD34, and Fcγ receptor (III/II). Expression of Sca-1 was strongly associated with LSC activity, with approximately 87% of detectable potential in the Sca-1+/Lin− fraction in comparison to Sca-1−/Lin− cells. Similarly, Flt3 also correlated with LSC potential, with 82% of LSCs in the Flt+/Lin− populations in comparison to Flt−/Lin− cells. In contrast, the HSC antigen CD15032 was not strongly expressed on primitive leukemia cells, with 96% of LSC potential found in the Sca+/CD150−/Lin− population. By combining these markers, we observed that Sca+/Flt+/CD150−/Lin− cells contained 1 in 7 LSCs by limiting dilution (2 independent experiments; n = 12 mice per experiment) and identified the majority of leukemia-initiating potential (Figure 4). Within this defined population, virtually all detectable cells were CD34+, thus the marker was not used for sorting strategies because it provided no further enrichment of LSCs. Interesting, c-kit was not a particularly useful marker for purification. Expression of c-kit is suppressed in bcCML leukemic cells, with the majority of the Lin− population possessing an intermediate level of antigen (Figure 4). Collectively, these antigenic features most closely resemble the phenotype of short-term HSCs (ST-HSCs) or possibly CMPs but are nonetheless aberrant and thereby distinguish the blast-crisis LSCs from normal HSCs.
In vitro CFU assays in the bcCML model indicated less-rigid developmental boundaries for primitive cells (Table 2). For example, while all the markers found on LSCs were associated with substantial CFU potential, there was also significant enrichment of colony-forming ability observed for the Lin−/Sca− and Lin−/Flt− populations. These data indicate that a feature of blast-crisis disease may be significantly elevated levels of self-renewal/growth within various progenitor populations, a finding consistent with similar studies from human CML.33
Cell-cycle analyses
Cell-cycle regulation is an important aspect of stem-cell biology and is likely related to tissue homeostasis and relative sensitivity to chemotherapy drugs. We thus performed a detailed analysis of stem and progenitor populations in the CML models. A notable feature of the experimental system is that coresident populations of normal versus leukemic cell types can be analyzed within the same animal. In addition, appropriate nonleukemic control transplants were analyzed. Table 3 shows the results of a representative flow cytometry study in which cell-cycle rates and the frequency of various phenotypically defined subpopulations were determined. Several features of the data are worth noting. First, within the stem- cell compartment of the chronic-phase model, cycle rates are nearly double for GFP+ versus GFP− cells. Thus, expression of BCR/ABL is strongly mitogenic and dramatically increases the growth rate of the stem-cell compartment. However, despite the elevated cycle rate, the frequency of cells in the primitive compartment is very similar to normal stem cells. This finding indicates that self-renewal frequency is not altered and that normal homeostatic regulation of stem-cell pool size is intact. Notably, elevated cycle rate but normal compartment size is also observed for progenitor cells in the chronic-phase model. In contrast, coexpression of BCR/ABL + Nup98/HoxA9 creates a malignant stem-cell population in which cell-cycle rates are no different than normal stem cells. However, the relative frequency of bcCML LSCs is elevated over 10-fold in comparison to GFP controls (0.3% vs 0.02%), suggesting an increase in self-renewal but relatively normal growth rates for stem cells in the blast-crisis model. Taken together, these findings indicate that the pathogenic mechanisms by which chronic versus acute forms of CML overwhelm normal hematopoiesis may be fundamentally different. The data also suggest that the consequences of drug treatment may vary substantially as a function of disease state and LSC cycle status.
Stem and progenitor cell analyses of frequency and cell-cycle status
| BM source . | Subpopulation . | Stem cells . | Progenitors . | |||||
|---|---|---|---|---|---|---|---|---|
| Phenotype . | Frequency, % . | Cycle, % . | Phenotype . | Frequency, % . | Cycle, % . | CFU/25K . | ||
| Naive control | NA | Sca+/Kit+/Lin− | 0.08 | 10 | Kit+/Lin− | 1.70 | 27 | 30 |
| GFP control | GFP+ | Sca+/Kit+/Lin− | 0.02 | 14 | Kit+/Lin− | 1.50 | 35 | nd |
| GFP control | GFP− | Sca+/Kit+/Lin− | 0.03 | 14 | Kit+/Lin− | 1.90 | 34 | nd |
| BCR only | GFP+ | Sca+/Kit+/Lin− | 0.02 | 32 | Kit+/Lin− | 1.90 | 43 | 48 |
| BCR only | GFP− | Sca+/Kit+/Lin− | 0.04 | 17 | Kit+/Lin− | 1.60 | 25 | nd |
| BCR + Nup | GFP+ | Sca+/Flt+/Lin−* | 0.30 | 10 | Kit+/Lin− | 8.40 | 28 | 442 |
| BCR + Nup | GFP− | Sca+/Kit+/Lin− | 0.07 | 13 | Kit+/Lin− | 1.90 | 21 | nd |
| BM source . | Subpopulation . | Stem cells . | Progenitors . | |||||
|---|---|---|---|---|---|---|---|---|
| Phenotype . | Frequency, % . | Cycle, % . | Phenotype . | Frequency, % . | Cycle, % . | CFU/25K . | ||
| Naive control | NA | Sca+/Kit+/Lin− | 0.08 | 10 | Kit+/Lin− | 1.70 | 27 | 30 |
| GFP control | GFP+ | Sca+/Kit+/Lin− | 0.02 | 14 | Kit+/Lin− | 1.50 | 35 | nd |
| GFP control | GFP− | Sca+/Kit+/Lin− | 0.03 | 14 | Kit+/Lin− | 1.90 | 34 | nd |
| BCR only | GFP+ | Sca+/Kit+/Lin− | 0.02 | 32 | Kit+/Lin− | 1.90 | 43 | 48 |
| BCR only | GFP− | Sca+/Kit+/Lin− | 0.04 | 17 | Kit+/Lin− | 1.60 | 25 | nd |
| BCR + Nup | GFP+ | Sca+/Flt+/Lin−* | 0.30 | 10 | Kit+/Lin− | 8.40 | 28 | 442 |
| BCR + Nup | GFP− | Sca+/Kit+/Lin− | 0.07 | 13 | Kit+/Lin− | 1.90 | 21 | nd |
Frequency indicates percentage of stem or progenitors cells as defined by the indicated phenotype; cycle %, percentage of cells in S + G2 phase of cell cycle; and CFU/25K, no. of colony-forming units per 25 000 cells plated in methylcellulose assay.
LSC phenotype (Sca+, Flt3+, kit−lo, CD34+, CD150−).
nd indicates not done.
In vivo effects of leukemia drugs
To assess the in vivo biology of primitive cells with regard to drug insult, we first examined the agent Ara-C (cytosine arabinoside) as a model of conventional cytotoxic chemotherapy. For these studies, cohorts of nonirradiated mice received a transplant of BCR/ABL-GFP + Nup98/HoxA9-YFP–infected cells from primary donors. At 10 to 11 days after transplantation, tumor burden was rapidly increasing (high white blood cell count [WBC], splenomegaly, and approximately 25% to 50% leukemic cells in marrow), thus approximating advanced stages of human disease. Leukemic animals were then challenged with a single bolus of Ara-C (100 mg/kg) and killed 4 or 20 hours later for cell-cycle analysis (n = 3 animals per time point, 2 independent experiments). These 2 time points were chosen based on previous studies of normal HSCs in which nadir and rebound of the stem cell compartment occurred at 4 and 20 hours after treatment, respectively.34 As shown in Table 4, Ara-C has a significant and rapid effect on the stem cell compartment, with approximately 50% reduction in the cycling pool (S + G2 populations) within 4 hours (P = .013 and P = .03 for HSCs and LSCs, respectively). Notably, there is no significant difference between normal and leukemia stem cells, indicating that the effects of Ara-C toward primitive cells are nonspecific. By 20 hours, both populations have returned to near steady-state cycle levels, indicating an equivalent ability of normal versus leukemic stem cells to respond to insult and recover following drug challenge. Similar effects were noted for the progenitor population. To further corroborate the cell-cycle data obtained in Figure 4 using conventional DAPI staining, we also performed studies using incorporation of BrdU as a means of determining the proportion of cells undergoing DNA synthesis. As shown in Figure S1 (available on the Blood website; see the Supplemental Figure link at the top of the online article), the BrdU uptake data are in good agreement with the DAPI analysis in Table 4.
Cell-cycle analyses of stem and progenitor cells after drug challenge
| . | Stem cells, % . | Progenitors, % . | ||
|---|---|---|---|---|
| Normal . | bcCML . | Normal . | bcCML . | |
| Untreated | 19 | 18 | 27 | 29 |
| Ara-C 4 h | 12* | 11* | 12* | 17* |
| Ara-C 20 h | 18 | 23 | 21 | 29 |
| Imatinib | 13 | 14 | 35* | 36* |
| . | Stem cells, % . | Progenitors, % . | ||
|---|---|---|---|---|
| Normal . | bcCML . | Normal . | bcCML . | |
| Untreated | 19 | 18 | 27 | 29 |
| Ara-C 4 h | 12* | 11* | 12* | 17* |
| Ara-C 20 h | 18 | 23 | 21 | 29 |
| Imatinib | 13 | 14 | 35* | 36* |
Percentage of cells in S + G2 phases of cell cycle for indicated populations. Analyses performed in triplicate for each condition, all results within 10% standard deviation.
Percentages with statistically significant increase or decrease relative to untreated controls (P < .05).
In a parallel series of experiments, we examined the widely used CML drug imatinib mesylate as a model of nontoxic targeted therapy. For these studies, animals were again allowed to progress to advanced stages of disease and then treated for 3 consecutive days before they were killed and analyzed (Table 4; n = 3 animals per time point, 2 independent experiments). Interestingly, treatment with imatinib has opposite effects on the stem versus progenitor populations. The more primitive cells exhibit a trend toward decreased cell-cycle activity (P = .07), suggesting that drug treatment is somewhat inhibitory for stem-cell growth, consistent with in vitro studies of human cells.35 In contrast, the progenitor pool shows an increase in cycle rate (P = .03 and P = .05 for normal and leukemic, respectively), possibly indicating a homeostatic response to systemic pressure exerted by the effects of imatinib. As noted for Ara-C treatment, the relative effects on cell-cycle rate are very similar between normal and leukemic populations. However, one notable leukemia-specific consequence of imatinib treatment was detected. Examination of the differentiation state of treated animals showed that the leukemic cells reduced the overall proportion of lin− cells by approximately 50%, with a concomitant increase in Lin+ cells. Presumably, this shift represents increased differentiation of the leukemic population, an effect consistent with the known clinical effects of imatinib. Thus, increased cycling of progenitor cells may be a homeostatic response of the leukemic population to increased differentiation pressure. Importantly, elevated cycle rates of the progenitor pool suggest that at least short-term imatinib exposure was not inhibitory to primitive cells in vivo.
Response of normal versus leukemic stem cells to irradiation
Finally, we examined treatments used for bone marrow transplantation (BMT) procedures. Prior to BMT, patients are commonly treated with total body irradiation as part of a conditioning regimen to ablate endogenous disease and enhance engraftment of donor hematopoietic cells. In many cases, patients undergoing such a procedure are in clinical remission. While this procedure is beneficial for engraftment of normal donor stem cells, the relative effects of irradiation on residual LSCs are unknown. Moreover, radiation is known to impair the viability/function of normal endogenous HSCs in the host.36,37 Thus, we employed our model system to investigate relative effects of radiation on LSCs versus HSCs in vivo. To this end, the system was modified to examine treatment of leukemic animals with low tumor burden. Cohorts of mice bearing the BCR/ABL + Nup98/HoxA9 leukemia were established and challenged during early stages of disease (2-3 days after transplantation). At this phase of disease evolution, leukemic cells are not yet evident in peripheral circulation and marrow levels are extremely low. Thus, irradiation at this point is roughly equivalent to patients with minimal residual disease. At 7 days following irradiation, animals were killed and analyzed with respect to stem and progenitor cell content (n = 3 animals per group, 2 independent experiments). As shown in Figure 5A, while the percentage of phenotypically defined normal HSCs was dramatically reduced, the LSC populations were only modestly suppressed in comparison to nonirradiated controls. Similarly, the frequency of progenitor cells as determined by CFU assay was suppressed over 10-fold in normal populations (P = .009) but was not significantly reduced in leukemic cells (P = .064; Figure 5B). These data indicate that the leukemic stem and progenitor populations in vivo are substantially resistant to radiation-induced damage.
Phenotypic and in vitro analyses of irradiated normal versus leukemic animals. (A) Bone marrow cells were isolated from control versus irradiated (5.5 Gy) animals and labeled with antibodies to detect stem-cell populations (indicated by the box in each panel). The top panels show GFP− (ie, normal) cells and the bottoms panels show GFP+ (ie, leukemic) cells. The data shown for each panel are gated on the Lin− population. (B) Colony-forming units (CFUs) were determined for sorted GFP+ (leukemic) and GFP− (normal) populations. Data shown are the absolute number of colonies measured per 5000 cells plated (triplicate assays). *Statistical significance(P = .009) as determined by Student t test. Error bars are SD.
Phenotypic and in vitro analyses of irradiated normal versus leukemic animals. (A) Bone marrow cells were isolated from control versus irradiated (5.5 Gy) animals and labeled with antibodies to detect stem-cell populations (indicated by the box in each panel). The top panels show GFP− (ie, normal) cells and the bottoms panels show GFP+ (ie, leukemic) cells. The data shown for each panel are gated on the Lin− population. (B) Colony-forming units (CFUs) were determined for sorted GFP+ (leukemic) and GFP− (normal) populations. Data shown are the absolute number of colonies measured per 5000 cells plated (triplicate assays). *Statistical significance(P = .009) as determined by Student t test. Error bars are SD.
Discussion
In much the same way that studies of murine hematopoiesis have provided important insights into normal human stem-cell biology, we suggest that mouse leukemia models have a similar role in the characterization of human LSCs. Our findings extend the work of Dash et al24 in establishing the mouse as a useful model of blast-crisis CML by providing the first detailed description of LSCs in this genetically defined system. With the initial characterization in place, the stage is now set for comprehensive studies to evaluate parameters relevant to the in vivo biology of normal versus malignant stem cells. Such efforts will likely take 2 main forms: (1) studies to better elucidate the mechanisms controlling malignant transformation of stem cells, and (2) studies designed to understand how LSCs can be selectively targeted for destruction. The latter line of investigation represents a particularly formidable challenge and is complicated by multiple factors. First, the LSC compartment displays a relatively quiescent cell-cycle profile, very similar to normal HSCs, thus strategies aimed at preferentially active populations are likely to fail. Second, the natural protective mechanisms of stem cells, such as expression of xenobiotic efflux pumps, may be retained by LSCs and make them relatively difficult to target by at least some drug classes. Third, the ways in which LSCs are modulated by the marrow microenvironment (ie, the stem-cell “niche”) are almost entirely uncharacterized. Hence, signals designed to protect normal stem cells from toxic insults may well be operative for LSCs. Fourth, as suggested by the radiation experiments in this study, LSCs may have enhanced mechanisms of DNA repair and/or the ability to evade senescence induction. Given these and other complicating factors, we suggest that the development and use of genetically and biologically accurate models of in vivo leukemogenesis will be essential for the design of better therapeutic regimens.
Findings from the present study suggest that LSCs can be effectively modeled in the mouse. Our data indicate that the LSCs are relatively rare, possess a defined cell-surface immunophenotype and cell-cycle profile, and display distinct responses to various forms of extrinsic stimuli. All of these features have either been described for human LSCs and/or are predicted by previously described components of leukemic disease.9,11 In addition, our data further corroborate concepts proposed to explain the genesis of LSCs. Expression of BCR/ABL provides a direct mitogenic signal, as evidenced by cell-cycle analysis of CML LSCs. Intriguingly though, the BCR/ABL-mediated proliferative signal is eclipsed by coexpression of Nup98/HoxA9. Accompanied by an increase in absolute LSC numbers, the activity of Nup98/HoxA9 appears to mediate a switch at the stem-cell level from outright proliferative advantage to increased self-renewal as the key pathogenic feature. These findings serve to help explain why coexpression of BCR/ABL + Nup98/HoxA9 is sufficient to induce LSC formation in myeloid progenitor populations (ie, CMPs and GMPs), which have typically lost intrinsic self-renewal potential. Moreover, the data are entirely consistent with the work of Jamieson et al33 in human disease, where CML arises in the stem-cell compartment but progression to acute disease can occur in myeloid progenitor populations. We note that our data on CML LSC frequency are in contrast to the previous studies of Koschmieder et al,38 who observed over a 20-fold increase in the LSK compartment using an inducible transgenic system to activate BCR/ABL expression. Due to major differences in the respective experimental systems, it is difficult to compare the 2 studies, but we suggest that the overall frequency of LSK cells may be dependent on multiple variables including target cell population, expression level of BCR/ABL, and the stage of disease.
Aside from the primary features mentioned above, several other aspects of the mouse model are worth noting. First, studies of human AML specimens have previously described down-regulation of c-kit as a feature of LSCs.39 Thus, the decreased expression of this common stem-cell marker we observe in the mouse model may parallel human disease. Second, studies by Somervaille and Cleary17 in mouse leukemia models have previously documented relatively promiscuous expression of cell-surface markers among primitive cell types. Therefore, finding some degree of leukemia-initiating potential and/or CFU potential in multiple compartments is not unprecedented. Indeed, one might imagine that less rigid developmental borders among early cell types may be an intrinsic feature of malignant populations. Third, the effects of both ara-C and imatinib mesylate on normal versus leukemic stem cells were entirely nonspecific, a finding that supports the concept that current therapeutic approaches do not selectively target LSCs. Fourth, primitive cells in the bcCML model display a striking degree of radioresistance. Given the well-documented properties of BCR/ABL in DNA repair,40 this finding is not surprising but does represent the first in vivo demonstration that leukemic stem/progenitors are selectively resistant to any form of therapy. Interestingly, radioresistance has also been recently observed for human glioma stem cells.41 If BCR/ABL activity is indeed the key component of our observations on LSCs, then pretreatment with imatinib prior to radiation should abrogate the protective effect and sensitize LSCs to radiation. If true, we suggest that this scenario may have relevance for bcCML patients undergoing preparative regimens for BMT.
Going forward, there are several lines of investigation to pursue with the mouse LSC model. For example, a variety of other cell-surface molecules have been implicated in stem-cell biology and should make interesting targets for analysis. Potential niche mediators such as CXCR4, VLA4, VLA5, Tie2, etc represent molecules that may preferentially modulate the biology of HSCs versus LSCs. Indeed, 2 recent studies indicate that targeting the CD44 antigen may induce LSC-specific loss of function.42,43 Similarly, sensitivity to therapeutic insult as a function of anatomic location may also be an important avenue to pursue. While HSCs appear to reside primarily in the niche, one consequence of transformation may be the ability of LSCs to successfully survive in extramedullary sites. If so, then LSCs outside the marrow niche may show increased sensitivity to certain drugs, and thereby suggest that mobilization of primitive leukemic cells could improve responses to at least some forms of therapy.
In summary, with the phenotypic and functional characterization of LSCs provided by this and other recent reports using mouse models,15,17,18 it should be possible to perform detailed in vivo characterization of primitive leukemia cells and develop more selective regimens for selective eradication of malignant stem cells.
An Inside Blood analysis of this article appears at the front of this issue.
The online vesion of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
These studies were supported by grants from the American Cancer Society (RSG-03–096-01-LIB; C.T.J.), the US Department of Defense (DAMD17–03-1–0263; C.T.J.), and the National Institutes of Health (NIH; R21-CA107355; A.B.). C.T.J. is a Scholar of the Leukemia and Lymphoma Society.
National Institutes of Health
Authorship
Contribution: S.J.N. led the laboratory studies. T.B. directed all flow-cytometry experiments. S.S. performed experiments and analyzed data for the CML model. J.A. performed radiation sensitivity experiments. R.M.R. assisted with numerous animal studies. P.-Y.W. assisted with animal studies. D.R.B. performed initial studies to establish the bcCML model. D.H. performed Southern-blot studies. A.B. designed and analyzed Southern-blot studies. C.T.J. was the principal investigator.
T.B. and S.S. contributed equally to this study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Craig T. Jordan, University of Rochester Medical Center, 601 Elmwood Ave, Box 703, Rochester, NY, 14642; e-mail:craig_jordan@urmc.rochester.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal