Abstract
CD56high acute myeloid leukemias (AMLs) have a poor prognosis, but it has been unclear how CD56 expression is controlled and how it relates to clinical aggressiveness. We show that CD56 expression on AML cells correlates with an abnormal expression pattern of runt-related transcription factor 1 (RUNX1) isoforms. Whereas full-length p48 RUNX1 (p48) up-regulated CD56 in AML cells, 3 previously unknown shorter RUNX1 isoforms, p38a, p30, and p24, suppressed CD56 expression. Both p48 and CD56 induced nuclear translocation of nuclear factor (NF)–κB and increased bcl2L12 expression, and inhibition of this pathway by small inhibitory RNA-mediated p48 knock down or NF-κB blockade substantially increased apoptosis in CD56+ AML cell lines. These findings indicate the potential for new therapy of CD56high AML by suppression of the “overactive” RUNX1/CD56/NF-κB signaling pathway(s).
Introduction
CD56 (NCAM-1) is a prototypic member of the family of Ca2+-independent cell adhesion molecules.1 During development, CD56 is abundantly expressed in the fetal nervous system,2 whereas after birth, various other tissues also express CD56.3,4 In acute myeloid leukemias (AMLs), CD56 expression occurs in 15% to 20% of cases, particularly in the M2, M4, and M5 French-American-British subtypes. CD56 positivity of leukemic blasts is an independent negative prognostic marker that is associated with a poor response to chemotherapy and with increased relapse rates, resulting in a shorter overall survival of affected patients.5–7 Furthermore, clonal evolution of leukemic blasts with CD56 expression has been observed in originally CD56− AMLs after relapse.6,8 To date, however, it has been unclear what regulates CD56 expression on AML cells and how it is linked to the aggressive AML phenotype
We recently reported that the runt-related transcription factor 1, RUNX1 (AML1), controls CD56 expression in ischemic heart failure.9 RUNX1 is also one of the most frequently mutated genes in AMLs.10 Therefore, we hypothesized that RUNX1 might also regulate CD56 expression in AMLs, thereby controlling functional features of AML cells with relevance to their malignant potential. We show that multiple isoforms of RUNX1 were expressed in AML cells and that a characteristic pattern of expressed RUNX1 isoforms correlated with CD56 expression. Abnormal overexpression of the full-length p48 isoform in AML cells stimulated CD56 transcription, whereas 3 previously unknown RUNX1 isoforms, p38a, p30, and p24, suppressed it to a variable extent. Moreover, small inhibitory RNA (siRNA) directed against p48 RUNX1 suppressed CD56 expression and nuclear factor (NF)-κB activation. Together, these findings suggest that strategies aimed at the balance between individual RUNX1 isoforms and/or NF-κB signaling could inhibit the survival of CD56high AML cells, thus providing promising new targets for therapy of this high-risk group of leukemias.
Patients, materials, and methods
Patients and control subjects
Myeloid blasts from 72 patients with acute myeloid leukemia, diagnosed according to the French-American-British study group (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) were isolated by fluorescent-activated cell sorting (CD33+/CD56+, CD33+/CD56−). Peripheral blood mononuclear cells from 12 normal healthy donors served as controls.
Informed consent in accordance with the Declaration of Helsinki was obtained from all subjects, and protocols were approved by the Institutional Review Boards of the participating institutions (Institute of Pathology, University of Wuerzburg, Germany; University Children's Hospital Hannover, Department of Pediatric Hematology and Oncology, Germany; Neurosciences Group, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, University of Oxford, United Kingdom; and Institute of Pathology, University of Heidelberg, University Hospital Mannheim, Germany).
cDNA library synthesis and screening
A human cDNA library derived from CD56−AML M2 blasts (Figure S3) was screened by colony hybridization using a 32P-labeled cDNA probe covering the RUNT domain of RUNX1. Sequencing analysis was performed on an ABI 3100 avant capillary sequencer (ABI, Weiterstadt, Germany).
RNase protection assay, Western blots, reverse transcriptase–polymerase chain reaction, and immunohistochemistry
Transfection of acute myeloid leukemia cell lines with mammalian expression vectors or small inhibitory RNA
Using the Amaxa nucleofection technology (Amaxa, Köln, Germany), K562, HL-60, and U937 cells (ATCC, Manassas, VA; Document S1) were transfected with RUNX1 isoforms p48, p38a, p30, and p24 as well as the CD56 isoforms CD56120kDa and CD56140kDa or 50 nM siRNA duplexes. Sequences of siRNAs are given in Figure S7.
Luciferase assay, preparation of nuclear and cytosolic proteins, and fluorescent-activated cell sorter analysis
HEK293 cells (2.5 × 105) were cotransfected with 1 μg each of pRSV-RUNX1-p48, -p38a, -p30, and -p24 expression vectors; pRSV-vector with .2 μg of a pGL3 basic/CD56 promoter luciferase vector; or pGL3 luciferase vector alone (SuperFect Transfection Reagent; Qiagen, Hilden, Germany). For CD56 promotor construct, the published human CD56/NCAM gene promotor region (accession number X53243, National Center for Biotechnology Information database) from nt211 to nt1413, including the transcription start of CD56/NCAM gene (nt1392), was amplified by polymerase chain reaction. Amplificates were cloned into the pGEMT vector (Promega, Heidelberg, Germany), and the identity of the nt211 to nt1413 sequence with published sequences was verified by complete sequence analysis (ABI avant 3100 capillary sequencer; Applied Biosystems, Foster City, CA). Analyses for luciferase activity as well as preparation of nuclear and cytosolic proteins and fluorescent-activated cell-sorting analysis were performed as described.12
Nuclear and cytoplasmatic proteins from RUNX1-p48, -p38a, -p30, and -p24 transfected cell lines K562 and HL-60 were prepared as described6 and used for subsequent Western blot analysis of NF-κB proteins.
Detection of apoptosis by annexin V staining and fluorescent-activated cell sorter analysis
Presence of phosphatidylserine in the outer leaflet of the plasma membrane was detected by flow cytometry using fluorescein isothiocyanate conjugated-coupled annexin V (annexin V) and 5 × 104 cells per experiment following the manufacturer's instructions (BD Biosciences, Heidelberg, Germany).
Statistical analysis
Data are expressed as means plus or minus a standard deviation (SD). Comparisons were made using the Mann-Whitney U test and the SPSS for Windows software package (SPSS, Chicago, IL). Differences were considered significant with P less than .05.
Results
RUNX1 p48 is overexpressed in CD56+ acute myeloid leukemias
We first analyzed blood-derived leukemic blasts from 72 patients with AML, and peripheral blood mononuclear cells from 12 healthy persons by Western immunoblots using antibodies to CD56 or RUNX1. Thirty-three patients' cells were CD56+, whereas 39 were CD56−. As shown in Figure 1A, CD56 positivity in AMLs was strongly correlated with overexpression of the known13 p48 RUNX1 isoform (designated as p48). By contrast, all CD56−/low? AMLs expressed p48 at barely detectable levels (Figure 1A). However, the antibody specific for the RUNX1 N-terminus, but not the one directed against the C-terminus (see “Patients, materials, and methods”) revealed an unexpected 30-kDa band (designated as p30), suggesting that p30 might represent a “novel” RUNX1 isoform containing an alternative C-terminal exon. This protein was found to be more strongly expressed in CD56− AML cases than in CD56+ AMLs (Figure 1A).
CD56 and RUNX1 expression in leukemic blasts of patients with acute myeloid leukemias (AMLs), defined as either CD56+ (n=4) or CD56− (n=4) by flow cytometry. (A) Top panel: Western blot analysis. Note much stronger expression of both CD56 and RUNX1 p48 in CD56+ than CD56− AMLs. RUNX1 N-terminal, but not RUNX1 C-terminal antibody, detects a strong additional 30-kDa band in CD56− AMLs, suggesting that p30 could harbor an alternative C-terminal exon. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), loading control. Top panel, right[b]: Reverse transcriptase–polymerase chain reaction quantification of CD56, p48, and p30 in of CD56+ and CD56− AMLs (n=72) (Figures S4,S5). There was significant concordant expression of CD56 and p48 (P < .001, Mann-Whitney U test). p30 expression was slightly higher in CD56− compared with CD56+ AMLs (P < .035). Bottom panel: RUNX1 isoforms and the antibodies used to discriminate them. RUNX1 (p48) and 3 new (p38a, p30, p24) RUNX1 isoforms isolated from a human CD56−cDNA library (Document S1). p30 and p24 harbor a new C-terminal exon, termed exon 5.4 (Ex. 5.4). Antibody symbols indicate binding regions of the commercial antisera to the N- and C-terminus of RUNX1 and of a custom-made rabbit antiserum raised against exon 5.4 (Figures S2 and 1B). TAD1 and TAD2, transactivating domains 1 and 2, respectively; RUNT (RUNT domain), DBD (DNA-binding domain). (B) Top panel: Semiquantitiative 32P-dCTP-based reverse transcriptase–polymerase chain reaction analysis of CD56 and RUNX1 isoforms in CD56+ and CD56− AMLs (Figure S2). CD56 and p48 expression are concordant, and there is slightly higher expression of p30 and p38a mRNA in CD56− AMLs. Expression levels are given as counts per minute (cpm) of phosphoimager analysis (see also Figure S2). Bottom panel: Confirmation of differential expression of p48 and p30 isoforms by immunohistochemistry in AMLs. Examples of staining of CD56+ (left row) and CD56− AMLs (right row) using RUNX1 C-terminal antibody (detecting p48 and p38a) and RUNX1 5.4 antibody (detecting p30 and p24). In the left row, AML M2 blasts infiltrating testis exhibit strong expression of CD56 and RUNX1 p48, whereas RUNX1 p30 expression is weak. By contrast, the right row depicts a bone marrow infiltration by CD56− AML M2 blasts with lack of RUNX1 p48 but strong expression of RUNX1 p30. Representative results of 4 experiments. Slides were viewed through an Olympus d×40 microscope (Olympus, Hamburg, Germany) with a UPlan APO lens and Xylol coverslipping films (Sekura, Heppenheim, Germany). Images were acquired with an Olympus BX50 camera and Olympus DP-Soft version 5.0 (Olympus), and processed using Adobe Photoshop version 3.0 (Adobe Systems, San Jose, CA).
CD56 and RUNX1 expression in leukemic blasts of patients with acute myeloid leukemias (AMLs), defined as either CD56+ (n=4) or CD56− (n=4) by flow cytometry. (A) Top panel: Western blot analysis. Note much stronger expression of both CD56 and RUNX1 p48 in CD56+ than CD56− AMLs. RUNX1 N-terminal, but not RUNX1 C-terminal antibody, detects a strong additional 30-kDa band in CD56− AMLs, suggesting that p30 could harbor an alternative C-terminal exon. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), loading control. Top panel, right[b]: Reverse transcriptase–polymerase chain reaction quantification of CD56, p48, and p30 in of CD56+ and CD56− AMLs (n=72) (Figures S4,S5). There was significant concordant expression of CD56 and p48 (P < .001, Mann-Whitney U test). p30 expression was slightly higher in CD56− compared with CD56+ AMLs (P < .035). Bottom panel: RUNX1 isoforms and the antibodies used to discriminate them. RUNX1 (p48) and 3 new (p38a, p30, p24) RUNX1 isoforms isolated from a human CD56−cDNA library (Document S1). p30 and p24 harbor a new C-terminal exon, termed exon 5.4 (Ex. 5.4). Antibody symbols indicate binding regions of the commercial antisera to the N- and C-terminus of RUNX1 and of a custom-made rabbit antiserum raised against exon 5.4 (Figures S2 and 1B). TAD1 and TAD2, transactivating domains 1 and 2, respectively; RUNT (RUNT domain), DBD (DNA-binding domain). (B) Top panel: Semiquantitiative 32P-dCTP-based reverse transcriptase–polymerase chain reaction analysis of CD56 and RUNX1 isoforms in CD56+ and CD56− AMLs (Figure S2). CD56 and p48 expression are concordant, and there is slightly higher expression of p30 and p38a mRNA in CD56− AMLs. Expression levels are given as counts per minute (cpm) of phosphoimager analysis (see also Figure S2). Bottom panel: Confirmation of differential expression of p48 and p30 isoforms by immunohistochemistry in AMLs. Examples of staining of CD56+ (left row) and CD56− AMLs (right row) using RUNX1 C-terminal antibody (detecting p48 and p38a) and RUNX1 5.4 antibody (detecting p30 and p24). In the left row, AML M2 blasts infiltrating testis exhibit strong expression of CD56 and RUNX1 p48, whereas RUNX1 p30 expression is weak. By contrast, the right row depicts a bone marrow infiltration by CD56− AML M2 blasts with lack of RUNX1 p48 but strong expression of RUNX1 p30. Representative results of 4 experiments. Slides were viewed through an Olympus d×40 microscope (Olympus, Hamburg, Germany) with a UPlan APO lens and Xylol coverslipping films (Sekura, Heppenheim, Germany). Images were acquired with an Olympus BX50 camera and Olympus DP-Soft version 5.0 (Olympus), and processed using Adobe Photoshop version 3.0 (Adobe Systems, San Jose, CA).
Identification of 3 novel RUNX1 isoforms in acute myeloid leukemias
To identify this hypothetical RUNX1 isoform, we screened a human cDNA library derived from a CD56− human AML M2 (“Patients, materials, and methods; cDNA library synthesis and screening”) with a cDNA probe encoding the RUNT domain of human RUNX1. In addition to cDNAs coding for p48, we isolated 3 unknown RUNX1 splice variants. The “new” p38a variant resembles the well-known p38 RUNX1 isoform with respect to the shared exons 3 through 613 that, however, are spliced in a previously unknown way to the known N-terminal exons 2a/2b. The 2 other “new” variants, p30 and p24, show splicing to an as-yet unknown alternative C-terminal exon that we propose to call exon 5.4 because it is located between the RUNX1 exons 5.3 and 613 (see Figure 1A). The intron/exon boundaries of exon 5.4 show RNA splice-donor and splice-acceptor consensus sites and a polyadenylation signal located 21 base pairs upstream from the polyA addition site (Document S1).
Predominant expression of RUNX1 p48 and the novel p30 isoform in acute myeloid leukemias
By RNAse protection assays, exon 5.4 mRNA could be detected in almost all human tissues investigated, albeit at quite variable levels (Figures S4,S5). Furthermore, semiquantitative reverse transcriptase–polymerase chain reaction amplification of the full-length coding sequences of p48, p38a, p30, and p24 (using oligonucleotide primers specific to the RUNT domain and either exon 6 or exon 5.4 [Figures S4,S5]) revealed expression of p48, p38a, p30, and p24 in all organs investigated (n=21), with a predominance of p48 and p30 expression compared with that of other RUNX1 isoforms (Figures S4,S5).
As shown by reverse transcriptase–polymerase chain reaction, the expression of individual RUNX1 transcripts differed markedly between CD56+ and CD56− AML cells (Figure 1B, Figures S4, S5). To correlate isoform-specific RUNX1 RNA levels with the respective protein levels, we raised rabbit antiserum against the p30 and p24 isoforms encoded by exon 5.4 of the RUNX1 gene (Document S1). Immunoblot analysis of normal human tissues and immunostaining of AMLs (Figures 1B and S1,S2) with exon- and isoform-specific antibodies showed that p48 and p30 protein levels reflect the expression levels of the respective transcripts. By contrast, p38a and p24 proteins could not be identified in either CD56+ or CD56− AML cells and normal tissues (Figure S6). Why detectable p38a and p24 mRNAs (Figure 1B) do not translate into detectable protein levels is unclear, as is the functional role of p38a and p24 in nontransfected cells (see “Results; Novel RUNX1 isoforms with dominant negative transcriptional activity control the expression of CD56 and other RUNX1 target genes” and “Discussion”).
Novel RUNX1 isoforms with dominant negative transcriptional activity control the expression of CD56 and other RUNX1 target genes
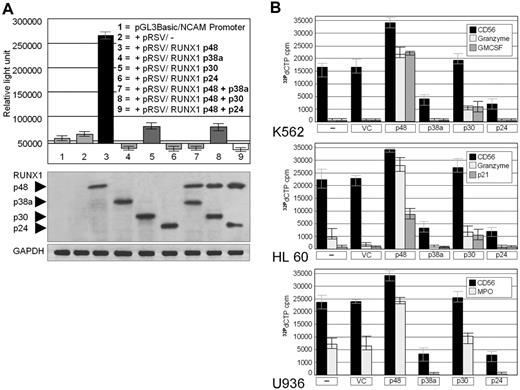
To show whether the RUNX1 isoforms control CD56 expression in AML (like in ischemic heart disease9 ), we cotransfected a luciferase reporter construct bearing the CD56 promoter and expression vector(s) coding for one or 2 RUNX1 isoform(s) into HEK293 cells. As shown in Figure 2A, p48 was a strong stimulator of the CD56 promoter, whereas 2 of the 3 novel RUNX1 isoforms, p38a and p24, strongly inhibited the CD56 promoter activity irrespective of the presence or absence of cotransfected p48. This dominant inhibitory function is probably related to molecular features of p38a and p24. Whereas both isoforms bear the DNA binding domain and recruitment sites for RUNX1 cofactors,14 the transactivating domain 2 (TAD2) is missing in p38a, and both TAD1 and TAD2 are absent in p24 (Figure 1A). This suggests that the transcriptionally inactive p38a and p24 isoforms compete for p48 binding to the CD56 promoter and/or sequester the RUNX1 cofactors that enhance RUNX1 activity. However, because p48 and the “new” p30 are by far the most abundant RUNX1 isoforms expressed in AMLs in vivo (Figure 1A), p30 rather than p24 and p38a might be of major importance for the regulation of CD56 expression in AMLs. Indeed, when HEK293 cells were exclusively transfected with p30 and the reporter, p30 showed only weak transcriptional activity on the CD56 promoter (column 5 in Figure 2A). However, when p30 and p48 were cotransfected (column 8 in Figure 2A), p30 strongly interfered with the p48-driven luciferase activity albeit slightly less than did p38a and p24.
Transcriptional activity of RUNX1 isoforms p48, p38a, p30, and p24 on the CD56 promoter. (A) Upper panel: HEK293 cells were cotransfected with a CD56 promoter/luciferase reporter gene and the RUNX1 isoforms. RUNX1 p48 strongly up-regulates CD56 expression. RUNX1 p48-induced expression of CD56 is inhibited by p38, p30, and p24 (columns 7-9), suggesting a dominant negative effect of the latter isoforms. Controls are NCAM/CD56 promoter/luciferase reporter gene only and NCAM/CD56 promoter/luciferase reporter gene plus “empty” RSV expression vector (columns 1 and 2). (Lower panel) Confirmation of RUNX1 expression by Western blot. Similar amounts of the various recombinant RUNX1 isoforms were detected in the transfected cells by a RUNT domain antibody. GAPDH, protein loading control. (B) Expression of RUNX1 target genes in acute myeloid leukemia cell lines. CD56, granzyme B, p21, interleukin-6, and granulocyte-macrophage colony-stimulating factor were detected by reverse transcriptase–polymerase chain reaction in human acute myeloid leukemia cell lines K562, HL60, and U937 transfected with RUNX1 isoforms p48, p38a, p30, and p24. All target genes were strongly induced by p48; p30 had approximately no effect on CD56 and minor inducing effects on the other targets. p38a and p24 inhibited expression of all target genes relative to endogenous expression in vector control (VC) or nontransfected cells (–). Data are means plus or minus SD.
Transcriptional activity of RUNX1 isoforms p48, p38a, p30, and p24 on the CD56 promoter. (A) Upper panel: HEK293 cells were cotransfected with a CD56 promoter/luciferase reporter gene and the RUNX1 isoforms. RUNX1 p48 strongly up-regulates CD56 expression. RUNX1 p48-induced expression of CD56 is inhibited by p38, p30, and p24 (columns 7-9), suggesting a dominant negative effect of the latter isoforms. Controls are NCAM/CD56 promoter/luciferase reporter gene only and NCAM/CD56 promoter/luciferase reporter gene plus “empty” RSV expression vector (columns 1 and 2). (Lower panel) Confirmation of RUNX1 expression by Western blot. Similar amounts of the various recombinant RUNX1 isoforms were detected in the transfected cells by a RUNT domain antibody. GAPDH, protein loading control. (B) Expression of RUNX1 target genes in acute myeloid leukemia cell lines. CD56, granzyme B, p21, interleukin-6, and granulocyte-macrophage colony-stimulating factor were detected by reverse transcriptase–polymerase chain reaction in human acute myeloid leukemia cell lines K562, HL60, and U937 transfected with RUNX1 isoforms p48, p38a, p30, and p24. All target genes were strongly induced by p48; p30 had approximately no effect on CD56 and minor inducing effects on the other targets. p38a and p24 inhibited expression of all target genes relative to endogenous expression in vector control (VC) or nontransfected cells (–). Data are means plus or minus SD.
To investigate whether the various RUNX1 isoforms affect the expression of endogenous target genes in AML cells, the cell lines K562, HL60, and U937 were transfected with vectors expressing the p48, p38a, p30, and p24 RUNX1 isoforms. Whereas p48 markedly enhanced the expression of endogenous CD56 mRNA, p30 had no significant positive effect. By contrast, p38a and p24 strongly suppressed CD56 expression (Figure 2B). The RUNX1 isoforms also differentially affected the expression of other known RUNX1 targets15 such as of MPO, granzyme B, p21, interleukin-6, and granulocyte-macrophage colony-stimulating factor genes (Figure 2B). In each case, p48 increased and p38a and p24 decreased expression of these target genes, whereas p30 again had no significant positive transcriptional effect.
Induction/inhibition of CD56 expression by small inhibitory RNA mediated knockdown of RUNX1 isoforms
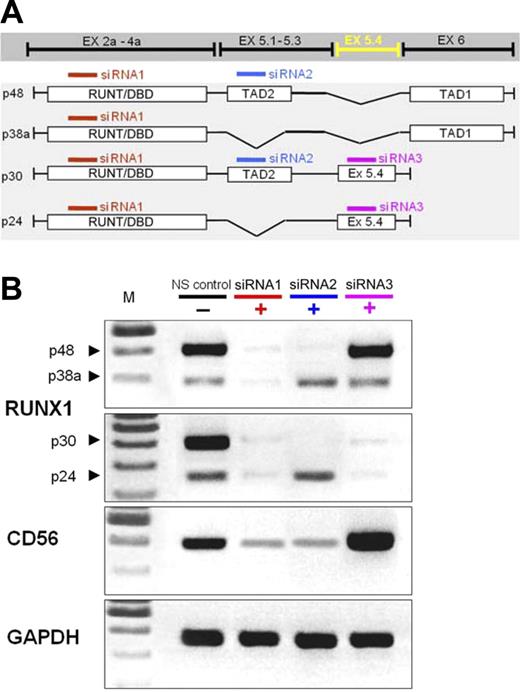
We confirmed the results on CD56 expression by siRNA-mediated transient knockdown experiments. Individual RUNX1 exons were targeted in K562 (Figure 3) and HL60 cells (not shown) by transfection with siRNA oligomers complementary to (1) the N-terminal RUNT/DBD domain, which is expressed by all RUNX1 isoforms; (2) exon 5.1 (harboring the TAD2, expressed in p48 and p30); or (3) exon 5.4 (as expressed in p30 and p24). These transfections caused a strong down-regulation of the respective RUNX1 mRNAs and changed CD56 mRNA expression by 50% to 90% of the steady-state levels (Figure 3). Notably, knock down of p48 reduced CD56 expression, whereas knockdown of p30 and p24 with siRNA for exon 5.4 increased CD56 expression above control levels.
Exon-specific knockdown of RUNX1 isoforms and effect on CD56 mRNA levels in K562 acute myeloid leukemia cells. Small inhibitory RNAs complementary to the RUNT-domain (affecting all 4 isoforms), exon 5a containing the transactivating domain 2 (affecting p48 and p30), the new exon 5.4 (affecting p30 and p24), and a nonsilencing control small inhibitory RNA (NS control) were used. Results confirm the expression of the different isoforms in the control cells and the appropriate down-regulation of each RUNX1 isoform with small inhibitory RNA. CD56 expression is down-regulated by inhibition of p48, but increased when only p30 and p24 are targeted. One representative experiment of 4.
Exon-specific knockdown of RUNX1 isoforms and effect on CD56 mRNA levels in K562 acute myeloid leukemia cells. Small inhibitory RNAs complementary to the RUNT-domain (affecting all 4 isoforms), exon 5a containing the transactivating domain 2 (affecting p48 and p30), the new exon 5.4 (affecting p30 and p24), and a nonsilencing control small inhibitory RNA (NS control) were used. Results confirm the expression of the different isoforms in the control cells and the appropriate down-regulation of each RUNX1 isoform with small inhibitory RNA. CD56 expression is down-regulated by inhibition of p48, but increased when only p30 and p24 are targeted. One representative experiment of 4.
RUNX1 p48 and CD56 inhibit apoptosis in acute myeloid leukemia cells through the NF-κB pathway: inhibition of NF-κB as a therapeutic option
To investigate whether the RUNX1 isoforms might differentially affect the fate of AML cells in terms of proliferation and apoptosis, we transfected either empty vectors, vectors expressing individual RUNX1 isoforms, or CD56 into K562 (Figure 4) and HL60 (not shown) AML cells and measured their numbers (after 4 days) and reactivity with annexin V (after 1 day in culture using fluorescent-activated cell sorter, Figure S9). As shown in Figure 4A, similar to CD56 expression, the expression of full-length p48 RUNX1 increased the number of K562 cells, whereas expression of p38a and p24 resulted in lower cell numbers. These findings are well correlated with the effect of RUNX1 proteins on apoptosis; although overexpressed p48 RUNX1 exerted an antiapoptotic effect, p38a and p24 increased spontaneous apoptosis of K562 cells (Figure 4B). Virtually identical results were obtained with HL60 cells (not shown).
Effect of RUNX1 and CD56 expression on K562 acute myeloid leukemia cells. (A) Effect on K562 cell growth. The cells were transfected with either the RUNX1 isoforms or the CD56 (120 kDa and 140 kDa) isoforms. After 4 days, RUNX1 p48 and CD56-transfected cells showed less than a 2-fold increase in total cell number compared with vector control [VC] or untransfected (–) cells. p38a and p24 transfected cells revealed a less than 50% growth inhibition, whereas p30 had no significant effect. Data are means plus or minus SD. (B) Detection of phosphatidylserine [PS] on the cell surface of RUNX1-p48, -p38a, -p30, and -p24 transfected K562 cells by fluorescent-activated cell sorter analysis using Annexin-V. There was a decreased number of apoptotic cells among the RUNX1-p48-transfected K562 cells compared with K562 cells transfected with the vector control. By contrast, there was clearly increased apoptosis among RUNX1-p38a and -p24-transfected K562 cells. Compared with vector control, the transfection of RUNX1-p30 had no significant impact on apoptosis. (C) NF-κB and bcl2L12 expression examined by reverse transcriptase–polymerase chain reaction and nuclear translocation of NF-κB/p65 visualized by Western blot of cytoplasmic [C] and nuclear [N] protein extracts (see also Figure S8) after transfection of the K562 acute myeloid leukemia cell line with either RUNX1 isoforms or CD56 (120 kDa and 140 kDa) compared with levels in vector-control [VC] or untransfected [–] cells. Increased expression of NF-κB and bcl2L12 as well as nuclear translocation of p65 in RUNX1 p48 and CD56-transfected cell lines. By contrast, p38a and p24-transfected K562 cells showed clear down-regulation of NF-κB and bcl2L12 and no nuclear translocation of p65.
Effect of RUNX1 and CD56 expression on K562 acute myeloid leukemia cells. (A) Effect on K562 cell growth. The cells were transfected with either the RUNX1 isoforms or the CD56 (120 kDa and 140 kDa) isoforms. After 4 days, RUNX1 p48 and CD56-transfected cells showed less than a 2-fold increase in total cell number compared with vector control [VC] or untransfected (–) cells. p38a and p24 transfected cells revealed a less than 50% growth inhibition, whereas p30 had no significant effect. Data are means plus or minus SD. (B) Detection of phosphatidylserine [PS] on the cell surface of RUNX1-p48, -p38a, -p30, and -p24 transfected K562 cells by fluorescent-activated cell sorter analysis using Annexin-V. There was a decreased number of apoptotic cells among the RUNX1-p48-transfected K562 cells compared with K562 cells transfected with the vector control. By contrast, there was clearly increased apoptosis among RUNX1-p38a and -p24-transfected K562 cells. Compared with vector control, the transfection of RUNX1-p30 had no significant impact on apoptosis. (C) NF-κB and bcl2L12 expression examined by reverse transcriptase–polymerase chain reaction and nuclear translocation of NF-κB/p65 visualized by Western blot of cytoplasmic [C] and nuclear [N] protein extracts (see also Figure S8) after transfection of the K562 acute myeloid leukemia cell line with either RUNX1 isoforms or CD56 (120 kDa and 140 kDa) compared with levels in vector-control [VC] or untransfected [–] cells. Increased expression of NF-κB and bcl2L12 as well as nuclear translocation of p65 in RUNX1 p48 and CD56-transfected cell lines. By contrast, p38a and p24-transfected K562 cells showed clear down-regulation of NF-κB and bcl2L12 and no nuclear translocation of p65.
Because CD56 is known to induce the antiapoptotic NF-κB pathway in neurons and astrocytes,16 we investigated the mRNA induction and nuclear translocation of NF-κB (p65, p50, and c-rel) in K562 and HL60 cells after transfection with p48, p38a, p30, and p24 RUNX1 isoforms as well as with CD56 (120-kDa and 140-kDa isoforms) (Figure S8). Compared with mock-transfected controls, p48 and CD56-transfected AML cells showed enhanced expression of p65/p50 and c-rel NF-κB, an increase in the nuclear translocation of p65 protein (Figure 4C), corresponding to the approximately 2-fold increase in cell numbers after a 4-day culture period (Figure 4A). Furthermore, the early response antiapoptotic protein bcl2L12, a new member of the bcl2 family, was clearly induced in AML cells transfected with p48 (Figure 4C). By contrast, expression of p38a and p24 RUNX1 isoforms led to an approximately 50% reduction of cell numbers (Figure 4A), a marked down-regulation of NF-κB mRNA expression, reduced NF-κB translocation to the nucleus, and a diminished expression of bcl2L12 mRNA (Figure 4C).
This functional correlation of RUNX1/CD56-dependent regulation of NF-κB p65/p50/c-rel mRNA expression was also confirmed by siRNA experiments (see previously and Figure 3) because siRNA driven knockdown of p30 and p24 (siRNA 3, Figure 3) caused a slight increase of NF-κB p65/p50/c-rel mRNA (not shown), whereas knockdown of p48 and p30 (siRNA 2, Figure 3) as well as all RUNX1 isoforms (siRNA 1, Figure 3) resulted in decreased expression of NF-κB p65/p50/c-rel mRNA as well as of bcl2L12 mRNA (not shown).
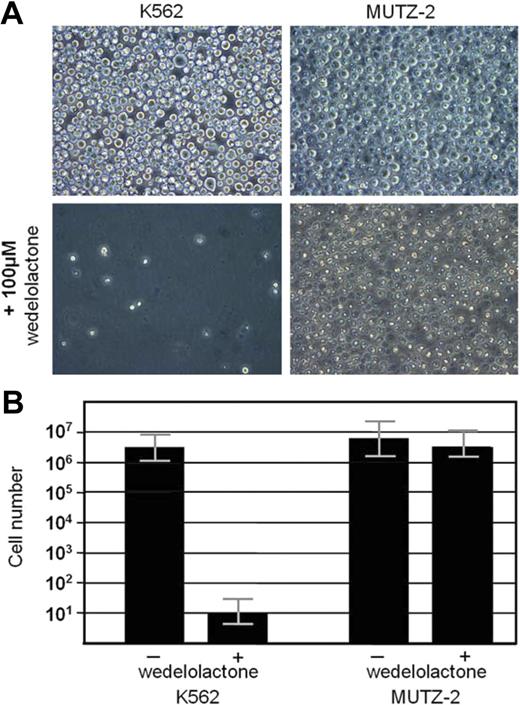
To see whether the activated, p48 RUNX1/CD56-driven NF-κB pathway in AML blasts is essential for their survival and, thus, might be a potential therapeutic target, we treated CD56-positive K562- and CD56-negative MUTZ-2 AML cell lines with the specific NF-κB inhibitor wedelolactone.17 Although K562 cells showed a concentration-dependent decrease in cell numbers and almost complete eradication of blasts after 4 days in culture, MUTZ-2 cells appeared to be resistant to the inhibitor (Figure 5).
Effect of specific NF-κB inhibition on cell survival. After 4 days in culture, the NF-κB inhibitor, wedelolactone (100 μM). strongly inhibited cell survival in the CD56+ cell K562 but had no significant effect on the CD56(-)MUTZ-2 acute myeloid leukemia cells (mean ; SEM, triplicates). One representative experiment of 3. Slides were viewed through a Leica HCX microscope (Leica, Wetzlar, Germany) with a PL Fluotar L lens at 40×/0.60 corr PH2. Images were acquired with a Leica DMIL camera (Leica) and Olympus DP-Soft version 5.0 (Olympus) and processed using Adobe Photoshop version 3.0 (Adobe Systems).
Effect of specific NF-κB inhibition on cell survival. After 4 days in culture, the NF-κB inhibitor, wedelolactone (100 μM). strongly inhibited cell survival in the CD56+ cell K562 but had no significant effect on the CD56(-)MUTZ-2 acute myeloid leukemia cells (mean ; SEM, triplicates). One representative experiment of 3. Slides were viewed through a Leica HCX microscope (Leica, Wetzlar, Germany) with a PL Fluotar L lens at 40×/0.60 corr PH2. Images were acquired with a Leica DMIL camera (Leica) and Olympus DP-Soft version 5.0 (Olympus) and processed using Adobe Photoshop version 3.0 (Adobe Systems).
Discussion
We have shown that CD56 expression by AML cells is positively regulated by RUNX1 p48 and negatively regulated by other, previously unknown, splice variants. At the protein level, it appears that the p48highp30low versus p48lowp30high RUNX1 isoform patterns are the major discriminators between CD56+ and CD56− AMLs, respectively. Moreover, both p48 and CD56 induce NF-κB and bcl2L12 in AML cells and protect them from apoptosis, potentially explaining the correlation between CD56 expression and the poor response of CD56+ AMLs to current treatments.5,18 However, considering the broad spectrum of p48 targets, other mechanisms contributing to the aggressive phenotype of CD56+ AMLs could be anticipated. Overexpression of macrophage colony-stimulating factor in t(8;21)(q22;q22) translocation-positive AMLs resulting from the interaction of wild-type RUNX1 with the RUNX1-ETO fusion protein19 is one example. Nevertheless, our findings suggest that p48 and RUNX1-driven NF-κB and bcl2 pathways are new candidates for targeted treatments in high-risk CD56+ AMLs. By contrast, the role of p38a and p24 in AMLs is not yet clear, because their variable mRNA levels in AML cells were not reflected by detectable levels of the respective proteins in any normal tissue or the AML samples.
The molecular basis of the differential, nonrandom expression patterns of RUNX1 isoforms in CD56+ and CD56− AMLs is unknown. Because the RUNX1 isoforms expressed in CD56+ AMLs are not qualitatively different from isoforms expressed by CD56− AMLs or normal tissues, it is tempting to speculate that the consistently imbalanced expression of p48 and p30 in AMLs results from skewed splicing. Indeed, changes in splice site selection resulting from genetic or epigenetic mechanisms have recently been found to be a common and sometimes functionally relevant phenomenon in malignant tumors.20,21 If verified in CD56+ AMLs, tipping the p48/p30 balance toward a p48low phenotype by targeting the RNA splicing machinery22 or applying RUNX1 isoform-specific siRNAs could be another therapeutic option in CD56+ AMLs.
Beyond the field of translational AML research, our results might help to explain the many recent reports that correlate CD56 overexpression with an aggressive clinical phenotype in a variety of nonmyeloid malignancies, including multiple myeloma23 and several common solid tumors such as colon carcinoma,24 renal cancer,25 and melanoma.26,27 Finally, because the regulatory properties of the new RUNX1 splice variants extend beyond CD56 to other RUNX1 target genes (Figure 2B), the results could be therapeutically relevant for RUNX1-related gene regulation in a broad spectrum of clinical settings, including heart diseases,9 hematology,5,28 and autoimmunity.29–31
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the Wilhelm-Sander-Stiftung (to S.G and A.M., grant 99.112.2). We thank Mrs Margrit Bonengel, Mrs Jaqueline Maar, Mrs Ilona Pietrovski, and Mr Erwin Schmitt for expert technical assistance.
Authorship
Contribution: S.G. and S.C. performed the research and analyzed the data; C.L. and D.R. contributed vital new reagents; H.-K.M.-H. and E.S. contributed to the design and data analysis; A.V. contributed to the data presentation and writing; and the work was initiated, designed, and coordinated by S.G. and A.M.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stefan Gattenloehner, Institute of Pathology, University of Wuerzburg, Josef-Schneiderstr.2, D-97080 Wuerzburg, Germany; e-mail: stefan.gattenloehner@mail.uni-wuerzburg.de.

![Figure 1. CD56 and RUNX1 expression in leukemic blasts of patients with acute myeloid leukemias (AMLs), defined as either CD56+ (n=4) or CD56− (n=4) by flow cytometry. (A) Top panel: Western blot analysis. Note much stronger expression of both CD56 and RUNX1 p48 in CD56+ than CD56− AMLs. RUNX1 N-terminal, but not RUNX1 C-terminal antibody, detects a strong additional 30-kDa band in CD56− AMLs, suggesting that p30 could harbor an alternative C-terminal exon. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), loading control. Top panel, right[b]: Reverse transcriptase–polymerase chain reaction quantification of CD56, p48, and p30 in of CD56+ and CD56− AMLs (n=72) (Figures S4,S5). There was significant concordant expression of CD56 and p48 (P < .001, Mann-Whitney U test). p30 expression was slightly higher in CD56− compared with CD56+ AMLs (P < .035). Bottom panel: RUNX1 isoforms and the antibodies used to discriminate them. RUNX1 (p48) and 3 new (p38a, p30, p24) RUNX1 isoforms isolated from a human CD56−cDNA library (Document S1). p30 and p24 harbor a new C-terminal exon, termed exon 5.4 (Ex. 5.4). Antibody symbols indicate binding regions of the commercial antisera to the N- and C-terminus of RUNX1 and of a custom-made rabbit antiserum raised against exon 5.4 (Figures S2 and 1B). TAD1 and TAD2, transactivating domains 1 and 2, respectively; RUNT (RUNT domain), DBD (DNA-binding domain). (B) Top panel: Semiquantitiative 32P-dCTP-based reverse transcriptase–polymerase chain reaction analysis of CD56 and RUNX1 isoforms in CD56+ and CD56− AMLs (Figure S2). CD56 and p48 expression are concordant, and there is slightly higher expression of p30 and p38a mRNA in CD56− AMLs. Expression levels are given as counts per minute (cpm) of phosphoimager analysis (see also Figure S2). Bottom panel: Confirmation of differential expression of p48 and p30 isoforms by immunohistochemistry in AMLs. Examples of staining of CD56+ (left row) and CD56− AMLs (right row) using RUNX1 C-terminal antibody (detecting p48 and p38a) and RUNX1 5.4 antibody (detecting p30 and p24). In the left row, AML M2 blasts infiltrating testis exhibit strong expression of CD56 and RUNX1 p48, whereas RUNX1 p30 expression is weak. By contrast, the right row depicts a bone marrow infiltration by CD56− AML M2 blasts with lack of RUNX1 p48 but strong expression of RUNX1 p30. Representative results of 4 experiments. Slides were viewed through an Olympus d×40 microscope (Olympus, Hamburg, Germany) with a UPlan APO lens and Xylol coverslipping films (Sekura, Heppenheim, Germany). Images were acquired with an Olympus BX50 camera and Olympus DP-Soft version 5.0 (Olympus), and processed using Adobe Photoshop version 3.0 (Adobe Systems, San Jose, CA).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/6/10.1182_blood-2007-02-074203/7/m_zh80140705070001.jpeg?Expires=1769104116&Signature=c1dxV89SC8rHhcsedvMvjoNjviPfqN~tThLladgsVoSCaa3HivUZwubIlxNw1epPzJ7YWHBFHC37QVu18YfYXwzfmgJPQp4JHd8AtOMTSLzfoDS8Uiidcjn~EcQz0I7lVuXoeVvN9zp4SXyOowCbJYqV5GBh613tFbyjX77ydpTsFRV6K7go9o2ys761AgfK89IGBR38Efgnw2LMlg6JMZkZl0oQBHMjeRKKpK8evUN4iM2IhHVQFgJLnxZ7zs00YXvU7T3-l9lbikk3QNH~0RfGJlnx1LeFmr9l8kmc9qyVjr~TbyxZRnryqMp4wQk01Smuuxx6kI-SpcP6CoTEuw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Effect of RUNX1 and CD56 expression on K562 acute myeloid leukemia cells. (A) Effect on K562 cell growth. The cells were transfected with either the RUNX1 isoforms or the CD56 (120 kDa and 140 kDa) isoforms. After 4 days, RUNX1 p48 and CD56-transfected cells showed less than a 2-fold increase in total cell number compared with vector control [VC] or untransfected (–) cells. p38a and p24 transfected cells revealed a less than 50% growth inhibition, whereas p30 had no significant effect. Data are means plus or minus SD. (B) Detection of phosphatidylserine [PS] on the cell surface of RUNX1-p48, -p38a, -p30, and -p24 transfected K562 cells by fluorescent-activated cell sorter analysis using Annexin-V. There was a decreased number of apoptotic cells among the RUNX1-p48-transfected K562 cells compared with K562 cells transfected with the vector control. By contrast, there was clearly increased apoptosis among RUNX1-p38a and -p24-transfected K562 cells. Compared with vector control, the transfection of RUNX1-p30 had no significant impact on apoptosis. (C) NF-κB and bcl2L12 expression examined by reverse transcriptase–polymerase chain reaction and nuclear translocation of NF-κB/p65 visualized by Western blot of cytoplasmic [C] and nuclear [N] protein extracts (see also Figure S8) after transfection of the K562 acute myeloid leukemia cell line with either RUNX1 isoforms or CD56 (120 kDa and 140 kDa) compared with levels in vector-control [VC] or untransfected [–] cells. Increased expression of NF-κB and bcl2L12 as well as nuclear translocation of p65 in RUNX1 p48 and CD56-transfected cell lines. By contrast, p38a and p24-transfected K562 cells showed clear down-regulation of NF-κB and bcl2L12 and no nuclear translocation of p65.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/6/10.1182_blood-2007-02-074203/7/m_zh80140705070004.jpeg?Expires=1769104116&Signature=U6GflDbCynFMzvyja3aDKZHf8uJqjjCuh4UpXGfpYPKMQJ~nDkENCcywCq55pOnPP6pITHBjqy8hrpNSQVIxpBqWe3CU~lnHD~YzPn~3vzo2x-tGgqsPi1H2DlztLVjXGKelAsPnBMISsUSxJOiHEONO8H1NcPYMS30CBB08Cej0G6kOV9MUCJBbnHTLyXwKbsUUyxN7ESEJtzZouuw4yL97Yky7IUXeSfsiy8lYNgTEeTVZbZnisQqA6nyV81ZgX2gEa1lfQp2mlooBLBauq~TrjH1BvjFbYFUT3Lfm-XFG0DWo-8OE0SRj-ofhZYjHDxvhpWzhCKZxOVzUb-65ng__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal