Abstract
CD4+CD25+Foxp3+ regulatory T cells (CD25+ Treg cells) direct the maintenance of immunological self-tolerance by active suppression of autoaggressive T-cell populations. However, the molecules mediating the anergic state and regulatory function of CD25+ Treg cells are still elusive. Using differential proteomics, we identified galectin-10, a member of the lectin family, as constitutively expressed in human CD25+ Treg cells, while they are nearly absent in resting and activated CD4+ T cells. These data were confirmed on the mRNA and protein levels. Single-cell staining and flow cytometry showed a strictly intracellular expression of galectin-10 in CD25+ Treg cells. Specific inhibition of galectin-10 restored the proliferative capacity of CD25+ Treg cells and abrogated their suppressive function. Notably, first identified here as expressed in human T lymphocytes, galectin-10 is essential for the functional properties of CD25+ Treg cells.
Introduction
A subset of CD4+ T cells, representing 5% to 10% of all peripheral CD4+ T cells in healthy individuals, constitutively expresses the IL-2 receptor α-chain (CD25).1 These CD4+CD25+ regulatory T cells (named CD25+ Treg cells) are hyporesponsive upon T-cell receptor (TCR) stimulation in vitro and suppress the proliferation of T effector cells.1,2 This suppression only occurs after activation of CD25+ Treg cells via their TCR, is strictly cell contact–dependent, mediated by yet-unknown surface molecules, and leads to inhibition of proliferation and cytokine production of cocultured CD4+ T cells.3,4 The main result of suppression appears to be the inhibition of IL-2 expression in the responder cell population.5 In addition to cell contact–dependent suppression, cell contact–independent mechanisms, mediated by a second generation of suppressor T cells via soluble factors like IL-10 and TGF-β (infectious tolerance), seem to be involved.6-8 Albeit the striking evidence that the transcriptional repressor Foxp3 seems to be essential for the development and function of CD25+ Treg cells,9,10 little is known about the molecular mechanisms and the proteins contributing to the in vitro hyporesponsive state and regulatory activity of these cells. A better understanding of their function on the protein level should provide more insights into a potential therapeutic exploitation of CD25+ Treg cells.
Using differential proteomics, we screened for proteins specifically expressed by human CD25+ Treg cells compared with CD4+CD25− T cells (in the following referred to as CD4+ T cells). The most differentially expressed protein was galectin-10, also known as Charcot-Leyden crystal protein,11,12 which has never been described in lymphocytes so far. Apart from its interaction with the eosinophil lysophospholipase,13 the regulation and function of galectin-10 is still unknown.
Here, we report that 3 isoforms of galectin-10 are constitutively expressed by human CD25+ Treg cells, but are nearly absent in CD4+ T cells. Because of its intracellular expression, galectin-10 seems not to be directly involved in the contact-dependent suppressive activity of CD25+ Treg cells. However, specific inhibition of galectin-10 restored the proliferative capacity of CD25+ Treg cells and abrogated their suppressive function.
Materials and methods
Isolation and stimulation of human T-cell populations
CD4+ T cells and CD25+ Treg cells were isolated from buffy coats and leukapheresis products (up to 1.5 × 1010 whole cells) of healthy volunteers as described before.7,8,14 Briefly, CD25+ cells were separated using limited amounts of CD25-microbeads (Miltenyi Biotec, Bergisch-Gladbach, Germany) resulting in CD25high cells. Afterward, contaminations of CD4− cells were depleted using CD14-, CD8-, and CD19-Dynabeads (Invitrogen/Dynal, Oslo, Norway), resulting in a purity of CD4+CD25high T cells greater than 95% (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). CD4+ T cells were isolated using CD4-microbeads; afterward, CD25+ T cells were depleted with CD25-Dynabeads (purity: CD4+ T cells greater than 98%). For isolation of CD25+ α4β1 Treg cells and CD25+ α4β7 Treg cells, CD4+ T cells were purified using CD4-Multisort beads (2 μL/107 peripheral blood mononuclear cells PBMCs; Miltenyi Biotec) as described before.7,8,14 For polyclonal activation of T cells, 1 μg/mL anti-CD3 (OKT-3) and 2 μg/mL anti-CD28 (CD28.2; BD Pharmingen, San Diego, CA) mAbs were used. For T-cell proliferation assays, T cells were suboptimally stimulated with 0.5 μg/mL anti-CD3 (OKT-3) in the presence of irradiated PBMCs as described before.14,16 If indicated, CD25+ Treg cells in coculture experiments were partially irradiated with 3000 rad (30 Gy) as indicated. All cultures were performed in serum-free X-VIVO-15 (Cambrex, Verviers, Belgium). For some experiments, CD25+ Treg cells and CD4+ T cells were labeled with Vybrant CFDA SE cell-tracker kit (Invitrogen, San Diego, CA) according to the manufacturer's protocol.
Proteome analysis
Up to 108 CD25+ Treg cells and CD4+ T cells were analyzed directly after isolation or after polyclonal activation for 48 hours. Proteomes of individual T-cell or eosinophil preparations13 (Figure S2) were studied using 2-dimensional gel electrophoresis (2D PAGE) and mass spectrometry in combination with bioinformatics. The gels cover a pI range from 4 to 10 and a molecular weight (MW) from 5 to 150 kDa according to a modified method by Klose and Kobalz.17,18
2D PAGE was performed using a combination of carrier ampholyte isoelectric focusing (IEF) and SDS-PAGE. IEF was performed in 400 mm × 0.9 mm rod gels containing 7 M urea and 2 M thiourea, 3.5% acrylamide, 0.3% piperazine diacrylamide, and 4% carrier ampholytes (pH 2-11). Approximately 80 μg of protein were loaded onto IEF gels focused under nonequilibrium pH gradient electrophoresis conditions. SDS-PAGE was performed in gels containing 15% acrylamide using the IEF gels as stacking gels. This method is mainly limited to soluble proteins while hydrophobic proteins and low abundance proteins are normally not displayed on a conventional two-dimensional gel. Two-dimensional gels were scanned as 16-bit grayscale images with 200 dpi image resolution and analyzed semiautomatically using the ProteomWeaver software (Definiens, Munich, Germany). Proteins were assumed as differentially produced when the fold change was greater than 2 or less than 0.5 in more than 66% of analyzed samples. For protein identification, protein spots were picked off the gels. After in-gel digestion with trypsin, the peptides were identified by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry using the Ultraflex mass spectrometer (Bruker Daltonics, Billerica, MA). Protein identification was achieved by using the search algorithms ProFound (Protometrix, Branford, CT) and Mascot (Matrix Science, Boston, MA).
Cloning, expression, and purification of galectin-10 fusion protein
Galectin-10 cDNA was amplified from human leukocyte Quick-Clone cDNA (BD Biosciences). The Echerichia coli strain BL21 (DE3) was transformed with the N-terminal His-tag galectin-10 construct (pET16b). Expression of His-galectin-10 fusion protein was induced with 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG; Sigma, Taufkirchen, Germany). Recombinant His-galectin-10 was purified using Ni-NTA affinity chromatography (Qiagen, Hilden, Germany). The purified protein was identified by MALDI mass spectrometry and used for the immunization of rabbits. For generation of the monoclonal galectin-10–specific B-F42, untagged galectin-10 was used.
RT-PCR and qRT-PCR
RNA was extracted from 106 cells using TRIZOL (GibcoBRL, Gaithersburg, MD) according to the manufacturer's instructions. RNA was reverse transcribed using RevertAid Moloney murine leukemia virus (M-MulV) reverse transcriptase (RT; MBI Fermentas, St Leon-Rot, Germany). Polymerase chain reaction (PCR) mixture contained 2.5 mM MgCl2, 0.2 mM dNTP, 0.5 μM forward and reverse primers, and 0.25 U of Biotherm DNA polymerase (GeneCraft, Lüdinghausen, Germany). To eliminate amplifications from contaminating genomic DNA, the following primers were designed to span an intron/exon boundary. Galectin-10 forward: 5′-TAC CCG TGC CAT ACA CAG AGG CTG-3′, reverse: 5′-CTT ATC TGG CAG CAC TGA GAT GCT C-3′; galectin-1 forward: 5′-CTG GTC GCC AGC AAC CTG AAT CTC-3′, reverse: 5′-AAG GTG ATG CAC ACC TCT GCA ACA C-3′; galectin-2 forward: 5′-CCG ATG GCA CTG ATG GCT TTG-3′, reverse: 5′-CTC AAA GGT CAC TGT GAA CTT G-3′; galectin-3 forward: 5′-CCA AAG AGG GAA TGA TGT TGC C-3′, reverse: 5′-TGA TTG TAC TGC AAC AAG TGA GC-3′; galectin-4 forward: 5′-TGT GCC TCC CAC AGG CAA GAG-3′, reverse: 5′-GCC ACA GCG AAT GGA CAG ATC-3′; galectin-7 forward: 5′-CTC CCA ATG CCA GCA GGT TCC-3′, reverse: 5′-CTT GAA GCC GTC GTC TGA CGC-3′; Galectin-8 forward: 5′-CTT TAA TGT TGA CCT ACT AGC AGG-3′, reverse: 5′-TTG TAC TCC AGG CTG TGT ACG C-3′; galectin-9 forward: 5′-CCG ATG CCT TTC ATC ACC ACC-3′, reverse: 5′-CAC CTT GAG GCA GTG AGC TTC-3′; galectin-12 forward: 5′-ACT GGT CTT GCA AGA GCC GAA G-3′, reverse: 5′-TCA GGA GTG GAC ACA GTA GAG C-3′; galectin-13 forward: 5′-AAT GAC CCA CAG CTG CAG GTG-3′, reverse: 5′-CGT AAA TGC GTA TGC CAT TGA CC-3′; 18S rRNA forward: 5′-TCG ATG CTC TTA GCT GAG TGT CC-3′, reverse: 5′-TGA TCG TCT TCG AAC CTC CG-3′, EF1a forward: 5′-GAT TAC AGG GAC ATC TCA GGC TG-3′, reverse: 5′-TAT CTC TTC TGG CTG TAG GGT GG -3′; Foxp3 forward: 5′-CTA CGC CAC GCT CAT CCG CTG G-3′, reverse: 5′-GTA GGG TTG GAA CAC CTG CTG GG-3′; β-actin forward: 5′-GAG CGG GAA ATC GTG CGT GAC ATT-3′, reverse: 5′-GAA GGT AGT TTC GTG GAT GCC-3′; and IL-2: Qiagen hIL-2 QuantiTect primer assay. Galectin-10 mRNA levels were quantified by quantitative RT-PCR (qRT-PCR) using the iCycler (Biorad, Munich, Germany) and the IQ SYBR Green Supermix Kit (Biorad). The relative expression levels of galectin-10 were normalized to 18S rRNA and EF1α.
Generation of polyclonal rabbit and monoclonal mouse anti–galectin-10 antibodies
Recombinant galectin-10 was used for immunization. A total of 50 μg protein solution was emulsified with an equal volume of complete Freund Adjuvant (CFA) and injected intradermally into several sites along the back of a rabbit. Booster injections were given subcutaneously 3 times in 3-week intervals in incomplete Freund Adjuvant (IFA). Antibody production was monitored by enzyme-linked immunosorbent assay (ELISA) and Western blotting. After the final bleedings, IgG was isolated from the antisera. A mouse mAb against galectin-10 (B-F42) was generated from BALB/cJ mice immunized 5 times with recombinant untagged galectin-10. Briefly, mice were injected intraperitoneally with 10 μg galectin-10 in 100 μL CFA. A total of 3 more intraperitoneal injections were given at 2-week intervals in IFA, followed by a final boost of the recombinant protein alone intravenously 96 hours before fusion.
Specificity of the mouse anti–galectin-10 mAb was confirmed by ELISA and by staining of galectin-10–transfected Jurkat cells. Isotypic mAb and rabbit preimmune serum served as controls.
Immunofluorescence staining and FACS methodology for detecting intracellular galectin-10
Cytospin preparations of freshly isolated CD4+ T cells and CD25+ Treg cells were air-dried and stored at −20°C until staining. For staining, slides were fixed in 4% paraformaldehyde for 15 minutes at room temperature. After washing and permeabilization with 0.2% Triton X-100, cytospins were stained with 15 μg/mL B-F42 for 1 hour, washed in PBS, and then detected with 10 μg/mL anti-mouse Oregon Green (Invitrogen) for an additional 30 minutes. Surface staining was performed with anti–CD3-PE (BD Pharmingen) for 1 hour, followed by nuclear staining with 100 ng/mL DAPI for 5 minutes. Coverslips were mounted on slides with a drop of mounting medium (Invitrogen). Immunefluorescence microscopy was carried out with a Spot insight camera (model no. 3.1.0; Diagnostic Instruments Inc, Sterling Heights, MI) mounted over an Axiovert S100 microscope (Zeiss, Göttingen, Germany) using a 32×/0.4 objective or a 63×/1.25 oil immersion objective (Zeiss). For image acquisition and processing, Meta Imaging Series 6.1 imaging software (Universal Imaging Corporation, Downingtown, PA) was used. For fluorescence-activated cell sorter (FACS) analysis, 5 × 105 PE–anti-human CD4 (BD Pharmingen) surface-stained cells were fixed in 1 mL PBS with 1% paraformaldehyde and 0.05% Tween-20 overnight at 4°C. Cells were washed with FACS buffer (PBS 1 ×, 3% fetal calf serum, 0.5% Tween-20) and incubated with an Oregon-Green–labeled (Molecular Probes/Invitrogen, Eugene, OR) mouse anti–galectin-10 (clone B-F42). After washing in FACS buffer cells were analyzed using a FACSCalibur with CELLQuest software (Becton Dickinson, San Jose, CA).
siRNA preparation and nucleofection
A total of 2 19-bp sequences were selected from the galectin-10 sequence and synthesized with an additional 2 bp overhang (Ambion, Huntington, United Kingdom). The dsRNA that displayed the highest suppressive activity to knock down the galectin-10 mRNA in CD25+ Treg cells was selected as follows: galectin-10 sense: GGA GGA AUC AGA CAU UGU CdTdT; galectin-10 antisense: GAC AAU GUC UGA UUC CUC CdTdT. Nucleofection was performed according to Amaxa's optimized protocol for T-cell nucleofection using the primary Human T Cell Nucleofector Kit (Amaxa, Gaithersburg, MD). Cells were immediately nucleofected and resuspended in prewarmed X-VIVO-15. Suppressive activity of siRNA was measured 24 to 48 hours after nucleofection by qRT-PCR and by galectin-10 staining. Transfection efficiency was controlled by transfection with fluoresencent siRNA and by transfection with a green fluorescent protein (GFP)–expressing vector (Amaxa). Cell recovery after transfection and incubation for an additional 48 hours was typically around 50% (as determined by propidium iodide PI staining). Dead cells were depleted using a dead cell removal kit (Miltenyi Biotec) according to the manufacturer's instructions, resulting in a viability greater than 92% (Figure S5).
Western blotting
Cells were lysed in SDS buffer, and protein concentration was determined using the DC protein assay (BioRad). Protein samples (5-10 μg) were resolved on 16% tricine SDS-PAGE and blotted on membranes. After blocking with Roti-Block solution (Roth, Karlsruhe, Germany) for 1 hour, membranes were incubated with anti–galectin-10 antibody or anti–β-actin in combination with anti-rabbit horseradish peroxidase conjugate. Membranes were stained with DAB.
Caspase and annexin V/PI assay of transfected CD4+CD25− T cells
T cells were resuspended in Nucleofector solution (Amaxa) and mixed with 2.5 μg of the galectin-10 expression vector or the empty vector pcDNA 3.1. Cells were immediately nucleofected and resuspended in prewarmed X-VIVO-15. Transfection efficiency was typically around 60% to 65%, controlled by transfection with a GFP-expressing vector and by immunostaining of galectin-10–transfected Jurkat cells. After dead cell removal via Annexin-coupled beads (Miltenyi Biotec), the activities of the active caspase-3 and caspase-7 were measured (Apo-ONE Homogeneous Caspase-3/7 Assay; Promega, Madison, WI) as specified by the manufacturer. The amount of fluorescent product generated is representative to the amount of active caspase-3/7 present in the sample. A total of 3 × 105 CD4+ T cells per sample were used. After 15 hours, a SpectraFluor reader (TECAM, Crailsheim, Germany) was used for detection of relative fluorescence intensities. In addition, cells were stained for Annexin V and PI using an AnnexinV-PI Kit (BD Pharmingen).
Cytokine assays
In order to analyze cytokine profiles of CD25+ Treg cells with and without suppressed galectin-10 expression in single culture and coculture with CD4+ T cells, supernatants of these cultures were analyzed by Cytometric Bead Array (BD Pharmingen) according to the manufacturer's protocol.
Statistical analysis
Statistical analysis was performed using StatView for Windows software (SAS Institute, Cary, NC). P values of .05 or less were considered significant (indicated by single asterisks in the figures). P values of .005 or less were considered highly significant (indicated by double asterisks in the figures).
Online supporting material
Figure S1 shows the isolation method and FACS analysis of freshly isolated CD25+ Treg cells. Figure S2 shows expression of galectin-10 isoforms A-C in CD25+ Treg cells and human eosinophils. Figure S3 displays the two-dimensional PAGE pattern of soluble proteins derived from human CD25+ Treg cells as well as the galectin-10 mRNA and protein expression in CD4+ T cell and CD25+ α4β1 and CD25+ α4β7 Treg cell subsets. Figure S4 shows expression of galectins in CD25+ Treg cells versus CD4+ T cells. Resting and stimulated CD25+ Treg cells and CD4+ T cells were analyzed for the expression of the displayed galectins by RT-PCR. The transfection rate and viabilities of siRNA-transfected T cells are shown in Figure S5.
Results
Identification of galectin-10 in human CD25+ Treg cells
Naturally occurring CD25+ Treg cells are characterized by their unique capacity to suppress the activation of CD4+ T cells (Figure 1). However, the proteins involved in this cell contact–dependent process are still unknown. In order to identify proteins, which are preferentially expressed by human CD25+ Treg cells and potentially involved in their functional properties, we isolated CD25+ Treg cells and CD4+CD25− T cells (in the following referred to as CD4+ T cells) with high purity (Figure 1; Figure S1) and performed differential proteome analysis of resting and activated CD25+ Treg cells compared with resting and activated CD4+ T cells (Figure S3A). Approximately 1600 protein spots were matched comparing these distinct T-cell populations. Serial paired comparison of these protein spots from individual healthy volunteers showed a high reproducibility in patterns and spot intensities. Differentially expressed proteins were identified by MALDI mass spectrometry. Regarding activated CD25+ Treg cells, analysis showed increased spot intensities for 9 proteins and decreased intensities for 14 proteins and their isoforms compared with activated CD4+ T cells (Table 1). Known differentially expressed surface molecules like CD25 or transcription factors like Foxp3 were not detectable with this technique, since hydrophobic transmembrane proteins are hardly resolved in 2D PAGE, and nuclei are separated during protein isolation.
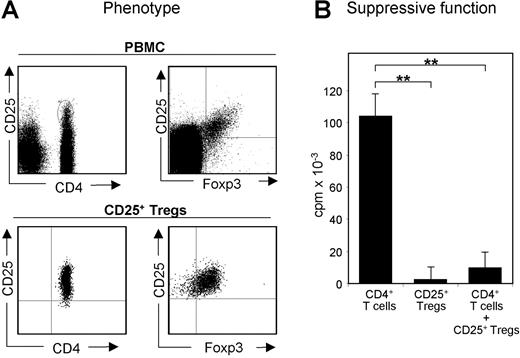
Phenotype and function of CD25+ Treg cells freshly isolated from leukapheresis products. CD4+ T cells and CD25+ Treg cells were isolated from buffy coats and leukapheresis products (up to 1.5 × 1010 whole cells).8 (A) PBMCs and freshly isolated CD25+ Treg cells were stained with the indicated markers using fluorescence-labeled mAb and analyzed by flow cytometry. (B) Functional testing of CD25+ Treg cells. A total of 1 × 105 freshly isolated human CD25+ Treg cells and CD4+ T cells were incubated with 3 × 105 irradiated (5000 rad, 50 Gy) PBMCs in the presence of 0.5 μg/mL anti-CD3. Proliferation was determined by 3H-Tdr incorporation on day 3; measurements were performed in triplicates.
Phenotype and function of CD25+ Treg cells freshly isolated from leukapheresis products. CD4+ T cells and CD25+ Treg cells were isolated from buffy coats and leukapheresis products (up to 1.5 × 1010 whole cells).8 (A) PBMCs and freshly isolated CD25+ Treg cells were stained with the indicated markers using fluorescence-labeled mAb and analyzed by flow cytometry. (B) Functional testing of CD25+ Treg cells. A total of 1 × 105 freshly isolated human CD25+ Treg cells and CD4+ T cells were incubated with 3 × 105 irradiated (5000 rad, 50 Gy) PBMCs in the presence of 0.5 μg/mL anti-CD3. Proliferation was determined by 3H-Tdr incorporation on day 3; measurements were performed in triplicates.
Differential expressed proteins: activated CD4+ T cells versus CD25+ Treg cells
| No. . | Protein . | Gene . | Accession no. . | Gene locus . | Expression ratio (n = 3) CD25+: CD4+ . |
|---|---|---|---|---|---|
| 8 | Galectin-10 | CLC | 17942629 | 19q13.1 | 42 |
| 12 | Tropomyosin 3 | TPM3 | 24119203 | 1q21.2 | 4 |
| 69 | Actin, beta, cytoplasmic isoform A | ACTB | 14250401 | 7p15-p12 | 3.3 |
| 14 | Actin, beta cytoplasmic isoform B | ACTB | 14250401 | 7p15-p12 | 3.2 |
| 15 | Actin, beta cytoplasmic isoform C | ACTB | 14250401 | 7p15-p12 | 2.4 |
| 42 | EH domain-containing 1 | EHD1 | 9956076 | 11q13 | 1.7 |
| 36 | Galectin 1 | LGALS1 | 4504981 | 22q13.1 | 1.6 |
| 89 | Actin gamma | ACTG1 | 178045 | 17q25.3 | 1.4 |
| 129 | Heterogeneous nuclear ribonucleoprotein A2/B1 isoform 2 | HNRPA2B | 4504447 | 7p15.2 | 1.2 |
| 112 | Tumor protein, translationally controlled 1 | TPT1 | 4507669 | 13q12-q14 | 0.8 |
| 135 | Homeobox prox 1 | PROX1 | 7706322 | 14q22.1 | 0.8 |
| 103 | FK506-binding protein 4 | FKBP4 | 4503729 | 12p13.33 | 0.8 |
| 26 | Stathmin 1 | STMN1 | 5031851 | 1p36.1-p35 | 0.6 |
| 2460 | MO25 protein | MO25 | 7706481 | 2q37.1 | 0.6 |
| 22 | Proteasome subunit beta type 7 | PSMB7 | 21465650 | 9q34.11-q34.12 | 0.6 |
| 115b | Nucleoside diphosphate kinase A isoform A | NME1 | 4557797 | 17q21.3 | 0.6 |
| 115a | Nucleoside diphosphate kinase A isoform B | NME1 | 4557797 | 17q21.3 | 0.6 |
| 39 | Ribonucleoside-diphosphate reductase isoform A | RRM1 | 4506749 | 11p15.5 | 0.5 |
| 38 | Ribonucleoside-diphosphate reductase isoform B | RRM1 | 4506749 | 11p15.5 | 0.5 |
| 48 | Flap structure-specific endonuclease 1 | FEN1 | 4758356 | 11q12 | 0.5 |
| 3 | dUTP pyrophosphatase–isoform A | DUT | 4503423 | 15q15-q21.1 | 0.4 |
| 30 | Eukaryotic translation initiation factor 5A | EIF5A | 4503545 | 17p13-p12 | 0.3 |
| 25 | dUTP pyrophosphatase–isoform B | DUT | 4503423 | 15q15-q21.1 | 0.3 |
| No. . | Protein . | Gene . | Accession no. . | Gene locus . | Expression ratio (n = 3) CD25+: CD4+ . |
|---|---|---|---|---|---|
| 8 | Galectin-10 | CLC | 17942629 | 19q13.1 | 42 |
| 12 | Tropomyosin 3 | TPM3 | 24119203 | 1q21.2 | 4 |
| 69 | Actin, beta, cytoplasmic isoform A | ACTB | 14250401 | 7p15-p12 | 3.3 |
| 14 | Actin, beta cytoplasmic isoform B | ACTB | 14250401 | 7p15-p12 | 3.2 |
| 15 | Actin, beta cytoplasmic isoform C | ACTB | 14250401 | 7p15-p12 | 2.4 |
| 42 | EH domain-containing 1 | EHD1 | 9956076 | 11q13 | 1.7 |
| 36 | Galectin 1 | LGALS1 | 4504981 | 22q13.1 | 1.6 |
| 89 | Actin gamma | ACTG1 | 178045 | 17q25.3 | 1.4 |
| 129 | Heterogeneous nuclear ribonucleoprotein A2/B1 isoform 2 | HNRPA2B | 4504447 | 7p15.2 | 1.2 |
| 112 | Tumor protein, translationally controlled 1 | TPT1 | 4507669 | 13q12-q14 | 0.8 |
| 135 | Homeobox prox 1 | PROX1 | 7706322 | 14q22.1 | 0.8 |
| 103 | FK506-binding protein 4 | FKBP4 | 4503729 | 12p13.33 | 0.8 |
| 26 | Stathmin 1 | STMN1 | 5031851 | 1p36.1-p35 | 0.6 |
| 2460 | MO25 protein | MO25 | 7706481 | 2q37.1 | 0.6 |
| 22 | Proteasome subunit beta type 7 | PSMB7 | 21465650 | 9q34.11-q34.12 | 0.6 |
| 115b | Nucleoside diphosphate kinase A isoform A | NME1 | 4557797 | 17q21.3 | 0.6 |
| 115a | Nucleoside diphosphate kinase A isoform B | NME1 | 4557797 | 17q21.3 | 0.6 |
| 39 | Ribonucleoside-diphosphate reductase isoform A | RRM1 | 4506749 | 11p15.5 | 0.5 |
| 38 | Ribonucleoside-diphosphate reductase isoform B | RRM1 | 4506749 | 11p15.5 | 0.5 |
| 48 | Flap structure-specific endonuclease 1 | FEN1 | 4758356 | 11q12 | 0.5 |
| 3 | dUTP pyrophosphatase–isoform A | DUT | 4503423 | 15q15-q21.1 | 0.4 |
| 30 | Eukaryotic translation initiation factor 5A | EIF5A | 4503545 | 17p13-p12 | 0.3 |
| 25 | dUTP pyrophosphatase–isoform B | DUT | 4503423 | 15q15-q21.1 | 0.3 |
The ratios of protein spot intensities comparing activated CD4+ T cells with activated CD25+ Treg cells are shown. Proteins from whole-cell lysates were separated by 2D gel electrophoresis. Corresponding protein spots were matched using the ProteomWeaver image analysis software, and intensities were acquired. Intensity ratios were calculated on the basis of 2D PAGE from sets of T cells derived from 3 individual human volunteers performed as triplicates.
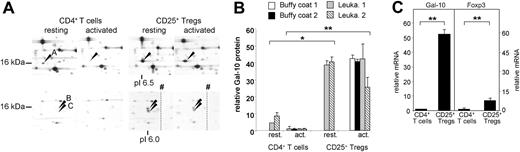
Among all differentially expressed proteins, galectin-10 showed the most striking difference in expression. 3 isoforms (A-C) of galectin-10 were identified on the protein level, which differ in their pI and MW (Figure 2A). All isoforms were detected in CD25+ Treg cells but were either marginally expressed or undetectable in CD4+ T cells. Resting as well as activated CD25+ Treg cells showed a strong expression of the prominent isoform A while in resting CD4+ T cells, its expression was low and even decreased after activation. When activated T-cell populations were compared, CD25+ Treg cells expressed 40 times more galectin-10 than did CD4+ T cells on a protein level (Figure 2B). The isoforms B and C were expressed in resting as well as activated CD25+ Treg cells with comparable intensities, but nearly absent in resting or activated CD4+ T cells. 2D PAGE analysis of human eosinophils also showed an expression of the previous unknown isoforms B and C in these cells (Figure S2). Taken together, in resting human T cells galectin-10 is predominantly expressed in CD25+ Treg cells and upon activation, CD25+ Treg cells almost exclusively express galectin-10.
Differential expression of galectin-10 protein and mRNA in CD4+CD25− T cells and CD25+ Treg cells. (A) Selected gel regions showing the different protein spot intensities of galectin-10 isoforms A-C in resting and activated human CD4+ T cells and CD25+ Tregs. Due to gel size, the IEF rod gels were cut in 2 halves at approximately pI 6.0 to 6.4 before further processing. Arrowheads show cutting edges, which were due to technical reasons. In this context, please also see Figure S6. (B) The ratios of protein spot intensities were calculated on the basis of 2D PAGE from sets of T cells derived from 4 individual human volunteers (performed as triplicates each; mean plus or minus SD from each triplicate shown; asterisks indicate P values as defined in “Materials and methods, Statistical analysis”). For proteome analysis, freshly isolated (leukapheresis products) as well as stimulated (leukapheresis products and buffy coats) T-cell populations were used. Due to limited recovery of CD25+ Treg cells isolated from buffy coats, only stimulated T-cell populations were analyzed. (C) Quantification of relative galectin-10 (Gal-10) mRNA levels compared with Foxp3 in freshly isolated CD4+ T cells and CD25+ Treg cells. cDNA samples were subjected to qRT-PCR. The relative quantities of galectin-10 and Foxp3 mRNA were normalized according to the expression of EF1-α mRNA. The qRT-PCR data are representative of 3 independent experiments; measurements were performed in triplicates; mean plus or minus SD from each triplicate shown; asterisks indicate P values as defined in “Materials and methods; Statistical analysis”.
Differential expression of galectin-10 protein and mRNA in CD4+CD25− T cells and CD25+ Treg cells. (A) Selected gel regions showing the different protein spot intensities of galectin-10 isoforms A-C in resting and activated human CD4+ T cells and CD25+ Tregs. Due to gel size, the IEF rod gels were cut in 2 halves at approximately pI 6.0 to 6.4 before further processing. Arrowheads show cutting edges, which were due to technical reasons. In this context, please also see Figure S6. (B) The ratios of protein spot intensities were calculated on the basis of 2D PAGE from sets of T cells derived from 4 individual human volunteers (performed as triplicates each; mean plus or minus SD from each triplicate shown; asterisks indicate P values as defined in “Materials and methods, Statistical analysis”). For proteome analysis, freshly isolated (leukapheresis products) as well as stimulated (leukapheresis products and buffy coats) T-cell populations were used. Due to limited recovery of CD25+ Treg cells isolated from buffy coats, only stimulated T-cell populations were analyzed. (C) Quantification of relative galectin-10 (Gal-10) mRNA levels compared with Foxp3 in freshly isolated CD4+ T cells and CD25+ Treg cells. cDNA samples were subjected to qRT-PCR. The relative quantities of galectin-10 and Foxp3 mRNA were normalized according to the expression of EF1-α mRNA. The qRT-PCR data are representative of 3 independent experiments; measurements were performed in triplicates; mean plus or minus SD from each triplicate shown; asterisks indicate P values as defined in “Materials and methods; Statistical analysis”.
Analysis of galectin-10 expression
Because expression of galectin-10 in CD25+ Treg cells was a novel finding, we determined whether other members of the galectin family could also be observed in human T-cell populations. As summarized in Figure S4, among the galectins detected, galectin-10 showed the strongest differences in expression comparing CD25+ Treg cells with CD4+ T cells. PCR analysis confirmed a dominant expression of galectin-10 by CD25+ Treg cells, which was even more pronounced compared with the protein level. Quantification of galectin-10 mRNA demonstrated an approximately 50-fold higher expression in CD25+ Treg cells (Figure 2C).
In order to analyze galectin-10 in CD25+ Treg cells on a single-cell basis, a rabbit antiserum and a mouse mAb (B-F42) against recombinant galectin-10 were generated. Specificity of both was verified by ELISA and staining of galectin-10–transfected Jurkat cells (not shown). The IgG fraction of the rabbit antiserum detects all 3 isoforms of galectin-10 in western blots (Figure S2), whereas B-F42 is suitable for flow cytometry.
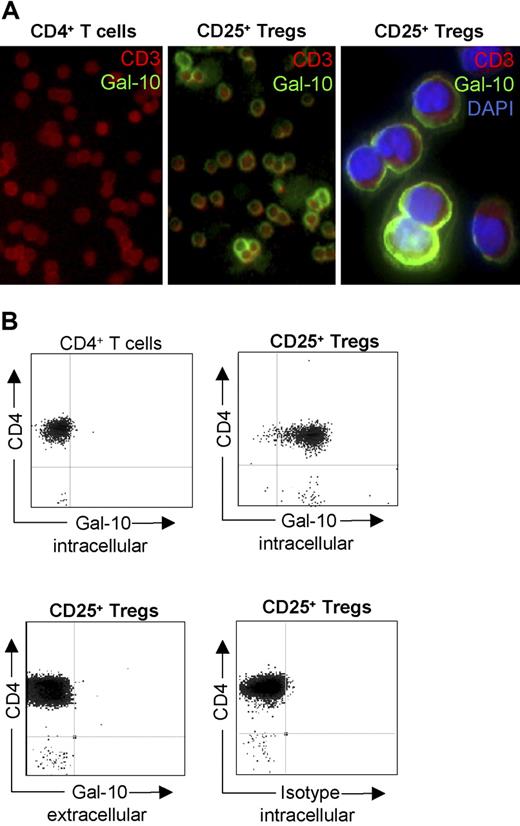
As shown in Figure 3A,B, galectin-10 expression was strictly located to the cytosol of human CD25+ Treg cells. Staining revealed different intensities of galectin-10 expression within this population, whereas it was not detectable in CD4+ T cells. Furthermore, no evidence for membrane expression or secretion of galectin-10 was found.
Galectin-10 is predominantly expressed in CD25+ Treg cells. (A) Cytospin preparations of freshly isolated T cells were incubated with the galectin-10–specific mAb B-F42 and detected with anti-mouse Oregon-Green. In addition, cells were stained with an anti-CD3 PE mAb (red fluorescence), and DAPI (blue fluorescence; right panel only). (B) Furthermore, freshly isolated T cells stained with galectin-10–specific mAb B-F42 Oregon-Green in combination with anti-CD4 PE were analyzed by flow cytometry. Representative results out of 6 independent experiments are shown.
Galectin-10 is predominantly expressed in CD25+ Treg cells. (A) Cytospin preparations of freshly isolated T cells were incubated with the galectin-10–specific mAb B-F42 and detected with anti-mouse Oregon-Green. In addition, cells were stained with an anti-CD3 PE mAb (red fluorescence), and DAPI (blue fluorescence; right panel only). (B) Furthermore, freshly isolated T cells stained with galectin-10–specific mAb B-F42 Oregon-Green in combination with anti-CD4 PE were analyzed by flow cytometry. Representative results out of 6 independent experiments are shown.
Staining of galectin-10 in CD25+ Treg cells revealed 2 subpopulations that differ in the intensity of galectin-10 expression. Since the pool of CD25+ Treg cells consists of different subsets, we also examined the expression of galectin-10 in the α4β1+ and α4β7+ Treg cells. Regarding galectin-10 we observed—in contrast to Foxp3 (not shown)—an up to 7-fold higher expression of galectin-10 in α4β7+ Treg cells compared with the α4β1+ subset (Figure S3B,C).
Functional significance of galectin-10 in human CD25+ Treg cells
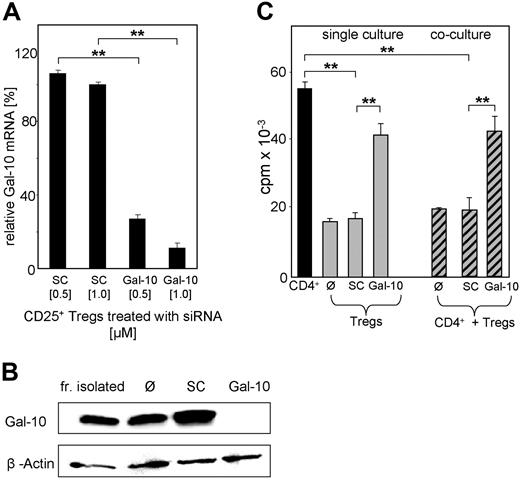
Functional consequences of galectin-10 expression in human CD25+ Treg cells were analyzed by different blocking experiments. Galectin-10–specific antibodies and recombinant galectin-10 protein had no effect on cultures of CD4+ T cells and/or CD25+ Treg cells, excluding a direct role for galectin-10 in the cell contact–dependent suppressive mechanism of CD25+ Treg cells (not shown). To uncover the functional role of strictly intracellular-expressed galectin-10, knock down experiments using galectin-10-specific siRNA were performed. Transfection with siRNA (transfection rates of up to 90%; Figure S5A) resulted in a dose-dependent suppression of galectin-10 mRNA (Figure 4B) and a homogenous down-regulation of galectin-10 protein in CD25+ Treg cells (Figure 4B). In contrast, siRNA-mediated knockdown did not affect the expression of surface molecules, such as CD25, CTLA-4, CD45RO, and CD62L, or the transcription factor Foxp3 (not shown). More importantly, galectin-10–specific siRNA treatment reversed the hyporesponsiveness of human CD25+ Treg cells in vitro, resulting in increased proliferation upon activation, virtually to the level of CD4+ T cells, while treatment with scrambled control siRNA had no effect (Figure 4C). Moreover, transfection of CD4+ T cells did not influence their proliferation (not shown). These data suggest that galectin-10 is essential for the anergic state of human CD25+ Treg cells in vitro. Moreover, the question arose whether this protein is also involved in the suppressive properties of CD25+ Treg cells. To evaluate this possibility, we analyzed the functional properties of siRNA-treated CD25+ Treg cells in suppression assays with CD4+ T cells. As shown in Figure 4C, inhibition of galectin-10 partially abrogated the suppressive activity of human CD25+ Treg cells, resulting in an enhanced proliferation of cocultured CD4+ T cells. CFSE labeling confirmed this finding (Figure 5A,B). Thus, inhibition of galectin-10 not only restored the proliferative capacity of human CD25+ Treg cells, but also abrogated their suppressive function. Furthermore, blockade of galectin-10 synthesis resulted in an up-regulated cytokine production by the CD25+ Treg cells themselves (Table 2).
Galectin-10 specific siRNA abolishes the anergic state and suppressive activity of human CD25+ Tregs. (A) Freshly isolated CD25+ Treg cells were nucleofected with 0.5 or 1 μM of siRNA targeted to galectin-10 (Gal-10) or with control siRNA (scrambled control SC). At 24 hours after nucleofection, RNA was prepared and used for qRT-PCR analysis. The expression level was normalized to expression of EF1-α. The qRT-PCR data are representative of 4 independent experiments; measurements were performed in triplicates (mean ± SD from each triplicate shown; asterisks indicate P values as defined in “Materials and methods, Statistical analysis”). (B) At 48 hours after nucleofection, galectin-10 protein expression was examined by galectin-10–specific Western blot analysis. (C) At 48 hours after nucleofection, T cells were stimulated with anti-CD3 mAb (0.5 μg/mL) and irradiated PBMCs. CD25+ Treg cells in coculture with CD4+CD25− T cells were irradiated. Proliferation was determined by 3H-Tdr incorporation. Representative results of 4 independent experiments are shown; measurements were performed in triplicates (mean ± SD) from each triplicate are shown; asterisks indicate P values as defined in “Materials and methods, Statistical analysis”).
Galectin-10 specific siRNA abolishes the anergic state and suppressive activity of human CD25+ Tregs. (A) Freshly isolated CD25+ Treg cells were nucleofected with 0.5 or 1 μM of siRNA targeted to galectin-10 (Gal-10) or with control siRNA (scrambled control SC). At 24 hours after nucleofection, RNA was prepared and used for qRT-PCR analysis. The expression level was normalized to expression of EF1-α. The qRT-PCR data are representative of 4 independent experiments; measurements were performed in triplicates (mean ± SD from each triplicate shown; asterisks indicate P values as defined in “Materials and methods, Statistical analysis”). (B) At 48 hours after nucleofection, galectin-10 protein expression was examined by galectin-10–specific Western blot analysis. (C) At 48 hours after nucleofection, T cells were stimulated with anti-CD3 mAb (0.5 μg/mL) and irradiated PBMCs. CD25+ Treg cells in coculture with CD4+CD25− T cells were irradiated. Proliferation was determined by 3H-Tdr incorporation. Representative results of 4 independent experiments are shown; measurements were performed in triplicates (mean ± SD) from each triplicate are shown; asterisks indicate P values as defined in “Materials and methods, Statistical analysis”).
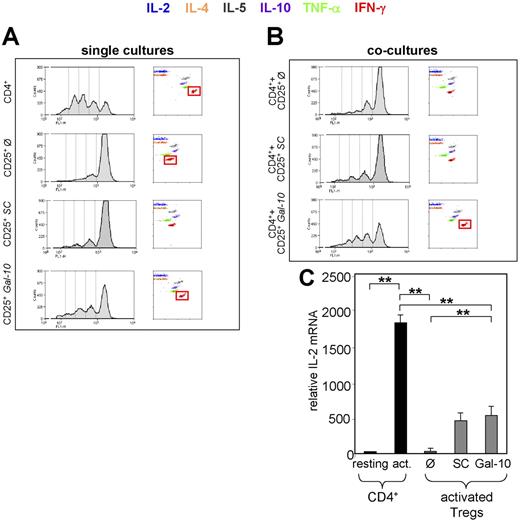
Suppressed expression of galectin-10 in CD25+ Treg cells alters their anergic phenotype, cytokine production, and suppressive activity. Freshly isolated CD25+ Treg cells were nucleofected with 1 μM siRNA targeted to galectin-10 (Gal-10) or control siRNA (scrambled control SC). At 48 hours after nucleofection, CFSE labeled and unlabeled T cells were stimulated with anti-CD3 mAb (0.5 μg/mL) and irradiated PBMCs. (A) Proliferation and cytokine release of the CFSE labeled T-cell populations in single culture. CFSE profile: FACS analysis of CFSE+ T cells determined proliferation on day 3 of stimulation. Cytokine release: cytokine release for the indicated cytokines was determined in supernatants derived from CFSE stimulation cultures on day 3, using human Cytometric Bead Array. (B) CFSE profile and cytokine release of CD4+ T cells in coculture with CD25+ Treg cells (untreated/siRNA-treated) in a ratio of 1:2. Cytokine release was determined as described. Results are representative of 4 independent experiments. (C) IL-2 mRNA expression in nucleofected CD25+ Treg cells and CD4+CD25− T cells. Freshly isolated CD25+ Treg cells were nucleofected with 1 μM siRNA targeted to galectin-10 (Gal-10) or control siRNA (scrambled control SC). At 48 hours after nucleofection, T cells were stimulated with anti-CD3 mAb (1 μg/mL) and anti-CD28 mAb (2 μg/mL) and after an additional 4 hours, RNA was prepared and used for qRT-PCR analysis. The expression level of IL-2 was normalized to the expression of EF1-α. Results are representative of 3 independent experiments; measurements were performed in triplicates (mean ± SD from each triplicate are shown; asterisks indicate P values as defined in “Materials and methods, Statistical analysis”).
Suppressed expression of galectin-10 in CD25+ Treg cells alters their anergic phenotype, cytokine production, and suppressive activity. Freshly isolated CD25+ Treg cells were nucleofected with 1 μM siRNA targeted to galectin-10 (Gal-10) or control siRNA (scrambled control SC). At 48 hours after nucleofection, CFSE labeled and unlabeled T cells were stimulated with anti-CD3 mAb (0.5 μg/mL) and irradiated PBMCs. (A) Proliferation and cytokine release of the CFSE labeled T-cell populations in single culture. CFSE profile: FACS analysis of CFSE+ T cells determined proliferation on day 3 of stimulation. Cytokine release: cytokine release for the indicated cytokines was determined in supernatants derived from CFSE stimulation cultures on day 3, using human Cytometric Bead Array. (B) CFSE profile and cytokine release of CD4+ T cells in coculture with CD25+ Treg cells (untreated/siRNA-treated) in a ratio of 1:2. Cytokine release was determined as described. Results are representative of 4 independent experiments. (C) IL-2 mRNA expression in nucleofected CD25+ Treg cells and CD4+CD25− T cells. Freshly isolated CD25+ Treg cells were nucleofected with 1 μM siRNA targeted to galectin-10 (Gal-10) or control siRNA (scrambled control SC). At 48 hours after nucleofection, T cells were stimulated with anti-CD3 mAb (1 μg/mL) and anti-CD28 mAb (2 μg/mL) and after an additional 4 hours, RNA was prepared and used for qRT-PCR analysis. The expression level of IL-2 was normalized to the expression of EF1-α. Results are representative of 3 independent experiments; measurements were performed in triplicates (mean ± SD from each triplicate are shown; asterisks indicate P values as defined in “Materials and methods, Statistical analysis”).
Altered IFN-γ and TNF-α release in CD25+ Treg cells after suppressed galectin-10 expression
| Population . | IFN-γ, median pg/mL (range) . | TFN-α, medan pg/mL (range) . |
|---|---|---|
| CD4+ | 3013* (929-<5000) | 538* (350-803) |
| CD25+ Ø | 128* (57-224) | 16* (10-20) |
| CD25+ SC | 177 (76-354) | 19 (12-25) |
| CD25+ Gal-10 | 825* (324-1558) | 55* (39-81) |
| CD4+ + CCD25+ Ø | 343 (117-558) | 17 (11-23) |
| CD4+ + CD25+ SC | 421 (200-684) | 21 (13-27) |
| CD4+ + CD25+ Gal-10 | 2290* (719-4518) | 89* (60-133) |
| Population . | IFN-γ, median pg/mL (range) . | TFN-α, medan pg/mL (range) . |
|---|---|---|
| CD4+ | 3013* (929-<5000) | 538* (350-803) |
| CD25+ Ø | 128* (57-224) | 16* (10-20) |
| CD25+ SC | 177 (76-354) | 19 (12-25) |
| CD25+ Gal-10 | 825* (324-1558) | 55* (39-81) |
| CD4+ + CCD25+ Ø | 343 (117-558) | 17 (11-23) |
| CD4+ + CD25+ SC | 421 (200-684) | 21 (13-27) |
| CD4+ + CD25+ Gal-10 | 2290* (719-4518) | 89* (60-133) |
Data are the results of 4 independent experiments performed as shown in Figure 5. Cytokine release was determined in supernatants derived from CFSE stimulation cultures on day 3, using human Cytometric Bead Array.
Restored proliferation and the increased cytokine production after down-regulation of galectin-10 emphasizes the thesis that the function of human CD25+ Treg cells is dependent on galectin-10 expression. The weak induction of IL-2 mRNA showed that nucleofection is not a neutral process (Figure 5C) but, on its own, did not reverse the functional activity of CD25+ Treg cells as shown by scrambled control siRNA.
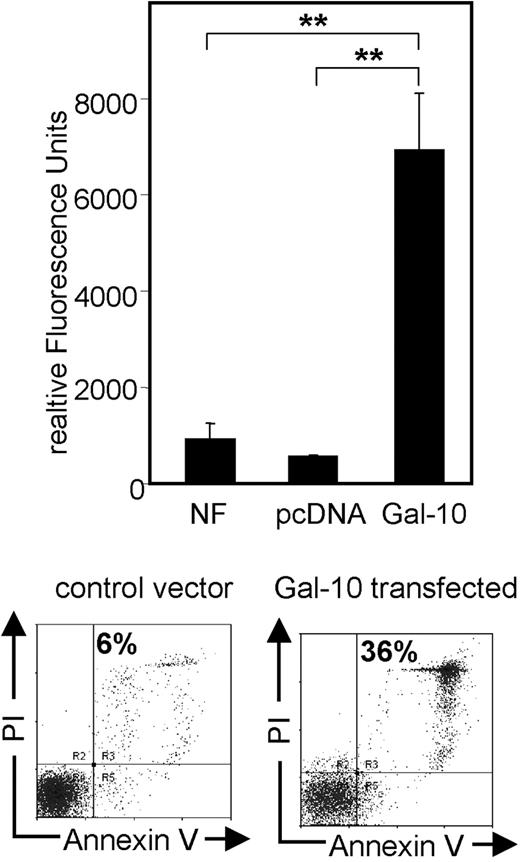
To investigate the functional properties of galectin-10 in more detail, we also tested different methods for functional transfection of CD4+ T cells with this protein. Transfection of CD4+ T cells with galectin-10—but not with control vectors—immediately induced apoptosis in combination with strong activation of caspase-3 and caspase-7 (Figure 6). Even though different viral and nonviral transfection methods were used, induction of apoptosis was the only observed result after ectopic expression of galectin-10 in CD4+ T cells.
Enhanced caspase activity and apoptosis in CD4+ T cells after transfection with galectin-10. T cells were resuspended in Nucleofector solution and mixed with 2.5 μg of the galectin-10 expression vector or the empty vector pcDNA 3.1. Cells were immediately nucleofected and resuspended in prewarmed X-VIVO-15. Transfection efficiency was typically around 60% to 65%, controlled by transfection with a GFP-expressing vector and by immunostaining of galectin-10–transfected Jurkat cells. After dead cell removal via Annexin-coupled beads (Miltenyi Biotec), the activities of the active caspase-3 and caspase-7 were measured (Apo-ONE Homogeneous Caspase-3/7 Assay; Promega) as specified by the manufacturer. The amount of fluorescent product generated is representative to the amount of active caspase-3/7 present in the sample. A total of 3 × 105 CD4+ T cells per sample were used. After 15 hours, a SpectraFluor reader was used for detection of relative fluorescence intensities. Results are representative of 3 independent experiments; measurements were performed in triplicates (mean ± SD from each triplicate are shown; asterisks indicate P values as defined in “Materials and methods, Statistical analysis”). In addition, cells were stained for Annexin V and PI and analyzed via FACS.
Enhanced caspase activity and apoptosis in CD4+ T cells after transfection with galectin-10. T cells were resuspended in Nucleofector solution and mixed with 2.5 μg of the galectin-10 expression vector or the empty vector pcDNA 3.1. Cells were immediately nucleofected and resuspended in prewarmed X-VIVO-15. Transfection efficiency was typically around 60% to 65%, controlled by transfection with a GFP-expressing vector and by immunostaining of galectin-10–transfected Jurkat cells. After dead cell removal via Annexin-coupled beads (Miltenyi Biotec), the activities of the active caspase-3 and caspase-7 were measured (Apo-ONE Homogeneous Caspase-3/7 Assay; Promega) as specified by the manufacturer. The amount of fluorescent product generated is representative to the amount of active caspase-3/7 present in the sample. A total of 3 × 105 CD4+ T cells per sample were used. After 15 hours, a SpectraFluor reader was used for detection of relative fluorescence intensities. Results are representative of 3 independent experiments; measurements were performed in triplicates (mean ± SD from each triplicate are shown; asterisks indicate P values as defined in “Materials and methods, Statistical analysis”). In addition, cells were stained for Annexin V and PI and analyzed via FACS.
Discussion
Different technologies, including differential display and microarray technology, have been used so far to compare gene and protein expression patterns of T-cell subpopulations.19,20 Here, we used the high-resolution 2D PAGE analysis combined with MALDI mass spectrometry in order to identify proteins differentially expressed in human CD25+ Treg cells. We compared expression patterns of separated T-cell protein preparations in which more than 1600 protein spots were detected per gel. The displayed proteins are mostly soluble and therefore mainly cytosolic, since hydrophobic proteins like transmembrane proteins and receptors are hardly resolved in 2D PAGE based on isoelectric focusing in the first and SDS PAGE in the second dimension. The 2D PAGE comparison of both resting and activated CD4+ T cells and CD25+ Treg cells showed a very similar protein pattern, with more than 90% consensus even when different sources like leukapheresis or buffy coats from different healthy volunteers were used in individual experiments. The most differentially expressed protein was identified as galectin-10. Resting as well as activated CD25+ Treg cells showed prominent expression of galectin-10, while this protein was nearly absent in CD4+ T cells.
Thus far, galectin-10 has been exclusively described in human eosinophils and basophils.21-23 Although galectin-1 was shown to be preferentially expressed in murine CD25+ Treg cells,19 there was no evidence for the expression of galectin-10 or any protein with comparable properties in the murine system.21-23 In human eosinophils, galectin-10 is a major constituent comprising an estimated 7% to 10% of total cellular protein, with comparable amounts expressed by basophils and forms hexagonal bipyramidal crystals (Charcot-Leyden crystal).21,24 Although we were able to confirm the dominant expression of galectin-10 in human CD25+ Treg cells, there was no evidence of crystal formation in these cells. Moreover, no galectin-10 could be detected on the cell surface (Figure 3A,B) or in the supernatants of resting and activated human CD25+ Treg cells (data not shown), suggesting a strictly intracellular function of galectin-10.
Apart from the interaction of galectin-10 with the eosinophil lysophospholipase,13 its regulation and function in human eosinophils and basophils has remained undefined. The observation that CD25+ Treg cells express galectin-10 is new, and provides a novel marker for the phenotypic characterization of CD25+ Treg cells. Unlike other markers described in human CD25+ Treg cells (eg, CD25 or CTLA-4), galectin-10 is not an activation marker. It therefore seems to be valuable to distinguish CD25+ Treg cells from CD4+ T cells, even when the immune system has been perturbed and effector cells are generated. More important, our data imply that galectin-10 is essentially involved in the suppressive properties of human CD25+ Treg cells.
A total of 3 isoforms of galectin-10, which differ in their pI and MW, were identified in CD25+ Treg-cell lysates. Expression of the prominent isoform (A: 16 kDa, pI 6.5) is up to 40-fold higher in CD25+ Treg cells compared with CD4+ T cells. The other 2 isoforms are almost exclusively expressed by CD25+ Treg cells.
Staining of galectin-10 in CD25+ Treg cells revealed 2 subpopulations, which differ in the intensity of galectin-10 expression. Since the pool of naturally occurring CD25+ Treg cells consist of different subsets, we also examined the expression of galectin-10 in the α4β1+ and α4β7+ Treg cells. Both subsets, to the same degree, contact-dependently suppress coactivated CD4+ T cells and convey their suppressive activity in the process of “infectious tolerance” by generating secondary T helper suppressor cells (Thsup cells).6-8 However, the properties of CD25+ Treg subsets are rather distinct: α4β7+ Treg cells induce IL-10–producing Thsup cells (Tr1-like), whereas α4β1+ Treg cells induce TGF-β–producing Thsup cells (Th3-like). Regarding galectin-10, we observed—in contrast to Foxp3 (not shown)—an up to 7-fold higher expression of galectin-10 in α4β7+ Treg cells compared with the α4β1+ subset (Figure 3B-C), suggesting that elevated levels of galectin-10 in CD25+ Treg cells might also play a role for the induction of distinct subsets of secondary Thsup cells.
To characterize a putative functional role of galectin-10 in CD25+ Treg cells, we inhibited galectin-10 expression in these cells by specific siRNA. Down-regulation abolished the anergic state of CD25+ Treg cells and partially abrogated their suppressive capacity. Moreover, the blockade of galectin-10 synthesis partially restored the IFN-γ and TNF-α production of CD25+ Treg cells themselves and in coculture with CD4+ T cells, emphasizing a general role for galectin-10 in the function of human CD25+ Treg cells. The fact that low amounts of IL-2 mRNA were also induced is a result of the nucleofection, detectable after transfection with scrambled control siRNA as well as after nucleofection without any siRNA (Figure 5C). Nevertheless, nucleofection with scrambled control siRNA—in contrast to galectin-10 siRNA—did not break the suppressive activities of human CD25+ Treg cells.
Concerning the regulation of galectin-10, it is described that in promyelocytic leukemia cells, expression of galectin-10 is induced by butyric acid.25,26 We assessed the effect of butyric acid on the expression and functional activity of galectin-10 in human CD4+ T cells and CD25+ Treg cells, but could not detect any functional alteration (data not shown). The most recent link to a functional role of galectin-10 in human T cells is the observation of an up-regulation in Th2 central memory T cells, after stimulation with thymic stromal lymphopoietin–activated dendritic cells,27 showing that maybe only specific T-cell subsets that possess a differentiated and activated phenotype are able to express galectin-10, while CD4+ T cells show only a marginal expression, which even decreases after activation.
Ectopic expression of galectin-10 in CD4+ T cells could possibly reveal its role in T-cell homeostasis. However, transfection of these cells with a vector encoding for galectin-10 immediately initiated the activation of caspases and subsequently induced apoptosis28,29 (Figure 6), which suggests that galectin-10 expressed in the environment of a specific transcriptional structure is involved in the functional properties of CD25+ Treg cells, whereas it possesses proapoptotic functions in the absence of these explicit requisites.
The fact that galectin-1 and galectin-3 were already found to participate in T-cell proliferation and regulation15 emphasizes a novel role of galectins in T-cell regulation. Perillo et al showed that dimeric galectin-1, expressed by stroma cells in thymus and lymph nodes, is involved in the induction of apoptosis in mature T cells.30 In contrast, up-regulation and secretion of monomeric galectin-1 switches off the effector function in T lymphocytes by arresting cell-cycle progression at the level of the S and G2/M stages, working as an autocrine negative-growth factor.31-33 Furthermore, galectin-1 is sufficient to suppress experimental autoimmune uveitis (EAU) by promoting concomitant Th2- and Treg-mediated anti-inflammatory responses.34 Consistently, adoptive transfer of CD4+ T cells obtained from galectin-1–treated mice prevented the development of active EAU in syngenic recipients, suggesting that galectin-1 favored the expansion of a subpopulation of IL-10– and/or TGF-β–secreting CD4+CD25− Treg cells. More recently, it was demonstrated that galectin-1 has a crucial role for Treg function.35 Blockade of galectin-1 by specific antibodies resulted in a decreased suppressive activity of human CD25+ Treg cells in vitro and CD25+ Treg cells of galectin-1–null−/− mice are significant less suppressive than their wild-type counterparts. In addition, recent evidence indicates that galectin-3 could also act in an ambivalent manner, either protecting T cells from apoptosis or inducing cell death, depending on whether the protein is expressed in the intracellular compartments36 or conveys its function extracellularly.37 Which galectin or galectin-like protein could be a functional analog for galectin-10 in other species such as mice, needs to be investigated.
In summary, galectin-10 is dominantly expressed in human CD25+ Treg cells, suggesting that a consistent expression of galectin-10 is necessary for the maintenance of Treg cell–specific functions. Down-regulation via siRNA abolishes their anergy and partially abrogates their suppressive capacity, indicating that galectin-10 is essential for the functional properties of human CD25+ Treg cells. However, further characterization of galectin-10 and interaction with binding partners, as well as the identification of analog proteins in murine CD25+ Treg cells, are necessary for better understanding of the physiologic role of galectins in T-cell regulation.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Drs S. J. Ackerman, D. Kabelitz, E. M. Shevach, M. Stassen, and K. Steinbrink for critical reading of the manuscript and helpful discussions. We also thank M. Schmidt-Habrich, L. Paragnik, S. Fondel, P. Hoelter, M. Milo, A. Dommermuth, and E. Schwitulla for their expert technical assistance, as well as S. Bailey for the protein identification, B. Bonin for generating the galectin-10–specific antibody B-F42, and Dr A. Baiker for the transfection of 293 T cells.
This work was supported by the Deutsche Forschungsgemeinschaft, grant A6 SFB548 (to E.S.) and grant A8 SFB548 (to H.J.), the European Fonds for Regional Development, and the government of Nordrhein-Westfalen, Germany (to Protagen AG).
Authorship
Contribution: J. Kubach designed and performed research, analyzed data, and wrote the paper; P.L. designed and performed research, and analyzed data; T.B. designed and performed research, and analyzed data; S.S. contributed new analytical tools, performed research, and analyzed data; C.B. contributed new reagents and performed research; E.H. performed research; C.R. performed research; P.W. designed and performed research; T.W. performed research; J. Knop designed research; S.M. designed research; J.W. contributed vital new reagents; H.S. designed research; E.S. designed research; and H.J. designed research and wrote the paper.
Conflict-of-interest disclosure: P.L. is an employee of Protagen AG, S.M. and P.W. are employees and shareholders of Protagen AG, and J.W. is an employee of Diaclone SAS. All other employees declare no competing financial interests.
Correspondence: Helmut Jonuleit, Department of Dermatology, Johannes Gutenberg-University, Langenbeckstr. 1, 55101 Mainz, Germany; e-mail: jonuleit@hautklinik.klinik.uni-mainz.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal