Abstract
AML1/RUNX1 is implicated in leukemogenesis on the basis of the AML1-ETO fusion transcript as well as somatic mutations in its DNA-binding domain. Somatic mutations in RUNX1 are preferentially detected in acute myeloid leukemia (AML) M0, myeloid malignancies with acquired trisomy 21, and certain myelodysplastic syndrome (MDS) cases. By correlating the presence of RUNX1 mutations with cytogenetic and molecular aberration in a large cohort of AML M0 (N = 90) at diagnosis, we detected RUNX1 mutations in 46% of cases, with all trisomy 13 cases (n = 18) being affected. No mutations of NRAS or KIT were detected in the RUNX1-mutated group and FLT3 mutations were equally distributed between RUNX1-mutated and unmutated samples. Likewise, a high incidence of RUNX1 mutations (80%) was detected in cases with trisomy 13 from other French-American-British (FAB) subgroups (n = 20). As FLT3 is localized on chromosome 13, we hypothesized that RUNX1 mutations might cooperate with trisomy 13 in leukemogenesis by increasing FLT3 transcript levels. Quantitation of FLT3 transcript levels revealed a highly significant (P < .001) about 5-fold increase in AML with RUNX1 mutations and trisomy 13 compared with samples without trisomy 13. The results of the present study indicate that in the absence of FLT3 mutations, FLT3 overexpression might be a mechanism for FLT3 activation, which cooperates with RUNX1 mutations in leukemogenesis.
Introduction
Acute myeloid leukemia (AML) is the consequence of a multistep process with the accumulation of multiple recurring genetic mutations in hematopoietic stem cells. Mutation targets in this process can be subdivided into two complementation groups.1 The first group confers a proliferation/survival advantage and is represented mainly by members of the RAS and the receptor tyrosine kinase families. Fms-like tyrosine kinase 3 (FLT3)-length mutations (FLT3-LMs or frequently called FLT3-internal tandem duplications [FLT3-ITDs]) in the juxtamembrane domain as well as mutations in the second tyrosine kinase domain of FLT3 (FLT3-TKD) confer a ligand-independent constitutive activation of downstream signaling pathways.2,3
Hematopoietic transcription factors belong to the second complementation group.1 Members of this group include AML1/RUNX1 and CBFB coding for the 2 components of the heterodimeric core binding factor (CBF).4 Both genes are involved in chromosomal translocations associated with acute leukemia.5 Approximately 8% to 15% of adult AML patients are affected by the translocation t(8;21)(q22;q22), which generates the AML1-ETO (RUNX1-RUNX1T1) fusion transcript.6 Less frequent are variant translocations of RUNX1 on chromosome 21q22 with other partners like EVI1 on 3q267 or ETV6 on 12p13.8 In transgenic mice, conditional expression of the AML1-ETO fusion gene per se was shown not to cause overt leukemia. However, the treatment of such mice with N-ethyl-N-nitrosourea induced hematopoietic neoplasms including AML, indicating that collaborating mutations that can be induced by this agent are necessary.9 Respectively, AML1-ETO in combination with a FLT3-LM/ITD, which is suspected to be one of the most frequent collaborating mutations in AML, induced aggressive acute leukemia in a murine bone marrow transplantation model.10
In addition to deregulated expression by translocations, RUNX1 gene function was found to be impaired by point mutations.11,12 Recently, haploinsufficiency of RUNX1 has been identified as the reason for autosomal familial platelet disorder (FPD), which predisposes to the development of AML.13 At the same time, somatic mutations of RUNX1 have been identified in various AML subtypes and myelodysplastic syndromes (MDSs).11 Somatic mutations in the N-terminal DNA-binding domain, the so-called Runt domain, of RUNX1 were reported to occur with the highest incidence in AML M0 as well as in myeloid malignancies displaying acquired trisomy 21.14–16 In MDSs, somatic mutations were found to be located in the N-terminal Runt domain as well as in the C-terminal transactivation domain of RUNX1 and were detected in refractory anemia with excess blasts (RAEB), RAEB in transformation (RAEBt), and AML following MDS (the 3 categories were collectively termed MDS/AML). In these cases, RUNX1 mutations were associated with a significantly worse prognosis compared with cases with wild-type RUNX1.17 A significant association of RUNX1 mutations with loss of the long arm of chromosome 7 was reported in therapy-related MDS (t-MDS) cases. Here, RUNX1 mutations were correlated with a predisposition to leukemic transformation.18
Consistent with the model of cooperation between mutations of genes of the two complementation groups, a recent report detected a significant association of inactivating RUNX1 point mutations with activating FLT3 mutations in the AML M0 subtype.16 Later this finding was extended to cooperation of RUNX1 mutations with activating mutations in the ras/receptor tyrosine kinase (rtk) signaling pathway in AML M019 but also in MDS/AML.20 An alternative to FLT3 activation by mutation, overexpression of FLT3 was recently proposed to be an additional mechanism that was associated with unfavorable prognosis.21,22 Gene amplification was suggested as one possible mechanism for FLT3 overexpression.22,23
In this study we analyzed the incidence of RUNX1 mutations in a large cohort of M0 AML (N = 90) and selected AML of other subtypes (n = 20) and correlated the results to cytogenetic aberrations as well as to FLT3 mutations of the samples. A significant association of RUNX1 mutation was detected to trisomy 13 but, in contrast to previous studies, not to FLT3 mutations. The presence of trisomy 13 was also found to be significantly associated with an AML1 mutation in other AML French-American-British (FAB) subtypes (n = 20). FLT3 expression was significantly increased in trisomy 13 samples, indicating that FLT3 expression levels might be the factor on chromosome 13 that collaborates with AML1 mutation in leukemia development.
Patients, materials, and methods
Patients
Fresh blood or bone marrow from 156 patients was analyzed after informed written consent was obtained in accordance with the Declaration of Helsinki. Diagnosis of AML was according to standard FAB and World Health Organization (WHO) criteria.24–26 Patients were referred to our laboratory for cytomorphologic, immunophenotypic, cytogenetic, and molecular analyses. Of the 156 patients, 115 were diagnosed as AML M0, 9 as AML M1, 15 as AML M2, 12 as AML M4, 1 as AML M5, and 1 was a secondary AML M2 after chronic myeloproliferative disorder (CMPD). Three AML cases were not further classified according to FAB criteria. The AML M0 cohort was unselected, whereas the samples of the other FAB subgroups were specifically selected for the presence of trisomy 13 or as the control group for the respective FLT3 analysis. Clinical sample characteristics are given in Table 1. Approval for this study was obtained from the Bayerische Landesärztekammer (Bavarian Medical Association).
Patients' characteristics
| AML patients . | N=156 . |
|---|---|
| Median age, y (range) | (63.3 (20–86) |
| Sex, M/F, n | 98/58 |
| AML M0 | 115 |
| AML M1 | 9 |
| AML M2 | 15 |
| AML M4 | 12 |
| AML M5 | 1 |
| s-AML M2 | 1 |
| AML (not further FAB classified) | 3 |
| Median WBC, /μL (range) | 13 300 (49–330 000) |
| Median platelet count, 109/L (range) | 65 000 (49–330 000) |
| Median hemoglobin level, g/dL (range) | 8.9 (3.0–13.8) |
| Median blast count, % (range) | 82 (21–99) |
| AML patients . | N=156 . |
|---|---|
| Median age, y (range) | (63.3 (20–86) |
| Sex, M/F, n | 98/58 |
| AML M0 | 115 |
| AML M1 | 9 |
| AML M2 | 15 |
| AML M4 | 12 |
| AML M5 | 1 |
| s-AML M2 | 1 |
| AML (not further FAB classified) | 3 |
| Median WBC, /μL (range) | 13 300 (49–330 000) |
| Median platelet count, 109/L (range) | 65 000 (49–330 000) |
| Median hemoglobin level, g/dL (range) | 8.9 (3.0–13.8) |
| Median blast count, % (range) | 82 (21–99) |
WBC indicates white blood cell count.
Mutation analysis of RUNX1
Mononucleated bone marrow cells were obtained by Ficoll density gradient centrifugation. The preparation of mRNA and cDNA was as previously described.27 The entire coding region of the RUNX1 gene was amplified from cDNA using 4 separate polymerase chain reactions (PCRs). For AML M0, amplicons 1 and 2 were analyzed surrounding the runt homology domain (RHD), whereas for the other FAB subtypes the analysis was extended to the entire coding region of RUNX1. Primer sequences are as previously described,16 with some modifications (Table 2). The PCR amplification of amplicons 1-3 (Table 2) was performed using the proofreading enzyme Optimase polymerase (Transgenomic, Omaha, NE) according to the instructions of the manufacturer. Amplicon 4 (Table 2) was amplified using the Taq PCR Master Mix Kit (Qiagen GmbH, Hilden, Germany). A touch-down protocol was applied for all PCR reactions, with preheating of the samples at 97°C for 2 minutes followed by 15 touch-down cycles, 20 cycles at constant annealing temperature, and a final elongation step at 72°C for 5 minutes. The touch-down cycles started with denaturation for 30 seconds at 97°C, annealing for 1 minute at 63°C, and elongation for 1 minute at 72°C, where the annealing temperature was decreased by 0.5°C after each cycle. The residual 20 PCR cycles were performed at 58°C annealing temperature.
Sequences of primers used for PCR amplification and sequencing of RUNX1
| Amplicon . | Forward primer . | Reverse primer . | Nucleotide position* . |
|---|---|---|---|
| 1 | 5′-tgcagggtcctaactcaatc-3′ | 5′-cattgccagccatcacagtgac-3′ | 1548-1906 |
| 2 | 5′-tttcaaggtggtggcccta-3′ | 5′-ctgagggttaaaggcagtggagt-3′ | 1842-2283 |
| 3 | 5′-ccgggagcttgtccttttcc-3′ | 5′-cggcaggtaggtgtggtag-3′ | 2144-2643 |
| 4 | 5′-caggcgccttcacctactc-3′ | 5′-tgacctacagcgagatcctg-3′ | 2546-3100 |
| Amplicon . | Forward primer . | Reverse primer . | Nucleotide position* . |
|---|---|---|---|
| 1 | 5′-tgcagggtcctaactcaatc-3′ | 5′-cattgccagccatcacagtgac-3′ | 1548-1906 |
| 2 | 5′-tttcaaggtggtggcccta-3′ | 5′-ctgagggttaaaggcagtggagt-3′ | 1842-2283 |
| 3 | 5′-ccgggagcttgtccttttcc-3′ | 5′-cggcaggtaggtgtggtag-3′ | 2144-2643 |
| 4 | 5′-caggcgccttcacctactc-3′ | 5′-tgacctacagcgagatcctg-3′ | 2546-3100 |
Nucleotide positions are according to GenBank entry D43968 of the human RUNX1 transcript.
DHPLC and DNA sequencing
The denaturing high-performance liquid chromatography (DHPLC) analysis was performed on a WAVE 3500 HT system (Transgenomic) with a DNASep Cartridge (Transgenomic) and Navigator Software 1.6.4 (Transgenomic). DNA was bound to the cartridge by 0.1 M triethylammonium acetate (TEAA) in H2O (pH 7.0; buffer A; Transgenomic) in the mobile phase at a flow rate of 1.5 mL/min. Increasing amounts of buffer B (0.1 M TEAA, 25% acetonitrile in H2O; pH 7.0; Transgenomic) were used to elute the DNA off the cartridge. DNA was detected by UV absorption (260 nm).
PCR products for amplicons 1-3 of the RUNX1 cDNA were obtained as described in “Mutation analysis of RUNX1.” For heteroduplex formation, the PCR products of the amplicons of each patient sample were mixed with equal amounts of the respective PCR product derived from a healthy control, denatured at 95°C for 5 minutes, followed by cooling to 4°C at a rate of 0.1°C/s. DHPLC analyses were done at the appropriate temperatures. Mutations were detected as aberrant elution profile of the PCR product from the cartridge and were verified by direct sequencing using BigDye chemistry (Applied Biosystems, Weiterstadt, Germany). To evaluate the sensitivity of mutation detection by DHPLC, we diluted PCR products of 2 homozygously mutated cases (case 34: R139X, 415C>T; case 40: R174Q, 521G>A) with increasing amounts of wild-type (wt) PCR products. While a dilution of the mutations down to 1:32 could be detected by DHPLC analysis, a 1:8 dilution already resulted in a background signal-to-noise ratio by direct sequencing. However, all mutations detected by DHPLC in this study could be verified by direct sequencing.
Screening of FLT3, NRAS, and KIT mutations and quantification of FLT3 transcripts
Cytogenetic analysis
Cytogenetic G-banding analysis was performed according to standard methods.30 The definition of a cytogenetic clone and description of karyotypes followed the International System for Human Cytogenetic Nomenclature (ISCN).31 The criteria for classification of a complex aberrant karyotype were as previously described.32
Statistical analysis
The correlation between RUNX1 mutation status and cytogenetics was assessed by chi-square test. The significance of the differences in FLT3 expression was evaluated by t test. For statistical analysis, SPSS (version 14.0) software (SPSS, Chicago, IL) was used.
Results
Frequency of RUNX1 mutations in the AML M0 cohort
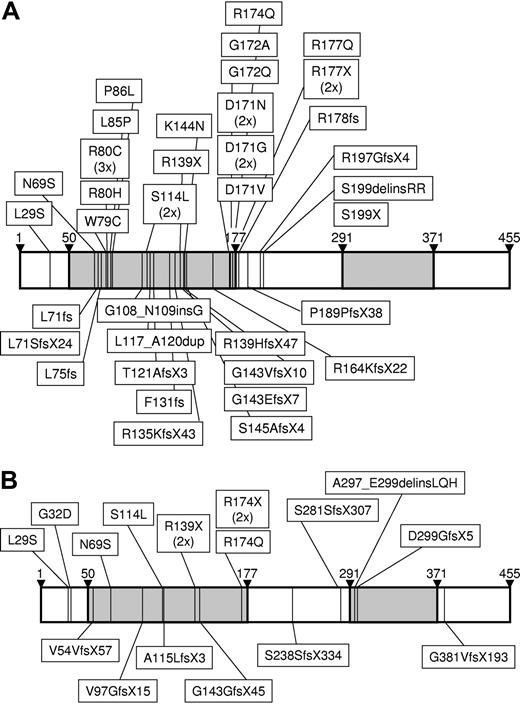
A total of 90 AML M0 samples were tested at diagnosis for RUNX1 mutations in the 227 N-terminal amino acids (aa's) of the RUNX1β transcript including the DNA-binding runt homology domain (RHD; aa's 50-177). RUNX1 mutations were detected in 41 (46%) of 90 cases and were mostly located inside the DNA-binding RHD (aa's 50-177), whereas only 6 were outside the RHD, displaying a missense mutation at Leu29 and frame-shift insertions/deletions at Arg178 (n = 1), Pro189 (n = 1), Arg197 (n = 1), and Ser199 (n = 2; Figure 1A; Table 3). The spectrum of mutations as summarized in Table 3 was composed of 21 missense mutations, 19 frame/stop mutations, and 3 in-frame deletions/insertions. Analysis of these mutations by direct sequencing identified a homozygous pattern in at least 19 of these cases (44%). In all cases referred to as heterozygous, homozygosity cannot be excluded due to the possibility of contaminated nonleukemic cells that may simulate a heterozygous pattern. In 2 patients, the combination of 2 heterozygous missense mutations was detected (Asn69Ser/Arg80His and Lys144Asn/Gly172Ala). Interestingly, amino acid residues that are involved in direct DNA binding34 were affected by 11 of 21 missense mutations (Arg80, n = 4; Asn171, n = 5; Arg174, n = 1; and Arg177, n = 1). Additional missense mutations targeted amino acids known to undergo conformational changes upon CBFB binding (Asn69; n = 1) or to be involved in protein-protein interaction between RUNX1 and CBFB (Ser114, n = 2).34
Schematic presentation of the location of RUNX1 mutations. The horizontal bars represent the full linear polypeptide sequence from amino acid 1 to 455 of RUNX1β. Amino acid numbering is given above the bars. The DNA-binding RHD (amino acids 50-177) and the TA domain (amino acids 291-371) are indicated in gray. The location of mutations is indicated by thin vertical lines with the type of mutation on the protein level given in text boxes for (A) the AML M0 cohort and for (B) the non-AML M0 cases.
Schematic presentation of the location of RUNX1 mutations. The horizontal bars represent the full linear polypeptide sequence from amino acid 1 to 455 of RUNX1β. Amino acid numbering is given above the bars. The DNA-binding RHD (amino acids 50-177) and the TA domain (amino acids 291-371) are indicated in gray. The location of mutations is indicated by thin vertical lines with the type of mutation on the protein level given in text boxes for (A) the AML M0 cohort and for (B) the non-AML M0 cases.
Molecular and cytogenetic characterization of the AML M0 cohort
| Patient . | Diagnosis . | Karyotype . | RUNX1 mutation* . | RUNX1 allele status . | FLT3 mutation . | NRAS mutation . | KIT D816 . |
|---|---|---|---|---|---|---|---|
| 1 | AML M0 | 46,XY [25] | Ser114Leu | mut/wt | FLT3-TKD | ND | ND |
| 2 | AML M0 | 46,XY,der(5)t(5;11)(q22;?) [9],46,XY [11] | Leu71fs | mut/wt | FLT3-LM | wt | wt |
| 3 | AML M0 | 46,XY [25] | [Asn69Ser(+)Arg80His] | mut/wt | FLT3-LM | wt | wt |
| 4 | AML M0 | 92,XXYY,+1,dic(1;5)(p21;q13),der(2)t(2;3)(q31;?),+8,-17 [5],46,XY [20] | Asp171Asn | mut/wt | FLT3-LM/TKD | wt | wt |
| 5 | AML M0 | 47,XX,+14 [6],46,XX [10] | Pro86Leu | mut/mut | ND | ND | ND |
| 6 | AML M0/M2 | 46,XX,del(5)(q15q31) [22] | Pro189ProfsX38 | mut/wt | ND | ND | ND |
| 7 | AML M0 | 92,XXYY [15] | Leu117_Ala120dup | mut/wt | wt | ND | ND |
| 8 | AML M0 | 46,XY [24] | Leu75fs | mut/mut | wt | ND | ND |
| 9 | AML M0 | 46,XX [20] | Asp171Gly | mut/wt | wt | wt | wt |
| 10 | AML M0 | 46,XX [25] | Arg80Cys | mut/mut | wt | ND | ND |
| 11 | AML M0 | 46,XX [25] | Leu71SerfsX24 | mut/wt | wt | wt | wt |
| 12 | AML M0 | 46,XY [22] | Gly108_Asn109insGly | mut/mut | wt | wt | wt |
| 13 | AML M0 | 46,XY [15] | Asn112LysfsX5 | mut/wt | wt | wt | wt |
| 14 | AML M0 | 46,XY [25] | Arg80Cys | mut/mut | wt | ND | ND |
| 15 | AML M0 | 47,XY,+21 [2],48,XY,+21,+21 [14],46,XY [1] | Asp171Asn | mut/wt | wt | wt | wt |
| 16 | AML M0 | 46,XX [25] | Ser199delinsArgArg | mut/wt | wt | wt | wt |
| 17 | AML M0 | 46,XY [25] | Arg80Cys | mut/mut | wt | wt | wt |
| 18 | AML M0 | 46,XY,der(2)t(2;5)(p23;q15)ins(2;12)(q23;q21q24), der(4)ins(4;17)(p14;??)t(4;5)(p15;?),del(5)(q15), der(12)t(2;12)(p23;p13)del(12)(q21),der(17)t(4;17)(p15;q11), del(17)(p13) [cp15],46,XY [5] | [Lys144Asn(+)Gly172Ala] | mut/wt | wt | wt | wt |
| 19 | AML M0 | 45,X,-Y [8],46,XY [17] | Arg177X | mut/wt | wt | wt | wt |
| 20 | AML M0 | 46,XX,del(7)(q11.2) [2],46,XX [23] | Arg139HisfsX47 | mut/wt | wt | wt | wt |
| 21 | AML M0 | 46,XY,i(17)(q10) [17] | Gly143ValfsX10 | mut/mut | wt | wt | wt |
| 22 | AML M0 | 46,XX,r(7)(p13q11.2) [19],46,XX [1] | Asp171Gly | mut/mut | wt | wt | wt |
| 23† | AML M0 | 46,XY.ish der(11)(MLL3′),der(10)(MLL5′) [24] | Leu29Ser | mut/wt | wt | ND | ND |
| 24 | AML M0 | 47,XY,+13 [11],46,XY [9] | Gly172Gln | mut/mut | FLT3-LM | ND | ND |
| 25 | AML M0 | 46,XX,der(19)t(13;19)(q11;p13) [6],46,XX [5] | Arg135LysfsX43 | mut/wt | FLT3-LM | ND | ND |
| 26 | AML M0 | 47,XY,+13 [16],46,XY [4] | Thr121AlafsX3 | mut/wt | ND | ND | ND |
| 27 | AML M0 | 94,XXYY,+13,+13 [13]46,XY [9] | Ser199X | mut/mut | wt | ND | ND |
| 28 | AML M0 | 47,XY,+13 [7],46,XY [8] | Arg197GlyfsX4 | mut/wt | wt | ND | ND |
| 29 | AML M0 | 47,XY,+13 [10],46,XY [2] | Phe131fs | mut/wt | wt | wt | wt |
| 30 | AML M0 | 47,XY,+13 [7],46,XY [5] | Ser114Leu | mut/wt | wt | wt | wt |
| 31 | AML M0 | 47,XY,+13 [5],45,X,-Y [8],46,XY [12] | Leu85Pro | mut/mut | wt | ND | ND |
| 32 | AML M0 | 47,XY,+13 [9],47,XY,del(7)(q22),+13 [2],46,XY [6] | Arg177Gln | mut/wt | wt | wt | wt |
| 33 | AML M0 | 47,XY,+13 [6],94,XXYY,+13,+13 [2],46,XY [6] | Arg178fs | mut/wt | wt | wt | wt |
| 34 | AML M0 | 47,XX,+13 [1],46,XX [2] | Arg139X | mut/mut | wt | wt | wt |
| 35 | AML M0 | 47,XY,+13 [10],94,XXYY,+13,+13 [1],46,XY [9] | Gly143GlufsX7 | mut/mut | wt | ND | ND |
| 36 | AML M0 | 47,XY,+13 [2],46,XY [12] | Trp79Cys | mut/mut | wt | wt | wt |
| 37 | AML M0 | 47,XY,+13 [18],46,XY [3] | Asp171Val | mut/mut | wt | wt | wt |
| 38 | AML M0 | 47,XY,+13 [6],46,XY [14] | Ser145AlafsX4 | mut/mut | wt | wt | wt |
| 39 | AML M0 | 47,XY,+13 [7],45,XY,-7 [5],46,XY [8] | Arg164LysfsX22 | mut/mut | wt | ND | ND |
| 40 | AML M0 | 47,XY,+13 [16],46,XY [2] | Arg174Gln | mut/mut | wt | ND | ND |
| 41 | AML M0 | 46,XX,-7,+13 [3],46,XX [17] | Arg177X | mut/mut | wt | ND | ND |
| 42 | AML M0 | 46,XY [11] | — | wt/wt | FLT3-TKD | wt | wt |
| 43 | AML M0 | 46,XY,t(11;19)(q13;p13) [17],46,XY [3] | — | wt/wt | FLT3-LM | ND | ND |
| 44 | AML M0 | 46,XY [25] | — | wt/wt | FLT3-LM | wt | wt |
| 45 | AML M0 | 46,XX [25] | — | wt/wt | FLT3-LM | wt | wt |
| 46 | AML M0 | 46,XX [20] | — | wt/wt | FLT3-LM | wt | wt |
| 47 | AML M0 | 46,XX,t(10;17)(p15;q21) [2],47,XX,+7,t(10;17)(p15;q21) [7],46,XX [16] | — | wt/wt | FLT3-LM | wt | wt |
| 48 | AML M0 | 47,XY,+8 [5],46,XY [15] | — | wt/wt | FLT3-LM | wt | wt |
| 49 | AML M0 | 43,XY,-3,der(3)t(3;7)(q11;q11),-5,der(7)t(5;7)(p11;q11), der(12)t(12;15)(p13;q22),-15,der(17)t(3;17)(q11;p13) [9],42,idem,-18 [7] | — | wt/wt | FLT3-LM | wt | wt |
| 50 | AML M0 | 55,XY,+X,+4,+5,+8,+10,+13,+14,+17,+18 [4],47,XY,+X [8] | — | wt/wt | FLT3-LM | ND | ND |
| 51 | AML M0 | 46,XY,del(11)(q13q21) [10],46,XY [15] | — | wt/wt | ND | ND | ND |
| 52 | AML M0 | 47,XY,+8,i(17)(q10) [20] | — | wt/wt | ND | ND | ND |
| 53 | AML M0 | 46,XX [25] | — | wt/wt | ND | ND | ND |
| 54 | AML M0 | 46,XX [7] | — | wt/wt | ND | ND | ND |
| 55 | AML M0 | 46,XX [12] | — | wt/wt | ND | ND | ND |
| 56 | AML M0 | 44,XY,add(3)(p13),-5,dic(11;?)(q14;?),add(11)(p15),-15,add(17)(p11) [13], 50–51,idem,+4,+5,+6,+6,+2–3mar [cp4],46,XY [3] | — | wt/wt | ND | ND | ND |
| 57 | AML M0 | 47,XX,+14 [19],46,XX [1] | — | wt/wt | ND | ND | ND |
| 58‡ | AML M0 | 46,XY [25],FISH-Screening: 65% Monosomie 13 oder grosse Deletion | — | wt/wt | ND | ND | ND |
| 59 | AML M0 | 47,XY,+22 [13],46,XY [20] | — | wt/wt | ND | ND | ND |
| 60 | AML M0 | 46,XX [15] | — | wt/wt | ND | ND | ND |
| 61 | AML M0 | 46,XY [25] | — | wt/wt | ND | ND | ND |
| 62 | AML M0 | 46,XY [20] | — | wt/wt | wt | mut | wt |
| 63 | AML M0 | 46,XX,-7,+19 [10],46,XX [14] | — | wt/wt | wt | ND | ND |
| 64 | AML M0 | 45,X,-Y,t(11;19)(q23;p13.3) [19],46,XY [1] | — | wt/wt | wt | wt | wt |
| 65 | AML M0 | 46,XX,t(9;11)(p22;q23) [12],46,XX [13] | — | wt/wt | wt | wt | wt |
| 66 | AML M0 | 45,XX,inv(3)(q21q26),-7 [20] | — | wt/wt | wt | wt | wt |
| 67 | AML M0 | 47,XY,+9 [2],54,XY,+4,+6,+8,+9,+11,+13,+13,+19 [4],46,XY [14] | — | wt/wt | wt | wt | wt |
| 68 | AML M0 | 46,XY [25] | — | wt/wt | wt | wt | wt |
| 69 | AML M0 | 46,XY [25] | — | wt/wt | wt | wt | wt |
| 70 | AML M0 | 46,XX [21] | — | wt/wt | wt | wt | wt |
| 71 | AML M0 | 46,XY [20] | — | wt/wt | wt | wt | wt |
| 72 | AML M0 | 46,XY [22] | — | wt/wt | wt | wt | wt |
| 73 | AML M0 | 42,XX,del(3)(p13),der(5)t(5;12)(q13;q15),dic(7;18)(p11;q11),i(8)(q10), dup(9)(q32q34),-12,der(13;15)(q10;q10), der(14)t(12;14)(p11;p10)t(1;14)(?;q32),-17 [15] | — | wt/wt | wt | wt | wt |
| 74 | AML M0 | 46,XY [20] | — | wt/wt | wt | wt | wt |
| 75 | AML M0 | 46,XY [21] | — | wt/wt | wt | mut | wt |
| 76 | AML M0 | 46,XX [25] | — | wt/wt | wt | ND | ND |
| 77 | AML M0 | 46,XY [20] | — | wt/wt | wt | wt | wt |
| 78 | AML M0 | 46,XY [22] | — | wt/wt | wt | wt | wt |
| 79 | AML M0 | 44–45,XX,del(4)(q23),der(4)t(4;7)(q23;?),del(5)(q13q31), der(7)del(7)(p15)t(4;7)(q23;q21),der(10)t(10;11)(q21;q13),-11, dic(12;17)(p13;p13),der(19)t(11;19)(?;q13)ins(11;19)(?;?)t(10;11) [cp12], 46,XX [2] | — | wt/wt | wt | wt | wt |
| 80 | AML M0 | 46,XX,del(5)(q13q31) [1],46,XX,t(1;3;9)(p11;q26;q34),del(5)(q13q31) [14] | — | wt/wt | wt | wt | wt |
| 81 | AML M0 | 46,XX,del(7)(q22q31) [19],46,XX [2] | — | wt/wt | wt | wt | wt |
| 82 | AML M0 | 45,XY,-7 [15] | — | wt/wt | wt | mut | wt |
| 83 | AML M0 | 45,XY,t(3;3)(q21;q26),-7 [11],46,XY [3] | — | wt/wt | wt | wt | wt |
| 84 | AML M0 | 46,XX,del(9)(q22) [4],46,XX [13] | — | wt/wt | wt | wt | wt |
| 85 | AML M0 | 46,XX,t(2;11)(p21;q23),del(5)(q13q33) [16],46,XX [6] | — | wt/wt | wt | ND | ND |
| 86 | AML M0 | 46,XY [15] | — | wt/wt | wt | ND | ND |
| 87 | AML M0 | 40–42,XY,-1,-5,-7,-8,-11,del(12)(p11),der(14)t(1;14)(p13;p11),-17, der(17)t(5;?;17)(q?;?;p12),-18,+2–4mar [cp16],46,XY [1],40,XY,-1,-5, add(7)(q11),der(8)t(1;8)(p13;p23),der(11)?del(11)(p13)del(11)(q21), del(12)(p11),add(14)(p11),der(17),add(1) | — | wt/wt | wt | ND | ND |
| 88 | AML M0 | 46,XY,t(3;4)(q25;q31),del(5)(q15),der(7)t(5;7)(q?;q11),del(15)(q15) [20] | — | wt/wt | wt | ND | ND |
| 89 | AML M0 | 46,XX,del(5)(q13q31),r(7)(p11q11),del(18)(q11) [20] | — | wt/wt | wt | wt | wt |
| 90 | AML M0 | 40–43,XY,del(5)(q13q31),der(8)t(8;12)(q21;q13),ins(11)(q13;?),-12, del(13)(q14),i(17)(q10) [cp6],46,XY [20] | — | wt/wt | wt | ND | ND |
| Patient . | Diagnosis . | Karyotype . | RUNX1 mutation* . | RUNX1 allele status . | FLT3 mutation . | NRAS mutation . | KIT D816 . |
|---|---|---|---|---|---|---|---|
| 1 | AML M0 | 46,XY [25] | Ser114Leu | mut/wt | FLT3-TKD | ND | ND |
| 2 | AML M0 | 46,XY,der(5)t(5;11)(q22;?) [9],46,XY [11] | Leu71fs | mut/wt | FLT3-LM | wt | wt |
| 3 | AML M0 | 46,XY [25] | [Asn69Ser(+)Arg80His] | mut/wt | FLT3-LM | wt | wt |
| 4 | AML M0 | 92,XXYY,+1,dic(1;5)(p21;q13),der(2)t(2;3)(q31;?),+8,-17 [5],46,XY [20] | Asp171Asn | mut/wt | FLT3-LM/TKD | wt | wt |
| 5 | AML M0 | 47,XX,+14 [6],46,XX [10] | Pro86Leu | mut/mut | ND | ND | ND |
| 6 | AML M0/M2 | 46,XX,del(5)(q15q31) [22] | Pro189ProfsX38 | mut/wt | ND | ND | ND |
| 7 | AML M0 | 92,XXYY [15] | Leu117_Ala120dup | mut/wt | wt | ND | ND |
| 8 | AML M0 | 46,XY [24] | Leu75fs | mut/mut | wt | ND | ND |
| 9 | AML M0 | 46,XX [20] | Asp171Gly | mut/wt | wt | wt | wt |
| 10 | AML M0 | 46,XX [25] | Arg80Cys | mut/mut | wt | ND | ND |
| 11 | AML M0 | 46,XX [25] | Leu71SerfsX24 | mut/wt | wt | wt | wt |
| 12 | AML M0 | 46,XY [22] | Gly108_Asn109insGly | mut/mut | wt | wt | wt |
| 13 | AML M0 | 46,XY [15] | Asn112LysfsX5 | mut/wt | wt | wt | wt |
| 14 | AML M0 | 46,XY [25] | Arg80Cys | mut/mut | wt | ND | ND |
| 15 | AML M0 | 47,XY,+21 [2],48,XY,+21,+21 [14],46,XY [1] | Asp171Asn | mut/wt | wt | wt | wt |
| 16 | AML M0 | 46,XX [25] | Ser199delinsArgArg | mut/wt | wt | wt | wt |
| 17 | AML M0 | 46,XY [25] | Arg80Cys | mut/mut | wt | wt | wt |
| 18 | AML M0 | 46,XY,der(2)t(2;5)(p23;q15)ins(2;12)(q23;q21q24), der(4)ins(4;17)(p14;??)t(4;5)(p15;?),del(5)(q15), der(12)t(2;12)(p23;p13)del(12)(q21),der(17)t(4;17)(p15;q11), del(17)(p13) [cp15],46,XY [5] | [Lys144Asn(+)Gly172Ala] | mut/wt | wt | wt | wt |
| 19 | AML M0 | 45,X,-Y [8],46,XY [17] | Arg177X | mut/wt | wt | wt | wt |
| 20 | AML M0 | 46,XX,del(7)(q11.2) [2],46,XX [23] | Arg139HisfsX47 | mut/wt | wt | wt | wt |
| 21 | AML M0 | 46,XY,i(17)(q10) [17] | Gly143ValfsX10 | mut/mut | wt | wt | wt |
| 22 | AML M0 | 46,XX,r(7)(p13q11.2) [19],46,XX [1] | Asp171Gly | mut/mut | wt | wt | wt |
| 23† | AML M0 | 46,XY.ish der(11)(MLL3′),der(10)(MLL5′) [24] | Leu29Ser | mut/wt | wt | ND | ND |
| 24 | AML M0 | 47,XY,+13 [11],46,XY [9] | Gly172Gln | mut/mut | FLT3-LM | ND | ND |
| 25 | AML M0 | 46,XX,der(19)t(13;19)(q11;p13) [6],46,XX [5] | Arg135LysfsX43 | mut/wt | FLT3-LM | ND | ND |
| 26 | AML M0 | 47,XY,+13 [16],46,XY [4] | Thr121AlafsX3 | mut/wt | ND | ND | ND |
| 27 | AML M0 | 94,XXYY,+13,+13 [13]46,XY [9] | Ser199X | mut/mut | wt | ND | ND |
| 28 | AML M0 | 47,XY,+13 [7],46,XY [8] | Arg197GlyfsX4 | mut/wt | wt | ND | ND |
| 29 | AML M0 | 47,XY,+13 [10],46,XY [2] | Phe131fs | mut/wt | wt | wt | wt |
| 30 | AML M0 | 47,XY,+13 [7],46,XY [5] | Ser114Leu | mut/wt | wt | wt | wt |
| 31 | AML M0 | 47,XY,+13 [5],45,X,-Y [8],46,XY [12] | Leu85Pro | mut/mut | wt | ND | ND |
| 32 | AML M0 | 47,XY,+13 [9],47,XY,del(7)(q22),+13 [2],46,XY [6] | Arg177Gln | mut/wt | wt | wt | wt |
| 33 | AML M0 | 47,XY,+13 [6],94,XXYY,+13,+13 [2],46,XY [6] | Arg178fs | mut/wt | wt | wt | wt |
| 34 | AML M0 | 47,XX,+13 [1],46,XX [2] | Arg139X | mut/mut | wt | wt | wt |
| 35 | AML M0 | 47,XY,+13 [10],94,XXYY,+13,+13 [1],46,XY [9] | Gly143GlufsX7 | mut/mut | wt | ND | ND |
| 36 | AML M0 | 47,XY,+13 [2],46,XY [12] | Trp79Cys | mut/mut | wt | wt | wt |
| 37 | AML M0 | 47,XY,+13 [18],46,XY [3] | Asp171Val | mut/mut | wt | wt | wt |
| 38 | AML M0 | 47,XY,+13 [6],46,XY [14] | Ser145AlafsX4 | mut/mut | wt | wt | wt |
| 39 | AML M0 | 47,XY,+13 [7],45,XY,-7 [5],46,XY [8] | Arg164LysfsX22 | mut/mut | wt | ND | ND |
| 40 | AML M0 | 47,XY,+13 [16],46,XY [2] | Arg174Gln | mut/mut | wt | ND | ND |
| 41 | AML M0 | 46,XX,-7,+13 [3],46,XX [17] | Arg177X | mut/mut | wt | ND | ND |
| 42 | AML M0 | 46,XY [11] | — | wt/wt | FLT3-TKD | wt | wt |
| 43 | AML M0 | 46,XY,t(11;19)(q13;p13) [17],46,XY [3] | — | wt/wt | FLT3-LM | ND | ND |
| 44 | AML M0 | 46,XY [25] | — | wt/wt | FLT3-LM | wt | wt |
| 45 | AML M0 | 46,XX [25] | — | wt/wt | FLT3-LM | wt | wt |
| 46 | AML M0 | 46,XX [20] | — | wt/wt | FLT3-LM | wt | wt |
| 47 | AML M0 | 46,XX,t(10;17)(p15;q21) [2],47,XX,+7,t(10;17)(p15;q21) [7],46,XX [16] | — | wt/wt | FLT3-LM | wt | wt |
| 48 | AML M0 | 47,XY,+8 [5],46,XY [15] | — | wt/wt | FLT3-LM | wt | wt |
| 49 | AML M0 | 43,XY,-3,der(3)t(3;7)(q11;q11),-5,der(7)t(5;7)(p11;q11), der(12)t(12;15)(p13;q22),-15,der(17)t(3;17)(q11;p13) [9],42,idem,-18 [7] | — | wt/wt | FLT3-LM | wt | wt |
| 50 | AML M0 | 55,XY,+X,+4,+5,+8,+10,+13,+14,+17,+18 [4],47,XY,+X [8] | — | wt/wt | FLT3-LM | ND | ND |
| 51 | AML M0 | 46,XY,del(11)(q13q21) [10],46,XY [15] | — | wt/wt | ND | ND | ND |
| 52 | AML M0 | 47,XY,+8,i(17)(q10) [20] | — | wt/wt | ND | ND | ND |
| 53 | AML M0 | 46,XX [25] | — | wt/wt | ND | ND | ND |
| 54 | AML M0 | 46,XX [7] | — | wt/wt | ND | ND | ND |
| 55 | AML M0 | 46,XX [12] | — | wt/wt | ND | ND | ND |
| 56 | AML M0 | 44,XY,add(3)(p13),-5,dic(11;?)(q14;?),add(11)(p15),-15,add(17)(p11) [13], 50–51,idem,+4,+5,+6,+6,+2–3mar [cp4],46,XY [3] | — | wt/wt | ND | ND | ND |
| 57 | AML M0 | 47,XX,+14 [19],46,XX [1] | — | wt/wt | ND | ND | ND |
| 58‡ | AML M0 | 46,XY [25],FISH-Screening: 65% Monosomie 13 oder grosse Deletion | — | wt/wt | ND | ND | ND |
| 59 | AML M0 | 47,XY,+22 [13],46,XY [20] | — | wt/wt | ND | ND | ND |
| 60 | AML M0 | 46,XX [15] | — | wt/wt | ND | ND | ND |
| 61 | AML M0 | 46,XY [25] | — | wt/wt | ND | ND | ND |
| 62 | AML M0 | 46,XY [20] | — | wt/wt | wt | mut | wt |
| 63 | AML M0 | 46,XX,-7,+19 [10],46,XX [14] | — | wt/wt | wt | ND | ND |
| 64 | AML M0 | 45,X,-Y,t(11;19)(q23;p13.3) [19],46,XY [1] | — | wt/wt | wt | wt | wt |
| 65 | AML M0 | 46,XX,t(9;11)(p22;q23) [12],46,XX [13] | — | wt/wt | wt | wt | wt |
| 66 | AML M0 | 45,XX,inv(3)(q21q26),-7 [20] | — | wt/wt | wt | wt | wt |
| 67 | AML M0 | 47,XY,+9 [2],54,XY,+4,+6,+8,+9,+11,+13,+13,+19 [4],46,XY [14] | — | wt/wt | wt | wt | wt |
| 68 | AML M0 | 46,XY [25] | — | wt/wt | wt | wt | wt |
| 69 | AML M0 | 46,XY [25] | — | wt/wt | wt | wt | wt |
| 70 | AML M0 | 46,XX [21] | — | wt/wt | wt | wt | wt |
| 71 | AML M0 | 46,XY [20] | — | wt/wt | wt | wt | wt |
| 72 | AML M0 | 46,XY [22] | — | wt/wt | wt | wt | wt |
| 73 | AML M0 | 42,XX,del(3)(p13),der(5)t(5;12)(q13;q15),dic(7;18)(p11;q11),i(8)(q10), dup(9)(q32q34),-12,der(13;15)(q10;q10), der(14)t(12;14)(p11;p10)t(1;14)(?;q32),-17 [15] | — | wt/wt | wt | wt | wt |
| 74 | AML M0 | 46,XY [20] | — | wt/wt | wt | wt | wt |
| 75 | AML M0 | 46,XY [21] | — | wt/wt | wt | mut | wt |
| 76 | AML M0 | 46,XX [25] | — | wt/wt | wt | ND | ND |
| 77 | AML M0 | 46,XY [20] | — | wt/wt | wt | wt | wt |
| 78 | AML M0 | 46,XY [22] | — | wt/wt | wt | wt | wt |
| 79 | AML M0 | 44–45,XX,del(4)(q23),der(4)t(4;7)(q23;?),del(5)(q13q31), der(7)del(7)(p15)t(4;7)(q23;q21),der(10)t(10;11)(q21;q13),-11, dic(12;17)(p13;p13),der(19)t(11;19)(?;q13)ins(11;19)(?;?)t(10;11) [cp12], 46,XX [2] | — | wt/wt | wt | wt | wt |
| 80 | AML M0 | 46,XX,del(5)(q13q31) [1],46,XX,t(1;3;9)(p11;q26;q34),del(5)(q13q31) [14] | — | wt/wt | wt | wt | wt |
| 81 | AML M0 | 46,XX,del(7)(q22q31) [19],46,XX [2] | — | wt/wt | wt | wt | wt |
| 82 | AML M0 | 45,XY,-7 [15] | — | wt/wt | wt | mut | wt |
| 83 | AML M0 | 45,XY,t(3;3)(q21;q26),-7 [11],46,XY [3] | — | wt/wt | wt | wt | wt |
| 84 | AML M0 | 46,XX,del(9)(q22) [4],46,XX [13] | — | wt/wt | wt | wt | wt |
| 85 | AML M0 | 46,XX,t(2;11)(p21;q23),del(5)(q13q33) [16],46,XX [6] | — | wt/wt | wt | ND | ND |
| 86 | AML M0 | 46,XY [15] | — | wt/wt | wt | ND | ND |
| 87 | AML M0 | 40–42,XY,-1,-5,-7,-8,-11,del(12)(p11),der(14)t(1;14)(p13;p11),-17, der(17)t(5;?;17)(q?;?;p12),-18,+2–4mar [cp16],46,XY [1],40,XY,-1,-5, add(7)(q11),der(8)t(1;8)(p13;p23),der(11)?del(11)(p13)del(11)(q21), del(12)(p11),add(14)(p11),der(17),add(1) | — | wt/wt | wt | ND | ND |
| 88 | AML M0 | 46,XY,t(3;4)(q25;q31),del(5)(q15),der(7)t(5;7)(q?;q11),del(15)(q15) [20] | — | wt/wt | wt | ND | ND |
| 89 | AML M0 | 46,XX,del(5)(q13q31),r(7)(p11q11),del(18)(q11) [20] | — | wt/wt | wt | wt | wt |
| 90 | AML M0 | 40–43,XY,del(5)(q13q31),der(8)t(8;12)(q21;q13),ins(11)(q13;?),-12, del(13)(q14),i(17)(q10) [cp6],46,XY [20] | — | wt/wt | wt | ND | ND |
mut indicates mutated; ND, not determined; and —, not applicable.
Nomenclature of the mutations is according to den Dunnen and Antonarakis.33
A cytogenetically cryptic MLL rearrangement; MLL-AF10 rearrangement was proven by RT-PCR.
A monosomy 13 detected by FISH.
Cytogenetic aberrations and molecular mutations in the AML M0 cohort
The AML M0 cohort (N = 90) was separated into 2 groups according to their RUNX1 mutation status. The results of the metaphase cytogenetic analysis in both groups are summarized in Tables 3 and 4.
RUNX1 mutation status is correlated to cytogenetic aberrations in the AML M0 cohort
| M0 . | RUNX1 mutation status . | P . | |
|---|---|---|---|
| Karyotype . | RUNX1-wt, n=49 . | RUNX1 mutated, n=41 . | |
| Normal | 21 | 11 | .114 |
| Complex | 10 | 2 | .031 |
| 11q23/MLL | 3 | 1 | ND |
| inv(3)t(3:3) | 2 | 0 | ND |
| -5/5q- | 0 | 1 | ND |
| -7/7q- | 2 | 2 | ND |
| +8 | 1 | 0 | ND |
| Others | 10 | 5 | .476 |
| +13 | 0 | 18* | <.001 |
| +21 | 0 | 1 | ND |
| M0 . | RUNX1 mutation status . | P . | |
|---|---|---|---|
| Karyotype . | RUNX1-wt, n=49 . | RUNX1 mutated, n=41 . | |
| Normal | 21 | 11 | .114 |
| Complex | 10 | 2 | .031 |
| 11q23/MLL | 3 | 1 | ND |
| inv(3)t(3:3) | 2 | 0 | ND |
| -5/5q- | 0 | 1 | ND |
| -7/7q- | 2 | 2 | ND |
| +8 | 1 | 0 | ND |
| Others | 10 | 5 | .476 |
| +13 | 0 | 18* | <.001 |
| +21 | 0 | 1 | ND |
ND indicates not determined.
In contrast to all other cases, the trisomy 13 subgroup was not entirely derived from a randomly selected cohort but was expanded from 9 (from the randomly selected group) to 18 samples (after the initial analysis) selected on the basis of a +13.
RUNX1 status was correlated to cytogenetics as summarized in Table 4. The distribution of RUNX1-mutated versus wt samples was not significantly different in AML M0 patients with normal karyotype (P = .114) or in samples with other abnormalities (classified as karyotype “others” in Table 4) including structural and numeric abnormalities not covered by the groups mentioned in Table 4 (P = .476; Table 4). In contrast, a complex aberrant karyotype was more likely to be associated with RUNX1-wt (P = .031), whereas trisomy 13 (+13) as the sole aberration (n = 18) was exclusively associated with a mutation in the RUNX1 gene (P < .001). However, only one case with +13 in the context of a complex aberrant karyotype was not RUNX1 mutated. Due to the size of some cohorts, the significance of distribution of the RUNX1 mutation was not calculated for the residual cytogenetic subgroups.
In addition, FLT3, NRAS (codons 12/1361), and KIT were analyzed for mutations (Table 3). In total, 38 (93%) of 41 RUNX1-mutated cases and 38 (78%) of 49 RUNX1-wt cases were available for analysis of FLT3-LMs as well as for mutations in the FLT3-TKD. We detected FLT3 mutations in 6 RUNX1-mutated cases (4 FLT3-LMs, 1 FLT3-TKD, 1 FLT3-LM + TKD). This number was not significantly different from the cohort with RUNX1-wt (7 FLT3-LMs and 1 FLT3-TKD). In addition, mutations in NRAS (codons 12/1361) and KIT (D816) were screened in RUNX1-mutated (n = 23) and RUNX1-wt cases (n = 29). Of the RUNX1-wt cases, 3 (10.3%) of 29 had mutations in NRAS in contrast to RUNX1-mutated cases that did not reveal any NRAS mutation. There was no overlap of NRAS with FLT3 mutations. KIT mutations were not detected in this cohort of 23 patients analyzed.
RUNX1 mutations in cases with trisomy 13 independent of FAB subtype
In the next step, we selected 20 further samples based primarily on the presence of trisomy 13 and independent of FAB AML subtypes and analyzed RUNX1 mutations within the entire coding region (Figure 1B; Table 5). Of these, 3 were diagnosed as AML M1, 7 as AML M2, 5 as AML M4, and 1 as AML M5. Three cases were not further morphologically classified according to FAB due to lacking bone marrow smears (1 acute leukemia, 2 AML). One case had a secondary AML (s-AML) after polycythemia vera. In this cohort, 17 samples carried +13 as the sole primary aberration, whereas 2 cases had 1 additional aberration and only 1 case had 2 additional cytogenetic aberrations, including +21. A RUNX1 mutation was detected in 16 (80%) of 20 and a FLT3 mutation in 3 (15%) of 20 (2 FLT3-LMs and 1 FLT3-TKD) of these samples (Table 5). Interestingly, 2 (1 FLT3-LM, 1 TKD) of the 3 FLT3 mutations were detected in cases with RUNX1-wt. The RUNX1 mutations could be divided into 5 missense and 12 out of frame/stop mutations (1 patient carried Gly32Asp/Ser238SerfsX334 in parallel). Three of the missense mutations were located inside the RHD and were already described for the AML M0 cohort (Asn69Ser, Ser114Leu, Arg174Gln), whereas two were located outside with Leu29Ser and Gly32Asp. Most of the 12 out of frame/stop mutations were also located inside the RHD (n = 8). The other 4 mutations, however, were N-terminal to or inside the transactivation (TA) domain (aa's 291-371), except for one, which was C-terminal to the TA domain (Gly381ValfsX193). The data on the non-AML M0 cases are summarized in Table 5.
Characterization of non–AML M0 cases according to karyotype and RUNX1 and FLT3 mutation
| Patient . | Diagnosis . | Karyotype . | RUNX1 mutation* . | RUNX1 allele status . | FLT3 mutation . |
|---|---|---|---|---|---|
| 1 | AL | 47,XX,+13 [20] | [Gly32Asp(+)Ser238SerfsX334] | mut/mut | wt |
| 2 | AML | 47,XX,+13 [10],46,XX [14] | R139X | mut/mut | wt |
| 3 | AML | 47,XX,+13 [3],48,XX,+13,+13 [6],46,XX [11] | Val54ValfsX57 | mut/wt | ND |
| 4 | AML M1 | 47,XX,+13 [2],46,XX [19] | wt | wt/wt | FLT3-LM |
| 5 | AML M1 | 47,XY,+13 [8],48,XY,+13,+13 [13],46,XY [8] | Asn69Ser | mut/wt | wt |
| 6 | AML M1 | 47,XY,+13 [5],46,XY [12] | Gly143GlyfsX45 | mut/mut | wt |
| 7 | AML M2 | 47,XY,+13 [20] | Arg174X | mut/mut | wt |
| 8 | AML M2 | 47,XY,+13 [20] | Leu29Ser | mut/wt | wt |
| 9 | AML M2 | 47,XY,+13 [4],46,XY [16] | wt | wt/wt | wt |
| 10 | AML M2 | 46,XY,+13,+15 [15],46,XY [2] | Val97GlyfsX15 | mut/wt | wt |
| 11 | AML M2 | 47,XY,+13 [2],47,XY,+13,i(17)(q10) [13],46,XY [5] | wt | wt/wt | ND |
| 12 | AML M2 | 94,XXYY,+13,+13 [2],93,XXYY,-7,+13,+13 [5],92,XXYY,-7,+13 [2],46,XY [11] | Ala115LeufsX3 | mut/mut | wt |
| 13 | AML M2 | 47,XY,+13 [12],46,XY [8] | Ser281SerfsX307 | mut/wt | wt |
| 14 | AML M4 | 47,XY,+13 [2],46,XY [18] | Arg174Gln | mut/mut | FLT3-LM |
| 15 | AML M4 | 47,XX,+13 [1],47,XX,+13,der(16)t(11;16)(q11;p13) [3],46,XX [20] | Gly381ValfsX193 | mut/wt | wt |
| 16 | AML M4 | 47,XX,+13 [16],46,XX [4] | Arg177X | mut/wt | wt |
| 17 | AML M4 | 47,XX,+13 [2],46,XX [25] | wt | wt/wt | FLT3-D835 |
| 18 | AML M4 | 48,XX,del(12)(p12),+13,+21 [20] | [Ala297_Glu299delinsLeuGlnHis]+[Asp299GlyfsX5] | mut/wt | wt |
| 19 | AML M5 | 47,XX,+13,der(18)t(11;18)(q23;q23) [20],46,XX [3] | Arg139X | mut/mut | wt |
| 20 | s-AML M2 | 47,XY,+13 [4],46,XY [9] | Ser114Leu | mut/wt | wt |
| Patient . | Diagnosis . | Karyotype . | RUNX1 mutation* . | RUNX1 allele status . | FLT3 mutation . |
|---|---|---|---|---|---|
| 1 | AL | 47,XX,+13 [20] | [Gly32Asp(+)Ser238SerfsX334] | mut/mut | wt |
| 2 | AML | 47,XX,+13 [10],46,XX [14] | R139X | mut/mut | wt |
| 3 | AML | 47,XX,+13 [3],48,XX,+13,+13 [6],46,XX [11] | Val54ValfsX57 | mut/wt | ND |
| 4 | AML M1 | 47,XX,+13 [2],46,XX [19] | wt | wt/wt | FLT3-LM |
| 5 | AML M1 | 47,XY,+13 [8],48,XY,+13,+13 [13],46,XY [8] | Asn69Ser | mut/wt | wt |
| 6 | AML M1 | 47,XY,+13 [5],46,XY [12] | Gly143GlyfsX45 | mut/mut | wt |
| 7 | AML M2 | 47,XY,+13 [20] | Arg174X | mut/mut | wt |
| 8 | AML M2 | 47,XY,+13 [20] | Leu29Ser | mut/wt | wt |
| 9 | AML M2 | 47,XY,+13 [4],46,XY [16] | wt | wt/wt | wt |
| 10 | AML M2 | 46,XY,+13,+15 [15],46,XY [2] | Val97GlyfsX15 | mut/wt | wt |
| 11 | AML M2 | 47,XY,+13 [2],47,XY,+13,i(17)(q10) [13],46,XY [5] | wt | wt/wt | ND |
| 12 | AML M2 | 94,XXYY,+13,+13 [2],93,XXYY,-7,+13,+13 [5],92,XXYY,-7,+13 [2],46,XY [11] | Ala115LeufsX3 | mut/mut | wt |
| 13 | AML M2 | 47,XY,+13 [12],46,XY [8] | Ser281SerfsX307 | mut/wt | wt |
| 14 | AML M4 | 47,XY,+13 [2],46,XY [18] | Arg174Gln | mut/mut | FLT3-LM |
| 15 | AML M4 | 47,XX,+13 [1],47,XX,+13,der(16)t(11;16)(q11;p13) [3],46,XX [20] | Gly381ValfsX193 | mut/wt | wt |
| 16 | AML M4 | 47,XX,+13 [16],46,XX [4] | Arg177X | mut/wt | wt |
| 17 | AML M4 | 47,XX,+13 [2],46,XX [25] | wt | wt/wt | FLT3-D835 |
| 18 | AML M4 | 48,XX,del(12)(p12),+13,+21 [20] | [Ala297_Glu299delinsLeuGlnHis]+[Asp299GlyfsX5] | mut/wt | wt |
| 19 | AML M5 | 47,XX,+13,der(18)t(11;18)(q23;q23) [20],46,XX [3] | Arg139X | mut/mut | wt |
| 20 | s-AML M2 | 47,XY,+13 [4],46,XY [9] | Ser114Leu | mut/wt | wt |
Nomenclature of the mutations is according to den Dunnen and Antonarakis.33
AL indicates acute leukemia; mut, mutated; and ND, not determined.
Of note is also the lack of RUNX1 mutations in 15 of 15 cases with +13, all seen in the context of a complex aberrant karyotype, indicating that these cases have completely different underlying molecular mechanisms (not shown).
Correlation of +13 to FLT3 expression
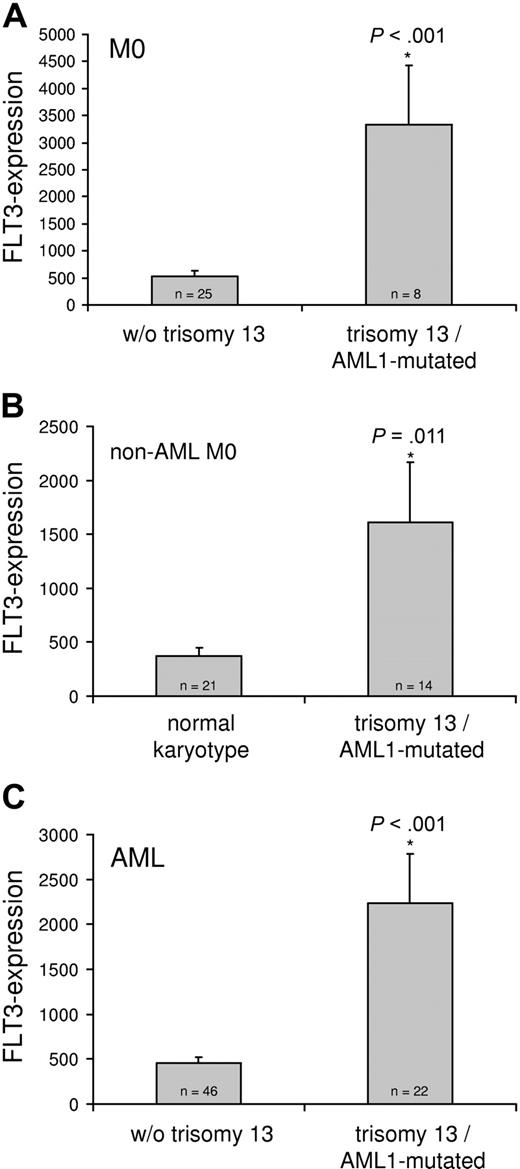
As there was a highly significant correlation of trisomy 13 with RUNX1 mutation independent of the AML FAB subtype, we suspected that factors on chromosome 13 might collaborate with RUNX1 mutations during development of myeloid leukemia. FLT3 is located on chromosome 13; however, there was no significant correlation of FLT3 mutation with +13 or with RUNX1 mutation. We hypothesized that trisomy 13 might be a mechanism to increase FLT3 expression in order to activate FLT3-mediated signal transduction.22 FLT3 transcript levels of 33 AML M0 cases were analyzed by quantitative real-time PCR (RQ-PCR) with a LightCycler instrument using ABL as a reference gene. This group consisted of 8 cases with trisomy 13 as the sole cytogenetic abnormality and RUNX1 mutation, and these cases were compared with 25 consecutive AML M0 cases without +13. The mean FLT3 expression level of the +13 cases with RUNX1 mutation (n = 8) was 3332 ± 1101 (mean ± SEM), which was significantly higher than in cases without +13 (mean value of 522 ± 109; P < .001, t test; Figure 2A). This increase was also significant (P = .024) when FLT3 expression of the AML M0 +13 cases (n = 8) was compared with AML M0 with normal karyotype (n = 9) in the same cohort with mean ± SEM values of 3332 ± 1101 and 659 ± 248, respectively.
FLT3 expression in AML cases with trisomy 13 and RUNX1 mutation compared with cases without trisomy 13.FLT3 transcripts were quantified by RQ-PCR on a LightCycler instrument. (A) Unselected AML M0 samples without trisomy 13 (w/o trisomy 13; n = 25) were compared with AML M0 with trisomy 13 plus a RUNX1 mutation (trisomy 13/RUNX1-mutated; n = 8). (B) AML samples from FAB subgroups other than AML M0 with normal karyotype (n = 21) were compared with samples with trisomy 13 and RUNX1 mutation (n = 14). (C) Unselected AML M0 samples together with AML samples from other FAB subgroups without trisomy 13 (w/o trisomy 13; n = 46) were compared with AML with trisomy 13 plus a RUNX1 mutation (trisomy 13/RUNX1-mutated; n = 22). FLT3 expression is given as 100*FLT3/ABL as previously described.21 The bars represent mean values ± SEM. Each sample was analyzed in duplicate.
FLT3 expression in AML cases with trisomy 13 and RUNX1 mutation compared with cases without trisomy 13.FLT3 transcripts were quantified by RQ-PCR on a LightCycler instrument. (A) Unselected AML M0 samples without trisomy 13 (w/o trisomy 13; n = 25) were compared with AML M0 with trisomy 13 plus a RUNX1 mutation (trisomy 13/RUNX1-mutated; n = 8). (B) AML samples from FAB subgroups other than AML M0 with normal karyotype (n = 21) were compared with samples with trisomy 13 and RUNX1 mutation (n = 14). (C) Unselected AML M0 samples together with AML samples from other FAB subgroups without trisomy 13 (w/o trisomy 13; n = 46) were compared with AML with trisomy 13 plus a RUNX1 mutation (trisomy 13/RUNX1-mutated; n = 22). FLT3 expression is given as 100*FLT3/ABL as previously described.21 The bars represent mean values ± SEM. Each sample was analyzed in duplicate.
To permit comparison of FLT3 expression of +13 samples to samples without +13 independent of FAB subtype, FLT3 expression was evaluated in FAB subtypes other than AML M0 (n = 35). This group consisted of 14 cases from various FAB subgroups with +13 and RUNX1 mutation as characterized in “Results,” “RUNX1 mutations in cases with trisomy 13 independent of FAB subtype,” and 21 AML cases with normal karyotype, which reflected the same FAB subtype distribution. Again, a highly significant increase of FLT3 expression of the +13/RUNX1-mutated cases versus the cases with normal karyotype was detected with 1612 ± 556 and 369 ± 78, respectively (mean ± SEM, P = .011; Figure 2B). These cases were combined with the previously analyzed AML M0 cases to a total number of 68 cases, consisting of 22 cases with trisomy 13 plus RUNX1 mutation and 46 cases without trisomy 13. The comparison of both groups showed a highly significant (P < .001, t test), about 4-fold, increase of FLT3 expression of the +13 samples (n = 22) versus the samples without trisomy 13 (n = 46) with mean ± SEM of 2237 ± 549 and 452 ± 69, respectively (Figure 2C).
Discussion
In this study we confirmed the high incidence of RUNX1 mutations in AML M0 that has been reported previously. For the first time we detected a highly significant correlation of RUNX1 mutation to trisomy 13 as the sole primary aberration, which is independent of the FAB subtype. However, trisomy 13 in the context of a complex aberrant karyotype was not correlated to RUNX1 mutation. FLT3 is located on chromosome 13 and quantification of FLT3 transcript levels indicated an increased FLT3 expression in samples with RUNX1 mutation together with trisomy 13 compared with samples without trisomy 13, arguing for cooperation of both events in leukemogenesis.
The RUNX1 gene is known to be involved in leukemogenesis since it was cloned as a fusion gene from the t(8;21) translocation.35 Morphologically most of the t(8;21)-carrying AML cells are classified as AML M2 and less frequently as AML M1. Recently, somatic mutations have been identified in the RUNX1 gene in myeloid leukemias.11 These mutations occurred with the highest frequency in the RHD of the RUNX1 gene in AML M0,14 in cases with acquired trisomy 21,14 and in certain MDS cases,17,18,36 indicating that leukemogenesis is effected by different signaling pathways by AML1-ETO and by RUNX1 somatic mutations. No mutations in RUNX1 were detected outside the RHD (aa's 50-177) in AML M0 in 2 previous studies when the entire cDNA of RUNX1β was analyzed.11,16 Therefore, most subsequent studies relied on screening of exons 3-5 in AML M0 containing the RHD up to R177.14,19 In the present study we analyzed RUNX1 mutations on the cDNA level (codons 1-227) in a large cohort of AML M0 samples at diagnosis (N = 90) to correlate the results to cytogenetic aberrations as well as to gene mutations in receptor-tyrosine kinase pathways. The percentage of AML M0 with RUNX1 mutations in our study, 46%, is high compared with previous studies with values ranging between 16% and 27%.14,16,19 Three possible explanations might account for this finding. First, our AML M0 patient population is characterized by a high incidence of trisomy 13 (20%; Table 3), a cytogenetic finding which is 100% correlated to RUNX1 mutation in our study. Second, some of the previous studies screened only exons 3-5 of the RUNX1 gene,14,19 where the 3′-end of exon 5 represents the codon for R177, the extreme C-terminus of the RHD. In our study, we screened codons 1-227 of the RUNX1β cDNA of the AML M0, and 5 of the 41 RUNX1-mutated cases had mutations C-terminal to R177 (Table 3). Third, in contrast to previous reports, which applied single-strand conformation polymorphism (SSCP) analysis, our screening method for the detection of RUNX1 mutations relied on the use of the more sensitive DHPLC technology.37
From structural analysis of the RHD of RUNX1,34 many of the missense mutations detected in the AML M0 population in our study are supposed to disrupt DNA binding or have an impact on CBFB binding (Asn69, Arg80, Ser114, Asn171, Arg174, Arg177) while leaving the TA domain of RUNX1 intact. On the other hand, the large majority of the residual mutations in our study displayed frame-shift or stop mutations in the RHD, which probably lead to a complete loss of RUNX1 protein function. Opposed to this are frame-shift mutations in 5 AML M0 samples that are C-terminal to the RHD, which are supposed to disrupt the TA domain while leaving the DNA-binding domain intact, indicating that different functional effects might result from these different classes of mutations. The next level of complexity is caused by the presence of these mutations on one or both alleles. In a previous study, almost all AML M0 patients with a RUNX1 mutation carried the mutation on both alleles.14 In the present study, homozygous and heterozygous mutations were about equally distributed in AML M0. However, homozygosity cannot be excluded because 20% to 30% of normal cells in the leukemic samples may mimic a pattern of heterozygosity.
Different from AML M0, a considerable portion of mutations were described to be located C-terminal to the RHD in therapy-related MDS as well as in MDS/AML.17,18 Therefore, the entire coding region of RUNX1 was screened for mutations in the non-AML M0 cases with trisomy 13 in this study. The mutation pattern in the RUNX1 gene in the non-AML M0 cases was similar to the AML M0 cases, with most of the mutation clustered inside the RHD (Table 5). However, we also detected 4 frame-shift mutations C-terminal to the RHD. These mutations would have escaped detection in our AML M0 screening, as they were located at codons 238, 281, 296, and 381 of the RUNX1 transcript. The latter mutation is even located C-terminal to the TA domain (aa's 291-371) and the functional significance of this mutation remains to be determined.
RUNX1 mutations in AML M0 in our study were not restricted to trisomy 13, but all trisomy 13 samples (n = 18) were RUNX1 mutated. In a second set of samples that were selected for trisomy 13 in different FAB subtypes other than AML M0, again an extremely high portion of cases was RUNX1 mutated (n = 16/20, 80%).
Overall, trisomy 13 is a rare recurring clonal chromosomal aberration with an incidence of 2.4% in de novo AML,38,39 and in agreement with our study these cases are mainly clustered in the AML M0 subtype.40 The correlation of trisomy 13 and RUNX1 mutations suggests that there might be a cooperating event between these two aberrations. Cooperation between hematopoietic transcription factors and members of the RAS or the RTK family has been proposed as a mechanism for leukemia development.1 Mutations of NRAS and KIT have not been detected in the RUNX1-mutated samples in our study in the AML M0 cohort. Activating mutations of the RTK FLT3, which is located on chromosome 13, have been suggested as a cooperating event previously.41 In de novo AML, these FLT3 mutations have the highest incidence in normal karyotype (39%) as well as in t(15;17) with the PML-RARA fusion gene (36%).29 Less frequent is the coincidence of FLT3-LMs with the AML1-ETO fusion gene from t(8;21) (9%).29 FLT3 mutations in our study did not correlate to RUNX1 somatic mutations or to trisomy 13. Another mechanism of FLT3 activation in addition to mutation is FLT3 overexpression, which results in FLT3 autophosphorylation and a decreased overall survival.22 However, the mechanism of FLT3 overexpression remains yet unknown.22 In the present study, we hypothesized that trisomy 13 might be a mechanism to increase FLT3 expression by means of a gene-dosage effect. This hypothesis is confirmed by quantitative real-time PCR, which detected a highly significant increase of FLT3 transcript levels in AML with trisomy 13 and RUNX1 mutation compared with samples without trisomy 13 (Figure 2). Trisomies as a gain-of-function mechanism by selection of a gain-of-function mutation or by gene dosage have been also implicated in other instances. Chromosome 4 harbors the KIT protooncogene, and AML patients with trisomy 4 as well as patients with t(8;21) both have the highest incidence of the activating KIT-D816 mutation,6 which lead to the amplification of the mutated allele by trisomy 4.42 Similar to this, amplification of chromosome 21, the locus of RUNX1, by acquired trisomy 21 in myeloid malignancies is associated with RUNX1 somatic mutations and duplication of the mutated allele.14 Beyond hematologic malignancies, the same phenomenon is known to occur from solid tumors in hereditary renal carcinoma, where trisomy 7 amplifies activating mutations of the MET oncogene,43,44 which belongs to the family of RTKs like KIT. In contrast to our study, trisomies in these studies effected the amplification of a mutated allele with an activating mutation. However, as shown by Ozeki et al,22 overexpression of FLT3 might already be sufficient for FLT3 activation, and this activation by overexpression might be a general phenomenon in RTK signaling, as t(8;21)-induced overexpression of KIT was also associated with KIT autophosphorylation.45 The gain of chromosomes (ie, trisomy 8, 11, and 13) as a mechanism for increased expression of genes located in the respective regions has been also demonstrated by gene-expression profiling in a previous study.46
Taken together, the data of the present study show a highly significant correlation of RUNX1 mutation with trisomy 13 in various AML FAB subgroups but not with FLT3, N-RAS, or KIT mutations. Trisomy 13 correlates with an increase of FLT3 transcript levels, which might cooperate with RUNX1 mutation in leukemogenesis. As FLT3 overexpression is associated with FLT3 activation,22 the subgroup of patients with RUNX1 mutation plus trisomy 13 might benefit from treatment with FLT3 inhibitors.47,48
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank all coworkers at the Munich Leukemia Laboratory for approaching together many aspects in the field of leukemia diagnostics and research. Our special thanks are directed at all referring centers for sending patient material for this analysis.
Authorship
Contribution: F.D. was responsible for the molecular biology, was in part responsible for the study design, and wrote the paper; C.H. was responsible for cytogenetic analysis; W.K. was responsible for immunophenotyping; T.H. was responsible for cytomorphology; and S.S. was responsible for study design and contributed in writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Frank Dicker, Munich Leukemia Laboratory GmbH, Max-Lebsche-Platz 31, 81377 Munich, Germany; e-mail: frank.dicker@mll-online.com; URL: www.mll-online.com.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal