Abstract
Derivative chromosome 9 deletions are seen in 10% to 15% of patients with chronic myelogenous leukemia and have been associated with a poor prognosis; however, no studies have been performed in the context of a randomized clinical trial. We developed a DNA-based deletion screen and investigated 339 chronic phase patients treated with interferon-α as first-line therapy in 3 controlled German studies with a median observation time of 7 years. Deletions were detected in pretreatment DNA of 59 of 339 (17%) patients. Of these, 21 spanned the ABL/BCR junction and 38 were centromeric (n = 20) or telomeric (n = 18) of the breakpoint. There was no significant difference in overall survival between deleted and nondeleted patients. Patients with breakpoint-spanning deletions had poorer survival compared with patients without deletions (4.7 versus 7.8 years; P = .003), but this was not significant when censored at allogeneic stem cell transplantation (n = 129) or imatinib (n = 62) treatment in the first chronic phase (P = .078). Unexpectedly, deletions that did not span the breakpoint were associated with improved survival compared with cases without deletions (P = .001). Multiple Cox regression analysis indicated that deletion status (P = .007), age (P = .018), and spleen enlargement (P < .001) were significant independent indicators of survival and confirmed that only deletions spanning the ABL/BCR breakpoint were associated with an adverse prognosis (P = .039).
Introduction
Chronic myelogenous leukemia (CML) is a clonal neoplastic disorder characterized by the presence of the BCR/ABL fusion gene, the oncogenic product of t(9;22)(q34;q11). This translocation also generates a reciprocal ABL/BCR fusion on the derivative chromosome 9 [der(9)] that is transcribed in approximately 70% of cases, although its biologic significance is not known.1,2 Several studies have found deletions at or encompassing the ABL/BCR junction in approximately 10% to 15% of CML cases that almost certainly arise during the translocation process.3–10 Importantly, deletions have been associated with an adverse prognosis, at least for cases treated with hydroxyurea or interferon-α (IFN)-based therapies.5,8 However, not all studies have found that der(9) deletions are an indicator of inferior outcome5 and no systematic studies have been performed in the context of a randomized clinical trial. To provide more accurate information about the significance of deletion status with regard to IFN therapy, we have developed a rapid DNA-based deletion screen based on multiplex ligation-dependent probe amplification (MLPA) and investigated the prognostic significance of deletion status in a large number of patients enrolled in 3 consecutive German CML Study Group clinical trials with long-term follow-up data
Patients, materials, and methods
Study group
The prospective, randomized German CML Studies 1-3 have been described in detail elsewhere.11–13 Of the 1435 chronic-phase patients recruited between 1983 and 2000, 843 were treated with IFN as first-line therapy as a single agent or in combination with hydroxyurea. In this investigation, we analyzed the der(9) deletion status in all 339 IFN-treated cases for whom adequate DNA was available before treatment. The median duration of IFN-based treatment was 17 months (range, 1-115 months) and clinical characteristics of the study group are summarized in Table 1. Baseline data were available to determine Sokal and Hasford (also known as New CML or Euro) scores for 338 cases, the latter having been developed specifically for patients treated with IFN.14,15
Characteristics of patients with or without derivative chromosome 9 deletions
| Clinical characteristics . | Total n = 339 . | Cases without deletions n = 280; 83% . | Cases with deletions n = 59; 17% . | ABL/BCRspanning deletionsn = 21; 6% . | Deletion of ABLonly or BCRonlyn = 38; 11% . |
|---|---|---|---|---|---|
| Age, y (range) | 50 (10-83) | 50 (10-83) | 49 (11-72) | 56 (21-71) | 45 (11-65) |
| Male sex, no. | 58 | 59 | 54 | 52 | 55 |
| Median observation time, y (range) | 7.1 (0.3-16) | 6.9 (0.3-16) | 8.0 (0.5-16) | 5.7 (0.5-9.8) | 8.4 (0.5-16) |
| No. alive (%) | 158 (47) | 125 (45) | 33 (56) | 3 (14) | 30 (79) |
| Median time on IFN, mon (range) | 17 (1-115) | 18 (0.1-115) | 14 (1-106) | 14 (3.8-81) | 13 (0.2-106) |
| Treatment after IFN failure, % | |||||
| Imatinib | 62 (18) | 49 (18) | 13 (22) | 4 (19) | 9 (24) |
| In first chronic phase | 49 (14) | 39 (14) | 10 (17) | 2 (10) | 8 (21) |
| Allogeneic stem cell transplantation | 129 (38) | 109 (39) | 20 (34) | 6 (29) | 14 (37) |
| In first chronic phase | 105 (31) | 87 (31) | 18 (31) | 6 (29) | 12 (32) |
| Sokal risk groups, no. (%) | |||||
| Evaluable | 338 (99) | 279 (99) | 59 (100) | 21 (100) | 38 (100) |
| Low | 133 (39) | 108 (39) | 25 (42) | 10 (48) | 15 (39) |
| Intermediate | 112 (33) | 91 (33) | 21 (36) | 9 (43) | 12 (32) |
| High | 93 (28) | 80 (28) | 13 (22) | 2 (9) | 11 (29) |
| Hasford risk groups, no. (%) | |||||
| Evaluable | 338 (99) | 279 (99) | 59 (100) | 21 (100) | 38 (100) |
| Low | 132 (39) | 106 (38) | 26 (44) | 11 (52) | 15 (39) |
| Intermediate | 166 (49) | 142 (51) | 24 (41) | 10 (48) | 14 (37) |
| High | 40 (12) | 31 (11) | 9 (15) | 0 | 9 (24) |
| Clinical characteristics . | Total n = 339 . | Cases without deletions n = 280; 83% . | Cases with deletions n = 59; 17% . | ABL/BCRspanning deletionsn = 21; 6% . | Deletion of ABLonly or BCRonlyn = 38; 11% . |
|---|---|---|---|---|---|
| Age, y (range) | 50 (10-83) | 50 (10-83) | 49 (11-72) | 56 (21-71) | 45 (11-65) |
| Male sex, no. | 58 | 59 | 54 | 52 | 55 |
| Median observation time, y (range) | 7.1 (0.3-16) | 6.9 (0.3-16) | 8.0 (0.5-16) | 5.7 (0.5-9.8) | 8.4 (0.5-16) |
| No. alive (%) | 158 (47) | 125 (45) | 33 (56) | 3 (14) | 30 (79) |
| Median time on IFN, mon (range) | 17 (1-115) | 18 (0.1-115) | 14 (1-106) | 14 (3.8-81) | 13 (0.2-106) |
| Treatment after IFN failure, % | |||||
| Imatinib | 62 (18) | 49 (18) | 13 (22) | 4 (19) | 9 (24) |
| In first chronic phase | 49 (14) | 39 (14) | 10 (17) | 2 (10) | 8 (21) |
| Allogeneic stem cell transplantation | 129 (38) | 109 (39) | 20 (34) | 6 (29) | 14 (37) |
| In first chronic phase | 105 (31) | 87 (31) | 18 (31) | 6 (29) | 12 (32) |
| Sokal risk groups, no. (%) | |||||
| Evaluable | 338 (99) | 279 (99) | 59 (100) | 21 (100) | 38 (100) |
| Low | 133 (39) | 108 (39) | 25 (42) | 10 (48) | 15 (39) |
| Intermediate | 112 (33) | 91 (33) | 21 (36) | 9 (43) | 12 (32) |
| High | 93 (28) | 80 (28) | 13 (22) | 2 (9) | 11 (29) |
| Hasford risk groups, no. (%) | |||||
| Evaluable | 338 (99) | 279 (99) | 59 (100) | 21 (100) | 38 (100) |
| Low | 132 (39) | 106 (38) | 26 (44) | 11 (52) | 15 (39) |
| Intermediate | 166 (49) | 142 (51) | 24 (41) | 10 (48) | 14 (37) |
| High | 40 (12) | 31 (11) | 9 (15) | 0 | 9 (24) |
Cytogenetic follow-up data were available for 286 patients (84%). A major cytogenetic response (MCR) on IFN therapy was achieved by 76 patients (27%) and a complete cytogenetic response (CCR) by 35 patients (12%). Sixty-two patients subsequently received imatinib (IM), whereas 129 patients underwent allogeneic stem cell transplantation (SCT) after a median of 1.5 years (range, 0.2-8.8 years), 105 of them in the first chronic phase. In addition to overall survival, survival times were censored at the start of IM or allogeneic SCT in the first chronic phase, because after the start of those treatments, survival probabilities of chronic-phase patients can no longer be linked directly to IFN. The Hasford score was developed under the same censoring principle, which is important for a meaningful assessment of potential prognostic factors in a multiple model. The study was approved by the ethics committees of the participating institutions (Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) and informed consent was obtained in accordance with the Declaration of Helsinki.
Detection of deletions by multiplex ligation probe amplification
Multiplex ligation-dependent probe amplification oligonucleotide probes were designed according to the manufacturer's instructions (MRC Holland B.V., Amsterdam, The Netherlands). Each probe consisted of adjacent 5′ and 3′ oligonucleotides that, after amplification with universal primers, generated products of a unique size.16 All oligonucleotides were designed to unique sequences as determined by BLAST searching17 and were also designed to avoid known single nucleotide polymorphisms.18 The 3′ oligonucleotides for each probe pair were modified with a phosphate group at their 5′ end. All oligonucleotides were obtained from biomers.net GmbH (Ulm, Germany) and had a tripartite structure consisting of a tag sequence (corresponding to the MLPA Salsa oligonucleotides subsequently), a variable length stuffer sequence, and a region complementary to the target sequence. Probe sequences and positions are shown in Table 2 and Figure 1.
Sequences of MLPA amplification probes
| Chromosome . | Gene . | Exon . | Amplicon . | Sequence . |
|---|---|---|---|---|
| 5′ MLPA oligonucleotide probe | ||||
| der (9) | ABL | 1b | 95 bp | GGG TTC CCT AAG GGT TGG AGT GCT GCA TTT TAT CAA AGG AGC AGG |
| der (9) | BCR | 16 | 100 bp | GGG TTC CCT AAG GGT TGG ACG TTG CAA GAC GAA GAT CCC CAA GGA GGA |
| der (9) | FLJ31568 | 2 | 105 bp | GG GTT CCC TAA GGG TTG GAC GTG ATG CAA CCA CAA TCG CAG CCT GTC CTA |
| der (9) | PRDM12 | 2 | 110 bp | GG GTT CCC TAA GGG TTG GAC CAT GCA TGC CAC GTG GAC ATC TGC AAG AAC AAC AAC |
| der (9) | ASS | 4 | 115 bp | G GGT TCC CTA AGG GTT GGA CCG CCG GAT CGG TGC CTA CTT CTT CCT TCT GGG CTC |
| der (9) | EXOSC2 | 7 | 135 bp | GGG TTC CCT AAG GGT TGG ACG GCA TAT GCC GCT TTA AGA GAT ATA GTT TTG GTC CAG GTT TCC CCC TCC |
| der (22) | BCR | 1 | 90 bp | GGG TTC CCT AAG GGT TGGATA CCA GAG CAT CTA CGT CGG GGG |
| der (22) | BCR | 2 | 120 bp | G GGT TCC CTA AGG GTT GGA GCA TGC GAC AGC TGC ACC AAG ATG GGC TGC CCT ACA TTG |
| der (22) | ABL | 6 | 125 bp | GGG TTC CCT AAG GGT TGG AGT CGA TCT TCC AGC TGC CCC CCG TTC TAT ATC ATC ACT GAG |
| der (22) | ABL | 11 | 130 bp | G GGT TCC CTA AGG GTT GGA CGT AGG CTA GGG ATG TAC GCG CTG AAT GAA GAT GAG CGC CTT CTC |
| 15 | UBR1 | 2 | 105 bp | GGG TTC CCT AAG GGT TGG ACG GCA TAC CAC AGT GGA GCA TTT CAG CTT TGT |
| 15 | NDNL2 | 110 bp | GG GTT CCC TAA GGG TTG GAC GGC ATA TGC CAG TAC GTC TTC GGG TAT AAG CTG | |
| 3′ MLPA oligonucleotide probe | ||||
| der (9) | ABL | 1b | 95 bp | GAA GAA GGA ATC ATC GAG GCA TGG GCG TCT AGA TTG GAT CTT GCT GGC AC |
| der (9) | BCR | 16 | 100 bp | CGG CGA GAG CAC GGA CAG ACT CAT GAG CCT CTA GAT TGG ATC TTG CTG GCA C |
| der (9) | FLJ31568 | 2 | 105 bp | CAA TAA TGT GCT CAA CCC TGG CTC GAT CGC TCT CTA GAT TGG ATC TTG CTG GCA C |
| der (9) | PRDM12 | 2 | 110 bp | CTC ATG TGG GAG GTA CGC GCG CCG TAT CTA GTC TAG ATT GGA TCT TGC TGG CAC |
| der (9) | ASS | 4 | 115 bp | CTC TTC CCG TAG GTG TTC ATT GAG CGT AGT CAT GAC CTC TAG ATT GGA TCT TGC TGG CAC |
| der (9) | EXOSC2 | 7 | 135 bp | CTG GTG AAA CGG CAG AAG ACC CAC GAT GGA ACT ATA CAT ACG CTC TAG ATT GGA TCT TGC TGG CAC |
| der (22) | BCR | 1 | 90 bp | CAT GAT GGA AGG GGA GGG CAA GGGCTC TAG ATT GGA TCT TGC TGG CAC |
| der (22) | BCR | 2 | 120 bp | ATG ACT CGC CCT CCT CAT CGC CCT CGA TCG CCA GCA TGC TCT AGA TTG GAT CTT GCT GGC AC |
| der (22) | ABL | 6 | 125 bp | TTC ATG ACC TAC GGG AAC CTC CTG CGT ACT ACA GCT GAG CTC TC TAG ATT GGA TCT TGC TGG CAC |
| der (22) | ABL | 11 | 130 bp | CCC AAA GAC AAA AAG ACC AAC TTG GAT GCG ATC GCA TAC AGC CTC TAG ATT GGA TCT TGC TGG CAC |
| 15 | UBR1 | 2 | 105 bp | GGG AGG GTT TTC AAA AGT GGA GAG GGT GAT GTC TAG ATT GGA TCT TGC TGG CAC |
| 15 | NDNL2 | 110 bp | GTG GAA CTT GAA CCC AAG AGC AAC GGT GAT CGT CTC TAG ATT GGA TCT TGC TGG CAC | |
| Chromosome . | Gene . | Exon . | Amplicon . | Sequence . |
|---|---|---|---|---|
| 5′ MLPA oligonucleotide probe | ||||
| der (9) | ABL | 1b | 95 bp | GGG TTC CCT AAG GGT TGG AGT GCT GCA TTT TAT CAA AGG AGC AGG |
| der (9) | BCR | 16 | 100 bp | GGG TTC CCT AAG GGT TGG ACG TTG CAA GAC GAA GAT CCC CAA GGA GGA |
| der (9) | FLJ31568 | 2 | 105 bp | GG GTT CCC TAA GGG TTG GAC GTG ATG CAA CCA CAA TCG CAG CCT GTC CTA |
| der (9) | PRDM12 | 2 | 110 bp | GG GTT CCC TAA GGG TTG GAC CAT GCA TGC CAC GTG GAC ATC TGC AAG AAC AAC AAC |
| der (9) | ASS | 4 | 115 bp | G GGT TCC CTA AGG GTT GGA CCG CCG GAT CGG TGC CTA CTT CTT CCT TCT GGG CTC |
| der (9) | EXOSC2 | 7 | 135 bp | GGG TTC CCT AAG GGT TGG ACG GCA TAT GCC GCT TTA AGA GAT ATA GTT TTG GTC CAG GTT TCC CCC TCC |
| der (22) | BCR | 1 | 90 bp | GGG TTC CCT AAG GGT TGGATA CCA GAG CAT CTA CGT CGG GGG |
| der (22) | BCR | 2 | 120 bp | G GGT TCC CTA AGG GTT GGA GCA TGC GAC AGC TGC ACC AAG ATG GGC TGC CCT ACA TTG |
| der (22) | ABL | 6 | 125 bp | GGG TTC CCT AAG GGT TGG AGT CGA TCT TCC AGC TGC CCC CCG TTC TAT ATC ATC ACT GAG |
| der (22) | ABL | 11 | 130 bp | G GGT TCC CTA AGG GTT GGA CGT AGG CTA GGG ATG TAC GCG CTG AAT GAA GAT GAG CGC CTT CTC |
| 15 | UBR1 | 2 | 105 bp | GGG TTC CCT AAG GGT TGG ACG GCA TAC CAC AGT GGA GCA TTT CAG CTT TGT |
| 15 | NDNL2 | 110 bp | GG GTT CCC TAA GGG TTG GAC GGC ATA TGC CAG TAC GTC TTC GGG TAT AAG CTG | |
| 3′ MLPA oligonucleotide probe | ||||
| der (9) | ABL | 1b | 95 bp | GAA GAA GGA ATC ATC GAG GCA TGG GCG TCT AGA TTG GAT CTT GCT GGC AC |
| der (9) | BCR | 16 | 100 bp | CGG CGA GAG CAC GGA CAG ACT CAT GAG CCT CTA GAT TGG ATC TTG CTG GCA C |
| der (9) | FLJ31568 | 2 | 105 bp | CAA TAA TGT GCT CAA CCC TGG CTC GAT CGC TCT CTA GAT TGG ATC TTG CTG GCA C |
| der (9) | PRDM12 | 2 | 110 bp | CTC ATG TGG GAG GTA CGC GCG CCG TAT CTA GTC TAG ATT GGA TCT TGC TGG CAC |
| der (9) | ASS | 4 | 115 bp | CTC TTC CCG TAG GTG TTC ATT GAG CGT AGT CAT GAC CTC TAG ATT GGA TCT TGC TGG CAC |
| der (9) | EXOSC2 | 7 | 135 bp | CTG GTG AAA CGG CAG AAG ACC CAC GAT GGA ACT ATA CAT ACG CTC TAG ATT GGA TCT TGC TGG CAC |
| der (22) | BCR | 1 | 90 bp | CAT GAT GGA AGG GGA GGG CAA GGGCTC TAG ATT GGA TCT TGC TGG CAC |
| der (22) | BCR | 2 | 120 bp | ATG ACT CGC CCT CCT CAT CGC CCT CGA TCG CCA GCA TGC TCT AGA TTG GAT CTT GCT GGC AC |
| der (22) | ABL | 6 | 125 bp | TTC ATG ACC TAC GGG AAC CTC CTG CGT ACT ACA GCT GAG CTC TC TAG ATT GGA TCT TGC TGG CAC |
| der (22) | ABL | 11 | 130 bp | CCC AAA GAC AAA AAG ACC AAC TTG GAT GCG ATC GCA TAC AGC CTC TAG ATT GGA TCT TGC TGG CAC |
| 15 | UBR1 | 2 | 105 bp | GGG AGG GTT TTC AAA AGT GGA GAG GGT GAT GTC TAG ATT GGA TCT TGC TGG CAC |
| 15 | NDNL2 | 110 bp | GTG GAA CTT GAA CCC AAG AGC AAC GGT GAT CGT CTC TAG ATT GGA TCT TGC TGG CAC | |
Tag sequences are in italics, stuffer sequences in plain type, and sequences corresponding to the region on interest are in bold.
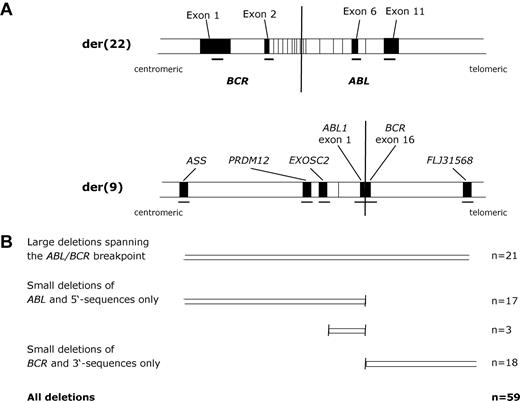
Summary of the multiplex ligation-dependent probe amplification assay to detect derivative chromosome 9 deletions. (A) Map showing positions of the der(9) and der(22) probes (not to scale). Probes on the der(9) were designed to detect deletions, whereas der(22) probes acted as controls. (B) Summary of patient results.
Summary of the multiplex ligation-dependent probe amplification assay to detect derivative chromosome 9 deletions. (A) Map showing positions of the der(9) and der(22) probes (not to scale). Probes on the der(9) were designed to detect deletions, whereas der(22) probes acted as controls. (B) Summary of patient results.
Reactions were performed in duplicate using the Salsa MLPA kit (MRC Holland B.V.) following the DNA detection and quantification protocol16 with 1.33 fmol/μL each MLPA oligonucleotide and 50 to 100 ng genomic DNA extracted from peripheral blood or bone marrow samples as described.19 The annealed and ligated probes were amplified by polymerase chain reaction with universal Salsa MLPA primers (Salsa MLPA forward primer sequence: 5′-6-FAM-GGGTTCCCTAAGGGTTGGA-3′; Salsa MLPA reverse primer sequence: 5′-GTGCCAGCAAGATCCAATCTAGA-3′). Amplification was performed for 35 cycles (30 seconds at 95°C, 30 seconds at 60°C, 1 minute at 72°C) followed by a final incubation at 72°C for 20 minutes. The amplified products were analyzed by capillary electrophoresis on an ABI Prism Genetic Analyzer 3100 (Applied Biosystems, Warrington, United Kingdom). Polymerase chain reaction products were processed by diluting 1 μL DNA in 8.9 μL Hi-Di-Formamide (Applied Biosystems) with 0.1 μL GeneScan-ROX 500 (Applied Biosystems) added as a size standard. The following run characteristics were used: 36-mm capillaries, POP-6 polymer, run temperature 60°C, capillary filling volume 184, prerun voltage 15 kV, prerun time 180 seconds, injection voltage 1 kV, injection time 12 seconds, run voltage 10 kV, data delay time 1 second, and run time 2100 seconds. Results were interpreted using Genotyper version 2.0 (Applied Biosystems) and peak heights from each patient were exported to an Excel spreadsheet, which was designed to assess the ratios of each peak relative to all other peaks for that patient. Ratios of test peaks to control peaks and control peaks to other control peaks in each patient sample were compared with the same ratios obtained for 2 healthy people that were included in each run. For normal sequences, a dosage quotient of 1.0 is expected; if a deletion or duplication is present, the dosage quotient should be 0.5 and 1.5, respectively. As previously described, we considered a particular marker to be deleted if the average dosage quotient of test to internal control peaks was less than 0.7,20 and we only scored patients as deletion positive if they were missing at least 2 consecutive der(9) markers. As controls, we analyzed 20 healthy people, the der(9) deletion positive cell line MC3,21 and 18 patients with CML with known der(9) deletions as determined by fluorescence in situ hybridization using the LSI BCR/ABL+9q34 Tricolor Dual Fusion Translocation Probe (Abbot GmbH KG, Wiesbaden, Germany).
Statistical analysis
Survival probabilities were estimated by the Kaplan-Meier method. For comparison of survival probabilities between different groups, the log rank test was applied using GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego, CA). To investigate the distribution of baseline values between groups, univariate tests were performed using the Mann-Whitney, Fisher exact, or χ2 tests as appropriate. The independent influence of breakpoint-spanning deletions was assessed by multiple Cox regression analysis using SAS version 9.1.3 (SAS Institute Inc., Cary, NC).
Results
Development of a multiplex ligation-dependent probe amplification assay to detect derivative chromosome 9 deletions
In most studies, der(9) deletions have been detected by fluorescence in situ hybridization (FISH).3–10,22–25 For our study group, however, fixed cells were generally not available and instead we sought to develop a methodology based on genomic DNA. Detection of deletions using genomic DNA has been performed in previous studies using real-time polymerase chain reaction9 or multiplex amplifiable probe hybridization.21 However, MLPA has rapidly become an established technique for the reliable detection of DNA copy number changes16,20,26 and therefore we focused on the development of an MLPA der(9) deletion test. We designed 6 probes to span the region on der(9) that is known to be deleted in most cases using commercially available FISH probes (Figure 1). Depending on the precise positions of the breakpoints, these probes are expected to span a region of 360 to 500 kb. As controls, we designed 4 probes within BCR and ABL that are retained on the der(22) and, in addition, we designed 2 additional control probes from chromosome 15. All samples were analyzed initially with the 10 chromosome 9 and 22 probes (BCR exon 1, ABL exon 1b, BCR exon 16, FLJ31568 exon 2, PRDM12 exon 12, ASS exon 4, BCR exon 2, ABL exon 6, ABL exon 11, EXOSC2 exon 7). Because this probe set would not distinguish between a der(9) deletion and a gain of the Philadelphia chromosome, samples were retested with a second set that included the 2 chromosome 15 control probes (BCR exon 1, SMARCB1 exon 2, UBR1 exon 2, NDNL2, BCR exon 2, ABL exon 6, ABL exon 11).
Sensitivity and validation of multiplex ligation-dependent probe amplification assay
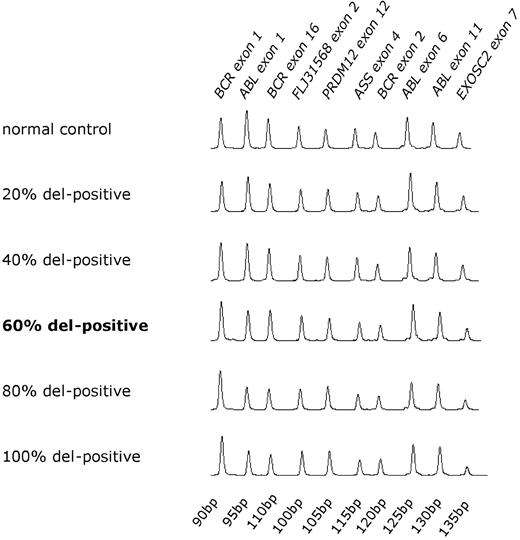
CML blood samples contain a variable background of Philadelphia chromosome-negative cells and therefore we initially established the sensitivity of MLPA to detect der(9) deletions using dilutions of the MC3 cell line or deleted patient DNA in normal DNA. The deletion was detected in 100%, 80%, and 60% dilutions of DNA from the 2 deleted patients with CML but was not detected at lower dilutions (Figure 2 and Table 3). MLPA is therefore capable of detecting deletions in the great majority of pretreatment CML samples, which we have previously shown by quantitative Southern blot analysis to harbor a median of 84% (range, 64%-120%) cells derived from the malignant clone.19
Sensitivity of multiplex ligation-dependent probe amplification assay. MLPA traces of chronic myelogenous leukemia DNA known to harbor a large der(9) deletion diluted in nondeleted DNA. The relative ratios of der(9) to control der(22) probes reduces as the proportion of patient DNA increases.
Sensitivity of multiplex ligation-dependent probe amplification assay. MLPA traces of chronic myelogenous leukemia DNA known to harbor a large der(9) deletion diluted in nondeleted DNA. The relative ratios of der(9) to control der(22) probes reduces as the proportion of patient DNA increases.
MLPA can detect deletions when the proportion of patient DNA known to harbor deletions is 60% or greater
| . | ASS . | PRDM12 . | EXOSC2 . | ABL exon 1 . | BCR exon 16 . | FLJ31568 . |
|---|---|---|---|---|---|---|
| 100% | del | del | del | del | del | del |
| 80% | del | del | del | del | del | del |
| 60% | del | del | del | del | del | del |
| 40% | del | + | del | del | + | + |
| 20% | + | + | del | + | + | + |
| 0% | + | + | + | + | + | + |
| . | ASS . | PRDM12 . | EXOSC2 . | ABL exon 1 . | BCR exon 16 . | FLJ31568 . |
|---|---|---|---|---|---|---|
| 100% | del | del | del | del | del | del |
| 80% | del | del | del | del | del | del |
| 60% | del | del | del | del | del | del |
| 40% | del | + | del | del | + | + |
| 20% | + | + | del | + | + | + |
| 0% | + | + | + | + | + | + |
At 60% patient DNA and over, all 6 markers are correctly scored as deleted, whereas at lower dilutions, deletions are often missed.
+ indicates not deleted (dosage quotient ≥ 0.7); del, deleted (dosage quotient < 0.7).
To validate the MPLA assay, we tested 20 samples from healthy people and 18 samples from patients with CML with der(9) deletions as determined by FISH. No deletions were detected in the healthy people; of the 480 individual measurements (6 test probes relative to 4 control probes for the 20 healthy people), the mean dosage quotient was 1.001 (standard deviation 0.07) and only a single measurement was outside the range of 0.7 to 1.3. In contrast, deletions were detected in 17 of the 18 CML samples. The CML case that was not detected had only 16% der(9)-positive cells and thus was well below the sensitivity of detection.
Derivative chromosome 9 deletions in the patient cohort
Der(9) deletions were detected in 59 of the 339 patients (17%) in the study group. Deletions encompassed both chromosome 9 and chromosome 22-derived sequences in 21 cases (36%) or were either upstream only (n = 20) or downstream only (n = 18) of the ABL/BCR fusion point. The median age of the 59 patients with deletions was 49 years (range, 11-72 years) with 54% being male. According to the Hasford score, 26 patients were low risk, 24 were intermediate risk, and 9 were high risk. Therapy subsequent to IFN included IM (n = 13) and allogeneic SCT (n = 20). After a median observation time of 8 years (range, 0.5-16 years), 33 (56%) of the der(9)-deleted cases were still alive.
Overall deletion status is not associated with a poor prognosis
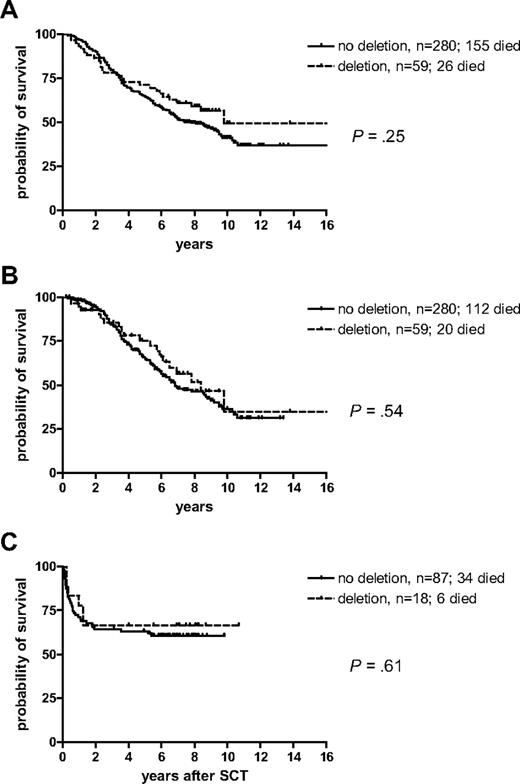
Of the 47 deleted cases for whom cytogenetic data were available, 10 (21%) achieved major cytogenetic response (MCR) and 5 (11%) achieved cytogenetic complete remission (CCR). These proportions were somewhat lower than that seen for the 239 evaluable case patients that did not have deletions, of whom 66 (28%) achieved MCR and 30 (13%) achieved CCR. However, no significant difference was seen between the overall survival of the 59 deleted cases compared with the 280 nondeleted patients (P = .25; median survival: 9.8 versus 7.8 years; Figure 3A). If chronic-phase patients were censored at the time of switchover to IM or allogeneic SCT, the difference between both groups further declined (P = .54; median survival 8.4 versus 6.8 years; Figure 3B). Furthermore, no impact of deletion status on survival was observed for the 105 patients who underwent SCT in the first chronic phase (P = .61; Figure 3C). In this group, the median survival was not reached. The 5-year survival for the 18 patients who harbored deletions was 0.67 compared with 0.63 for the 87 nondeleted cases.
Kaplan-Meier survival curves for the 59 patients with deletions compared with the 280 patients without deletions. (A) Overall survival, (B) survival censored at the time of switch to imatinib or allogeneic stem cell transplantation in the first chronic phase, (C) survival after allogeneic stem cell transplantation in the first chronic phase.
Kaplan-Meier survival curves for the 59 patients with deletions compared with the 280 patients without deletions. (A) Overall survival, (B) survival censored at the time of switch to imatinib or allogeneic stem cell transplantation in the first chronic phase, (C) survival after allogeneic stem cell transplantation in the first chronic phase.
Significance of deletions that span or do not span the ABL/BCR breakpoint
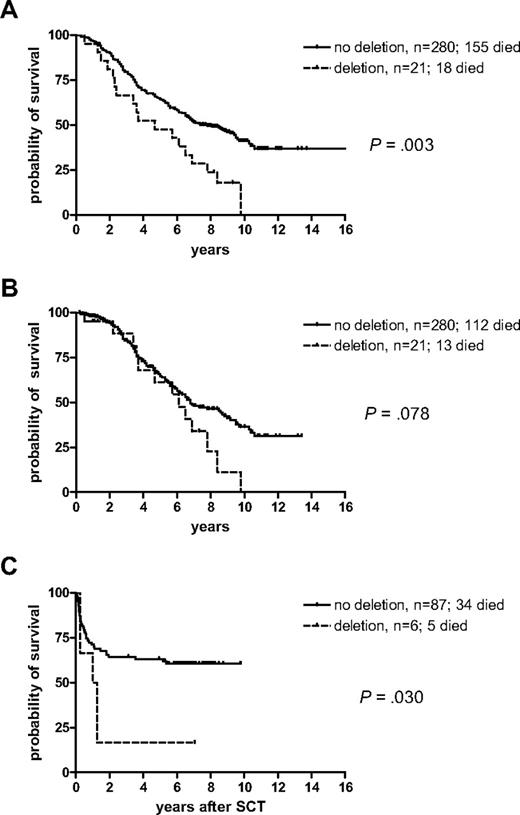
To determine whether the position or size of the deletion might be important, we considered those patients with deletions that spanned the ABL/BCR junction separately from those that had deletions on the ABL or BCR sides only. Of the 21 cases with breakpoint spanning deletions, 20 were missing all 6 markers and one lacked all markers apart from FLJ31568. Overall, survival for these 21 patients was significantly worse than for the nondeleted patients (P = .003; median survival 4.7 years versus 7.8 years; Figure 4A). Censorship at the time of IM or allogeneic SCT in the chronic phase leads to a loss of power and significance (P = .078; median survival: 6.1 versus 6.8 years), but the trend is still apparent (Figure 4B). Despite this adverse association, 5 of the 19 evaluable patients with junction-spanning deletions achieved MCR and 2 achieved CCR on IFN. Survival for the 6 patients with ABL/BCR-spanning deletions who underwent allogeneic SCT in the first chronic phase was significantly shorter compared with the 87 nondeleted patients (P = .030; Figure 4C).
Kaplan-Meier survival curves for the 21 patients with deletions that spanned the ABL/BCR fusion compared with all patients without deletions. (A) Overall survival, (B) survival censored at the time of switch to imatinib or allogeneic stem cell transplantation in the first chronic phase, (C) survival after allogeneic stem cell transplantation in the first chronic phase.
Kaplan-Meier survival curves for the 21 patients with deletions that spanned the ABL/BCR fusion compared with all patients without deletions. (A) Overall survival, (B) survival censored at the time of switch to imatinib or allogeneic stem cell transplantation in the first chronic phase, (C) survival after allogeneic stem cell transplantation in the first chronic phase.
Deletions of either ABL or BCR sequences only
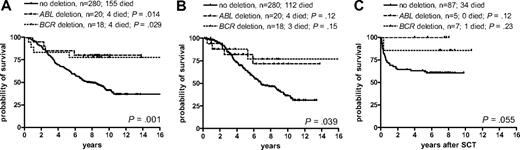
Of the 20 patients with deletions only on the ABL side of the der(9) fusion point, 17 included all 4 markers and 3 included only ABL exon 1b and EXOSC2. By definition, all 18 cases with deletions on the BCR side only included both markers because we only scored cases as being deletion-positive if they lacked 2 consecutive markers. Although no difference in survival was seen between cases with deletions either upstream only (ABL side: 4 patients died; 8-year survival = 0.80) or downstream only (BCR side: 4 patients died; 8-year survival = 0.78) of the breakpoint, unexpectedly we found that these deletions were associated with a superior survival compared with the 280 cases without deletions (P = .029 for BCR deletion only; P = .014 for ABL deletion only; P = .001, all 38 grouped together; Figure 5A). When patients with IM or SCT in the chronic phase were censored, the comparison between those without deletions and the 38 with one-sided deletions grouped together retained significance (P = .039; Figure 5B). The superior impact of the one-sided deletions was also not detectable with regard to outcome after allogeneic SCT in the first chronic phase (87 without deletions versus 12 patients with one-sided deletions; P = .055; 5-year survival: 0.63 and 0.92; Figure 5C). Of the 28 cases with one-sided deletions and cytogenetic data available, 5 (18%) achieved MCR and 3 (11%) achieved CCR.
Kaplan-Meier survival curves for the patients with deletions of either ABL (n = 20) or BCR sequences (n = 18), only, compared with all patients without deletions. (A) Overall survival, (B) survival censored at time of switch to imatinib or allogeneic stem cell transplantation in the first chronic phase, (C) survival after allogeneic stem cell transplantation in the first chronic phase.
Kaplan-Meier survival curves for the patients with deletions of either ABL (n = 20) or BCR sequences (n = 18), only, compared with all patients without deletions. (A) Overall survival, (B) survival censored at time of switch to imatinib or allogeneic stem cell transplantation in the first chronic phase, (C) survival after allogeneic stem cell transplantation in the first chronic phase.
Comparison between deletion status and hematologic parameters
To determine whether deletion status correlated with standard clinical and hematologic parameters (age, spleen enlargement, white blood cell count, hemoglobin, and the proportions of blasts, eosinophils, basophils, and platelets in peripheral blood), univariate analysis was performed for patients with (1) deletions that spanned the ABL/BCR breakpoint, (2) deletions upstream or downstream of ABL or BCR, or (3) patients without deletions. We found that spleen enlargement was larger in patients with deletions not spanning the breakpoint compared with either patients with spanning ABL/BCR deletions or no deletions (P = .002). Patients with ABL or BCR deletions only were younger (P = .030) and showed a higher proportion of blasts than others (P = .004). All other parameters showed no significant differences.
Independent influence of deletion status on survival probabilities
To determine whether deletion status was an independent influence with regard to survival, we performed multiple Cox regression analysis. To attribute survival probabilities to IFN treatment, survival times were censored at the start of IM therapy or the date of an allogeneic SCT for patients still in the first chronic phase. We also sought to establish the prognostic value of deletion status in relation to the Hasford score, which was developed and validated to differentiate 3 prognostic risk groups with regard to survival after treatment with IFN.14,27–30
Of 338 patients with deletion status and Hasford score available, 131 had died at the time of analysis. The low-risk group contained 132 patients (median survival, 8.6 years), the intermediate-risk group 166 (median survival, 6.8 years), and the high-risk group 40 patients (median survival, 5.6 years). The survival probabilities among the 3 prognostic groups were significantly different (P = .049). As prognostic factors in a common Cox model, deletion status and the Hasford score kept their statistical significance (P = .007 and P = .011, respectively), which hints that both are an independent prognostic influence on survival (model A, Table 4). By comparison, deletion status and Sokal score yielded slightly higher P values (P = .016 and P = .028, respectively).
Multiple Cox regression: deletion status and Hasford score as independent prognostic factors for survival probabilities
| Model A . | No./died† . | Estimation of coefficient β . | Standard deviation of estimated β . | Wald's χ2 statistic . | Pvalue . | Hazard ratio . |
|---|---|---|---|---|---|---|
| Deletion status | 338/131 | — | — | 10.084 (2 df) | .007 | — |
| No | 279/111 | Baseline | — | — | — | — |
| One side of breakpoint | 38/7 | −0.896 | 0.394 | 5.163 | .023 | 0.408 |
| Whole breakpoint | 21/13 | 0.620 | 0.297 | 4.343 | .037 | 1.859 |
| Hasford score | 338/131 | — | — | 8.966 (2 df) | .011 | — |
| Low | 132/36 | −0.279 | 0.204 | 1.876 | .17 | 0.756 |
| Intermediate | 166/73 | Baseline | — | — | — | — |
| High | 40/22 | 0.546 | 0.249 | 4.818 | .028 | 1.727 |
| Model A . | No./died† . | Estimation of coefficient β . | Standard deviation of estimated β . | Wald's χ2 statistic . | Pvalue . | Hazard ratio . |
|---|---|---|---|---|---|---|
| Deletion status | 338/131 | — | — | 10.084 (2 df) | .007 | — |
| No | 279/111 | Baseline | — | — | — | — |
| One side of breakpoint | 38/7 | −0.896 | 0.394 | 5.163 | .023 | 0.408 |
| Whole breakpoint | 21/13 | 0.620 | 0.297 | 4.343 | .037 | 1.859 |
| Hasford score | 338/131 | — | — | 8.966 (2 df) | .011 | — |
| Low | 132/36 | −0.279 | 0.204 | 1.876 | .17 | 0.756 |
| Intermediate | 166/73 | Baseline | — | — | — | — |
| High | 40/22 | 0.546 | 0.249 | 4.818 | .028 | 1.727 |
Survival times of patients with imatinib or stem cell transplantation in first chronic phase were censored.
df indicates degrees of freedom; —, not applicable.
For one patient, the Hasford score was not evaluable.
The Hasford score uses age, spleen enlargement, platelet count, and proportions of blasts, eosinophils, and basophils as covariates.14 The independent impact of these variables was assessed along with deletion status, age, hemoglobin, white blood cell count, and gender. This analysis yielded a final model with 3 independent variables: deletion status, age, and spleen enlargement with P = .007, P = .018, and P < .001, respectively (model B, Table 5). The P value for deletion status reflects both the adverse risk associated with breakpoint-spanning deletions (P = .026) and the beneficial effect of one-sided deletions (P = .039).
Multiple Cox regression: best prognostic model for survival probabilities
| Model B . | No./died† . | Estimation of coefficient β . | Standard deviation of estimated β . | Wald's χ† statistic . | P value . | Hazard ratio . |
|---|---|---|---|---|---|---|
| Deletion status | 335/130 | — | — | 9.784 (2 df) | .007 | — |
| No | 276/110 | Baseline | — | — | — | — |
| One side of breakpoint | 38/7 | −0.870 | 0.391 | 4.944 | .026 | 0.419 |
| Whole breakpoint | 21/13 | 0.623 | 0.302 | 4.251 | .039 | 1.864 |
| Age, fully completed years | 335/130 | 0.018 | 0.007 | 5.627 (1 df) | .018 | 1.018 |
| Spleen enlargement, cm | 335/130 | 0.058 | 0.016 | 13.519 (1 df) | < .001 | 1.059 |
| Model B . | No./died† . | Estimation of coefficient β . | Standard deviation of estimated β . | Wald's χ† statistic . | P value . | Hazard ratio . |
|---|---|---|---|---|---|---|
| Deletion status | 335/130 | — | — | 9.784 (2 df) | .007 | — |
| No | 276/110 | Baseline | — | — | — | — |
| One side of breakpoint | 38/7 | −0.870 | 0.391 | 4.944 | .026 | 0.419 |
| Whole breakpoint | 21/13 | 0.623 | 0.302 | 4.251 | .039 | 1.864 |
| Age, fully completed years | 335/130 | 0.018 | 0.007 | 5.627 (1 df) | .018 | 1.018 |
| Spleen enlargement, cm | 335/130 | 0.058 | 0.016 | 13.519 (1 df) | < .001 | 1.059 |
Survival times of patients with imatinib or stem cell transplantation in the first chronic phase were censored.
df indicates degrees of freedom; —, not applicable.
For the candidate variables deletion status, age, spleen enlargement, blasts, basophils, eosinophils, platelet count, haemoglobin, white blood cell count, and sex, 335 patients with complete cases were available of whom 130 patients died.
Discussion
Deletions of downstream BCR sequences at the der(9) fusion junction were first identified in a minority of patients with CML by Southern blot analysis.31 Development of FISH probes revealed that the deletions were larger than had been previously suspected and frequently included loss of SMARCB1, 500 kb telomeric of BCR.23 Subsequently, it was shown that the deletions were variable in size, often encompassed several megabases, and usually included both chromosome 9- and chromosome 22-derived sequence, that is, they spanned the reciprocal ABL/BCR junction.4 This study was also the first to suggest that the deletions may be associated with a poor prognosis, a hypothesis that was expanded in a larger analysis from the same group in which the median survival of the 14% of cases with deletions (most of which spanned the ABL/BCR junction) was significantly shorter than those without deletions.5 The adverse prognosis was confirmed in an independent large study in which deleted patients had a shorter duration of chronic phase, inferior survival, and increased probability of relapse after SCT.8 A more recent study of IFN-treated cases, however, failed to detect any difference in clinical outcome between deleted and nondeleted cases, although deleted patients were found to present with significantly lower hemoglobin levels and higher leukocyte counts.32 After imatinib therapy, der(9) deletions have not yet been reported to be a strong prognostic indicator. Huntly et al found no difference in survival between patients with and without deletions, although deleted cases had more rapid disease progression and exhibited poorer hematologic and cytogenetic responses.33 Quintas-Cardama et al, however, found no influence of der(9) deletions with regard to response, survival, or response duration.34 It seems that much longer follow up will be required to determine whether deletions do or do not have prognostic value for imatinib-treated patients.
In our study, we sought to (1) determine whether deletion status genuinely did predict a poor prognosis for IFN-treated cases and (2) determine the relationship of deletion status to the Hasford and Sokal risk scores. We developed and validated a novel DNA-based MLPA deletion assay and investigated 339 patients enrolled over 15 years in 3 trials of the German CML Study Group with an observation time up to 16 years. This is the largest group of IFN-treated cases to be analyzed for the impact of deletions. We found a similar proportion of deleted cases as other studies, but our series is unusual in that only a relatively small proportion of deletions (36% of deleted cases; 6% of all cases) spanned the ABL/BCR breakpoint.
Although we found no difference in survival between deleted and nondeleted cases, more detailed analysis indicated that deletions that spanned the breakpoint were a significant indicator of inferior prognosis (P = .039; Table 5). Unexpectedly, we found that deletions on one side of the breakpoint only were associated with improved survival (P = .026; Table 5), resulting in an overall multivariate P value of .007 for deletion status. Improved survival was seen for both deletions on the BCR side only and on the ABL side only. Because other published series only had a small proportion of cases with one-sided deletions, it is possible that any beneficial effect would have escaped notice. The biologic explanation for a beneficial effect of one-sided deletions is not immediately obvious. Current models suggest that the poor prognosis associated with deletions is probably the result of heterozygous loss of one or more loci rather than loss of ABL-BCR expression, general genomic instability, or effects on BCR-ABL.22,35,36 Our findings are consistent with the hypothesis that at least 2 separate loci may be targeted, one on each side of the fusion junction. However, it is not clear how deletion of both these loci could result in an adverse prognosis, whereas deletion of just one has the opposite effect.
Currently, the Hasford score is the best predictor of outcome for patients with CML treated with IFN and outperforms the Sokal score in this context.14,28 Information was available to calculate both scores for all but one of the cases in our study group, and overall there was no difference in clinical baseline data between deleted and nondeleted cases. It is noteworthy, however, that there were relatively few Hasford or Sokal high-risk patients (see Table 1) in the breakpoint-spanning deletion group and a relative excess of high-risk cases in patients with one-sided deletions. Given the overall good outcome of the latter group, this suggests that one-sided deletions may overcome or negate the adverse risk associated with having a high Hasford or Sokal score.
To examine the relationship between the Hasford score and deletion status in more detail, we first examined their contribution as prognostic factors in a common Cox model. Both kept their statistical significance, suggesting they are an independent prognostic influences on survival (Table 4). When individual components of the Hasford score were considered independently, age and spleen enlargement were the most important prognostic variables in addition to deletion status (Table 5). Our findings thus confirm that at diagnosis, deletion status provides independent, statistically significant prognostic information with respect survival probability. However, only deletions that span the der(9) breakpoint are an adverse risk factor.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by Deutsche Krebshilfe, the Leukaemia Research Fund (United Kingdom), the Wessex Cancer Trust, the Lady Tata Memorial Trust, the Competence Network “Acute and chronic leukemias,” sponsored by the German Bundesministerium für Bildung und Forschung (Projektträger Gesundheitsforschung, DLR e.V.-01 GI9980/6), and the European LeukemiaNet within the 6th European Community Framework Programme for Research and Technological Development.
We thank all those who contributed to the sample and data collection at the CML trial office in Mannheim, Germany.
Authorship
Contribution: S.K., K.W., C.H., and A.C. performed the laboratory work; S.K. and M.P. performed the statistical analysis; R.H., A.R., and A.H. provided clinical samples and data; A.H. and N.C.P.C. conceived, designed, and directed the study; and all authors contributed to writing the manuscript and approved the final version.
A complete list of the participating institutions of the German CML Study Group is provided in Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nicholas C. P. Cross, Wessex Regional Genetics Laboratory, University of Southampton, Salisbury District Hospital, Salisbury, SP2 8BJ, United Kingdom; e-mail: ncpc@soton.ac.uk.