Abstract
Frameshift mutations in exon 12 of the nucleophosmin gene (NPM1) result in aberrant cytoplasmic localization of the NPM protein (NPMc+) and occur in 25% to 35% of adult acute myeloid leukemia (AML). In adults with AML, NPMc+ has been associated with normal karyotype, FLT3/ITD mutations, high remission induction rates, and improved survival (particularly in patients lacking FLT3/ITD). NPMc+ has not been well characterized in childhood AML. This study examines the incidence and clinical significance of NPMc+ in 295 children with newly diagnosed AML treated on a large cooperative group clinical trial (POG-9421). We find that NPMc+ is relatively uncommon in childhood AML (23 of 295 patients, 8%); and is significantly associated with FLT3/ITD mutations (P = .046), female sex (P = .029), older age (P = .047), and normal cytogenetics (P < .001). There is a favorable impact of NPMc+ on survival in children lacking FLT3/ITD (5-year EFS, 69% vs 35%; hazard ratio, 0.39; P = .051), which is similar in magnitude to the favorable impact of t(8;21) and inv(16). We conclude that NPMc+ is relatively rare in childhood AML, particularly in younger children. NPMc+ does not abrogate the negative prognostic influence of FLT3/ITD mutations, but may contribute to risk stratification in children who lack FLT3/ITD mutations by identifying a group with superior prognosis.
Introduction
Acute myeloid leukemia (AML) is a clinically and genetically heterogeneous disease that accounts for 15% to 20% of childhood leukemia. Currently, cytogenetic analysis at diagnosis allows for risk stratification of childhood AML into favorable, adverse, and intermediate risk groups.1 Treatment protocols are, to some extent, risk adapted in an attempt to improve survival and decrease treatment-related toxicity. Unfortunately, prognostic implications have not been reliably established for AML in the intermediate risk category, a group that includes 60% to 70% of childhood AML. In recent years, molecular analysis has identified novel markers with prognostic relevance in this diverse group. For example, cases of AML with internal tandem duplications of fms-like tyrosine kinase-3 (FLT3/ITD) have a poor prognosis.2-5 Conversely, cases with a mutation in the transcription factor CCAAT/enhancer-binding protein-α (CEBPA) have a more favorable prognosis.6,7 Mutations in exon 12 of nucleophosmin (NPM1) have been investigated recently as another potentially useful molecular marker in AML.8
NPM1 encodes a phosphoprotein that is ubiquitously expressed and is highly conserved. The protein is localized primarily to the nucleolus, but shuttles rapidly between the nucleus and cytoplasm in its role as a molecular chaperone.9 The NPM1 protein has been shown to contribute to many basic cellular processes, including biosynthesis of ribosomes,10 preventing aggregation of proteins in the nucleolus,11 and regulating centrosome duplication through cyclin E/cyclin-dependent kinase 2 phosphorylation.12 Further, it has been found to be a stress-induced regulator of p53 function13 and is a nucleolar binding partner of ARF, involved in p53-independent cell cycle regulation.14 Abnormalities in NPM1 have been implicated in the pathogenesis of various hematopoietic malignancies, including anaplastic large cell lymphoma, where the t(2;5)(p23;q35) translocation occurs in the majority of cases, resulting in expression of the NPM1-ALK fusion protein.15
Frameshift mutations in exon 12 of NPM1 are the most common mutation in adult AML. Several variants of NPM1 mutations have been identified. All of the variants result in the insertion of 4 base pairs in the C-terminal region, causing loss of a nucleolar localization signal and aberrant localization of the protein to the cytoplasm (NPMc+).8 In adults, NPMc+ is present in 25% to 35% of all AML and up to 60% of normal karyotype AML.8,16-22 NPMc+ has been shown to be significantly associated with several genetic and clinical features including high frequency of FLT3/ITD mutations,8,16-20,22 higher white blood cell count (WBC) at diagnosis,16-22 higher percentage of blasts,17-19,22 and improved responsiveness to induction chemotherapy.8,17,19,20,22 NPM1 mutations are found in a broad spectrum of AML subtypes with the highest frequency in the myelomonocytic and monocytic leukemias.8,16,17,20-22 Adult studies have also suggested that patients with NPM1 mutations in the absence of FLT3/ITD mutation have better long-term outcomes.16,17,19,20,22 As age increases, the frequency of NPM1 mutations increases.8,16,18,19 A number of studies in adults have suggested that NPM1 mutations may also be useful as molecular markers for minimal residual disease monitoring.18,23,24
To date, there are only a few small studies specifically investigating NPM1 mutations in childhood AML. The AIEOP in Italy reported NPMc+ in 7 (6.5%) of 107 children treated on its AML02 protocol,25 a Taiwanese group reported NPMc+ in 1 (2.1%) of 47 children,18 and the BFM in Germany reported NPMc+ in 9 (12%) of 75 children and young adults treated on its AML98 and AML2004 protocols.26 The prognostic significance of NPMc+ in childhood AML is not known. The objective of our study was to determine the prevalence and prognostic significance of NPM1 mutations in childhood AML in a large, well-characterized cohort of homogenously treated children enrolled in a large, cooperative group clinical trial (POG-9421).
Patients, materials, and methods
Patient samples
The specimens used in this study were bone marrow samples archived at the Children's Oncology Group AML cell bank from Pediatric Oncology Group (POG) study 9421. The POG 9421 clinical trial enrolled patients younger than 21 years with de novo AML from 1995 to 1999, and the results of the trial were recently published.27 The total number of eligible patients enrolled in POG-9421 was 622. There were 353 specimens archived in the POG reference laboratory's leukemia cell bank. Of these 353 samples, 306 (87%) were diagnostic bone marrow specimens. RNA was isolated from these samples using previously described methods.28 Institutional review board (IRB) approval was obtained from each participating institution for all studies described. Informed consent was obtained in accordance with the Declaration of Helsinki.
Detection of NPMc+ mutations
RNA samples were reverse transcribed with random hexamer primers to generate cDNA. The cDNA corresponding to exon 12 on NPM1 was amplified by polymerase chain reaction (PCR) using the following primers: NPMF, 5′-CCATCATCAACACCAAGATCA and NPMR, 5′-CATGTCTGACCACCGCTACT. Reactions (50 μL each) were prepared for each sample using 45 μL PCR Super Mix (Invitrogen, Carlsbad, CA), 1 μL of a 5-mM solution of each primer, and approximately 100 ng cDNA. Samples were amplified using the following PCR conditions: 94°C for 3 minutes; 35 cycles of 94°C for 45 seconds, 52°C for 30 seconds, 72°C for 1 minute; 72°C for 10 minutes. PCR product (20 μL) was mixed with 5 μL 5X DNA gel loading solution (Quality Biological, Gaithersburg, MD) and loaded on a 5% nondenaturing polyacrylamide TBE gel. Electrophoresis was performed at approximately 5 V/cm for 4 to 6 hours at 4°C. Resolved PCR products were stained with ethidium bromide and visualized using a UV transilluminator. Mutant samples were identified, and the corresponding PCR products were purified using PureLink PCR Purification Kit (Invitrogen) following the kit insert. Purified PCR products were directly sequenced at the Genetic Resource Core Facility of Johns Hopkins University using forward and reverse primers. Resultant sequences were analyzed using Sequencher 4.6 software (Gene Codes, Ann Arbor, MI).
Detection of FLT3/ITD mutations
The methods for analysis of FLT3 genotype in the RNA specimens from POG 9421 have been previously described.28
Statistical methods
Statistical significance of the difference between groups for continuous variables was determined by the Mann-Whitney test. Statistical significance of the difference between groups for categoric variables was determined by Fisher exact test. Actuarial estimates of event-free survival (EFS) and overall survival (OS) were calculated using the Kaplan-Meier method.29 EFS is defined as the time from randomization to treatment failure (relapse, second malignancy, or remission failure) or death. Patients lost to follow-up were censored at their last known point of study. Statistical significance of the difference in survival curves was determined by log-rank test.30 Multivariate analyses were performed using Cox proportional hazards regression.31
Results
Study population
Of the 622 children with de novo AML enrolled on POG 9421 between February 15, 1995, and August 15, 1999, 306 had diagnostic marrow specimens available for analysis and were included in this study. We compared the characteristics of patients with marrow available (N = 306) and those without marrow available (N = 316). The 2 groups were similar in terms of race (P = .595), sex (P = .228), karyotype (P = .37), and induction response rates (P = .733). The study population was more likely to have an initial WBC higher than 50 × 109/L (147 [48%] of 306 versus 49 [16%] of 316, P < .001) and was more likely to be older than 2 years (256 [84%] of 306 versus 214 [68%] of 316, P < .001) After adjusting for the differences in WBC and age, there was no significant difference between the 2 groups in 5-year EFS (P = .33) or 5-year OS (P = .20).
NPM1 mutation (NPMc+) analysis
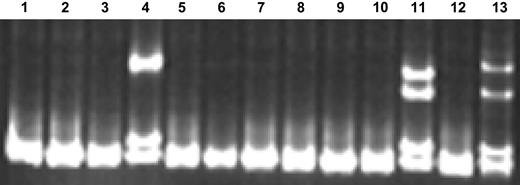
Of 306 patients with samples available for analysis, NPM1 mutation analysis was successful for 295 (96%). The unsuccessful cases (N = 11) were due to failure of either reverse transcription or PCR amplification. The prevalence of NPMc+ in this population of 295 patients was 8% (23 of 295) (Table 1). An example of 2 NPMc+ cases detected using single-stranded conformational polymorphism (SSCP) gel electrophoresis screening is provided in Figure 1. All mutations were confirmed by direct sequencing (Table 2). Of the 23 mutations, 15 (65%) have been previously reported and 8 (35%) are novel mutations. Of the 8 novel mutations, 7 involved typical 4–base pair insertions. One novel mutation consisted of 5 individual base pair substitutions (the bottom sample in Table 2), which predict changes in 4 amino acid residues in the NPM1 protein, including a change in the tryptophan at residue 290 (known to be critical for nucleolar localization) to glycine.
Characteristics of study population according to NPM1 mutation status
| . | All patients . | NPMc+ . | NPMc− . | P . | |||
|---|---|---|---|---|---|---|---|
| N . | % . | N . | % . | N . | % . | ||
| Total | 295 | 100 | 23 | 7.8 | 272 | 92.2 | |
| FLT3/ITD | .046‡ | ||||||
| Positive | 52 | 19 | 8 | 36 | 44 | 18 | |
| Negative | 218 | 81 | 14 | 64 | 204 | 82 | |
| Sex | .029‡ | ||||||
| Male | 144 | 49 | 6 | 26 | 138 | 51 | |
| Female | 151 | 51 | 17 | 74 | 134 | 49 | |
| Age, y | |||||||
| Median (range) | 9.5 (0-19.5) | 10.9 (3.1-18.8) | 9.4 (0-19.5) | .047‡ | |||
| 0 to younger than 3 | 71 | 24 | 0 | 0 | 71 | 26 | .002‡ |
| 3 to younger than 6 | 30 | 10 | 3 | 13 | 27 | 10 | .716 |
| 6 to younger than 9 | 39 | 13 | 4 | 17 | 35 | 13 | .523 |
| 9 to younger than 12 | 38 | 13 | 6 | 26 | 32 | 12 | .095 |
| 12 to younger than 15 | 60 | 20 | 4 | 17 | 56 | 21 | .999 |
| 15 to younger than 18 | 49 | 17 | 4 | 17 | 45 | 17 | .999 |
| 18 to younger than 21 | 8 | 3 | 2 | 9 | 6 | 2 | .122 |
| WBC, × 109/L, median (range) | 47.7 (1.3-667) | 38.2 (1.6-667) | 49.1 (1.3-633.6) | .942 | |||
| Cytogenetics | |||||||
| Normal | 87 | 29 | 17 | 74 | 70 | 26 | < .001‡ |
| 11q23* | 46 | 16 | 1 | 4 | 45 | 17 | .138 |
| inv(16) | 24 | 8 | 0 | 0 | 24 | 9 | .236 |
| t(8;21) | 26 | 9 | 0 | 0 | 26 | 10 | .142 |
| Miscellaneous† | 69 | 23 | 3 | 13 | 66 | 24 | .206 |
| Failed | 12 | 4 | 1 | 4 | 11 | 4 | .999 |
| Not received | 31 | 11 | 1 | 4 | 30 | 11 | .487 |
| Response to induction chemotherapy | .369 | ||||||
| CR | 248 | 85 | 18 | 78 | 230 | 85 | |
| Not CR | 45 | 15 | 5 | 22 | 40 | 15 | |
| . | All patients . | NPMc+ . | NPMc− . | P . | |||
|---|---|---|---|---|---|---|---|
| N . | % . | N . | % . | N . | % . | ||
| Total | 295 | 100 | 23 | 7.8 | 272 | 92.2 | |
| FLT3/ITD | .046‡ | ||||||
| Positive | 52 | 19 | 8 | 36 | 44 | 18 | |
| Negative | 218 | 81 | 14 | 64 | 204 | 82 | |
| Sex | .029‡ | ||||||
| Male | 144 | 49 | 6 | 26 | 138 | 51 | |
| Female | 151 | 51 | 17 | 74 | 134 | 49 | |
| Age, y | |||||||
| Median (range) | 9.5 (0-19.5) | 10.9 (3.1-18.8) | 9.4 (0-19.5) | .047‡ | |||
| 0 to younger than 3 | 71 | 24 | 0 | 0 | 71 | 26 | .002‡ |
| 3 to younger than 6 | 30 | 10 | 3 | 13 | 27 | 10 | .716 |
| 6 to younger than 9 | 39 | 13 | 4 | 17 | 35 | 13 | .523 |
| 9 to younger than 12 | 38 | 13 | 6 | 26 | 32 | 12 | .095 |
| 12 to younger than 15 | 60 | 20 | 4 | 17 | 56 | 21 | .999 |
| 15 to younger than 18 | 49 | 17 | 4 | 17 | 45 | 17 | .999 |
| 18 to younger than 21 | 8 | 3 | 2 | 9 | 6 | 2 | .122 |
| WBC, × 109/L, median (range) | 47.7 (1.3-667) | 38.2 (1.6-667) | 49.1 (1.3-633.6) | .942 | |||
| Cytogenetics | |||||||
| Normal | 87 | 29 | 17 | 74 | 70 | 26 | < .001‡ |
| 11q23* | 46 | 16 | 1 | 4 | 45 | 17 | .138 |
| inv(16) | 24 | 8 | 0 | 0 | 24 | 9 | .236 |
| t(8;21) | 26 | 9 | 0 | 0 | 26 | 10 | .142 |
| Miscellaneous† | 69 | 23 | 3 | 13 | 66 | 24 | .206 |
| Failed | 12 | 4 | 1 | 4 | 11 | 4 | .999 |
| Not received | 31 | 11 | 1 | 4 | 30 | 11 | .487 |
| Response to induction chemotherapy | .369 | ||||||
| CR | 248 | 85 | 18 | 78 | 230 | 85 | |
| Not CR | 45 | 15 | 5 | 22 | 40 | 15 | |
Percentages are calculated based on the available data.
NPMc+ indicates positive for NPM1 mutation; NPMc−, negative for NPM1 mutation; FLT3/ITD, internal tandem duplication of FLT3; and WBC, white blood cell count.
The 11q23 category included all cases with translocations involving the MLL gene at 11q23, most of which were t(9;11)(p22;q23), forming the MLL-AF9 fusion.
The miscellaneous category includes patients with cytogenetic abnormalities other than 11q23, inv(16), or t(8;21).
Significant P values.
SSCP gel demonstrating NPMc+ screening method. (Lane 12) NPM wild-type (NPMc−) control sample; (lane 13) NPMc+ control sample; (lanes 1-11) unknown samples. Lanes 4 and 11 contain samples confirmed to be NPMc+ by direct sequencing.
SSCP gel demonstrating NPMc+ screening method. (Lane 12) NPM wild-type (NPMc−) control sample; (lane 13) NPMc+ control sample; (lanes 1-11) unknown samples. Lanes 4 and 11 contain samples confirmed to be NPMc+ by direct sequencing.
NPM1 sequencing data for mutant samples
| Mutation . | No. (%) . | Sequence . |
|---|---|---|
| Wild type | — | GATCTCTG~~~~GCAG~~~~TGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| A | 10 (43) | GATCTCTGTCTGGCAG~~~~TGGA~~~~GGAACTCTCTTTAAGAAAATAG |
| B | 2 (9) | GATCTCTGCATGGCAG~~~~TGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| D | 1 (4) | GATCTCTGCCTGGCAG~~~~TGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| F | 1 (4) | GATCTCTG~~~~GCAG~~~~TGGACCCTGGAAGTCTCTTTAAGAAAATAG |
| J | 1 (4) | GATCTCTGTATGGCAG~~~~TGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| Novel | 1 (35) | GATCTCTGTACGGCAG~~~~TGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| Novel | 1 (35) | GATCTCTG~~~~GCAG~~~~CGGATGGCCGAAGTCTCTTTAAGAAAATAG |
| Novel | 1 (35) | GATCTCTGCGGAGCAG~~~~TGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| Novel | 2 (35) | GATCTCTG~~~~GCAG~~~~CGGATTCCGGAAGTCTCTTTAAGAAAATAG |
| Novel | 1 (35) | GATCTCTG~~~~GCAGCGTTCGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| Novel | 1 (35) | GATCTCTG~~~~GCAG~~~~TGGATGGAGGAAGTCTCTTTAAGAAAATAG |
| Novel* | 1 (35) | GATTTTTG~~~~GCAG~~~~GGGA~~~~GGAAGTTTTTTTAAGAAAATAG |
| Mutation . | No. (%) . | Sequence . |
|---|---|---|
| Wild type | — | GATCTCTG~~~~GCAG~~~~TGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| A | 10 (43) | GATCTCTGTCTGGCAG~~~~TGGA~~~~GGAACTCTCTTTAAGAAAATAG |
| B | 2 (9) | GATCTCTGCATGGCAG~~~~TGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| D | 1 (4) | GATCTCTGCCTGGCAG~~~~TGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| F | 1 (4) | GATCTCTG~~~~GCAG~~~~TGGACCCTGGAAGTCTCTTTAAGAAAATAG |
| J | 1 (4) | GATCTCTGTATGGCAG~~~~TGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| Novel | 1 (35) | GATCTCTGTACGGCAG~~~~TGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| Novel | 1 (35) | GATCTCTG~~~~GCAG~~~~CGGATGGCCGAAGTCTCTTTAAGAAAATAG |
| Novel | 1 (35) | GATCTCTGCGGAGCAG~~~~TGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| Novel | 2 (35) | GATCTCTG~~~~GCAG~~~~CGGATTCCGGAAGTCTCTTTAAGAAAATAG |
| Novel | 1 (35) | GATCTCTG~~~~GCAGCGTTCGGA~~~~GGAAGTCTCTTTAAGAAAATAG |
| Novel | 1 (35) | GATCTCTG~~~~GCAG~~~~TGGATGGAGGAAGTCTCTTTAAGAAAATAG |
| Novel* | 1 (35) | GATTTTTG~~~~GCAG~~~~GGGA~~~~GGAAGTTTTTTTAAGAAAATAG |
Insertions are shown in underlined type; substitutions are shown in bold italic type.
This sample contains multiple point mutations rather than a 4–base pair insertion (see “NPM1 mutation [NPMc+] analysis”).
FLT3/ITD analysis
Of 306 patients with samples available for analysis, FLT3/ITD mutation analysis was successful for 281 (92%). The unsuccessful cases (N = 25) were due to failure of either reverse transcription or PCR amplification. FLT3/ITD was present in 19% of cases (53 of 281). Of the 295 cases with known NPM1 mutation status, FLT3/ITD mutation analysis was successful for 270 (92%), and FLT3/ITD was present in 19% (52 of 270) (Table 1). The incidence of NPMc+ was higher in FLT3/ITD-positive cases (8 of 52, 15%) than in FLT3/ITD-negative cases (14 of 218, 6%; P = .046; odds ratio [OR] = 2.7; 95% confidence interval [CI] = 1.1-6.7).
Relationship between NPMc+ and other features
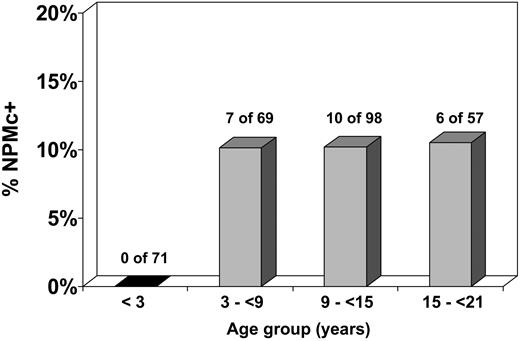
The demographic, laboratory, and cytogenetic characteristics of patients with and without NPM mutations were compared (Table 1). NPMc+ was significantly associated with certain cytogenetic and demographic features. The karyotype was more likely to be normal in NPMc+ cases (17 of 21, 81%) than in NPMc− cases (70 of 231, 30%; P < .001; OR = 9.8; 95% CI = 3.2-30.1). Of cases with normal karyotype, the prevalence of NPMc+ was 20% (vs 2% for cases with karyotypic abnormalities). NPMc+ cases demonstrated a female predominance, with a prevalence of NPMc+ of 11% in females versus 4% for males (P = .029; OR = 2.9; 95% CI = 1.1-7.6). Patients with NPMc+ were older, with a median age of 10.9 years for NPMc+ cases compared with 9.4 years for NPMc− cases (P = .047). Strikingly, NPMc+ was noted in 0 of 71 patients younger than 3 years at diagnosis, versus 23 (10%) of 224 patients at least 3 years old (P = .002; OR = 0.06; 95% CI = 0.003-1.001). There was no significant relationship between increasing age and NPMc+ incidence in patients 3 years of age or older (Figure 2). Median white blood cell count at diagnosis did not significantly differ between NPMc+ and NPMc− cases (P = .942).
Incidence of NPMc+ according to age. NPMc+ is extremely rare in children younger than 3 years. In children 3 years or older, the incidence of NPMc+ is approximately 10% and does not vary significantly by age group.
Incidence of NPMc+ according to age. NPMc+ is extremely rare in children younger than 3 years. In children 3 years or older, the incidence of NPMc+ is approximately 10% and does not vary significantly by age group.
Given the notable lack of NPMc+ in children less than 3 years, we compared the relative prevalence of NPMc+ and other molecular and cytogenetic abnormalities in this age group versus patients at least 3 years old (Table 3). Rearrangements of MLL at 11q23 were significantly more common in children younger than 3 years old (22 of 71, 31%) versus older children (24 of 224, 11%; P = .001). The cytogenetic abnormality t(8;21) was significantly less common in children younger than 3 years (0 of 71, 0%) versus older children (26 of 224, 12%; P = .001). Otherwise, there were no significant differences in the incidences of cytogenetic abnormalities in these 2 groups of patients.
Comparative prevalence of molecular and cytogenetic abnormalities by age group
| . | Younger than 3 y . | 3 y or older . | P . | ||
|---|---|---|---|---|---|
| N . | % . | N . | % . | ||
| Total | 71 | 100 | 224 | 100 | — |
| Molecular | |||||
| NPMc+ | 0 | 0 | 23 | 10 | .002* |
| FLT3/ITD | 4 | 6 | 49 | 22 | .001* |
| Cytogenetic | |||||
| Normal | 15 | 21 | 72 | 32 | .1 |
| 11q23 | 22 | 31 | 24 | 11 | .001* |
| inv(16) | 3 | 4 | 21 | 9 | .216 |
| t(8;21) | 0 | 0 | 26 | 12 | .001* |
| Miscellaneous | 21 | 30 | 48 | 21 | .198 |
| . | Younger than 3 y . | 3 y or older . | P . | ||
|---|---|---|---|---|---|
| N . | % . | N . | % . | ||
| Total | 71 | 100 | 224 | 100 | — |
| Molecular | |||||
| NPMc+ | 0 | 0 | 23 | 10 | .002* |
| FLT3/ITD | 4 | 6 | 49 | 22 | .001* |
| Cytogenetic | |||||
| Normal | 15 | 21 | 72 | 32 | .1 |
| 11q23 | 22 | 31 | 24 | 11 | .001* |
| inv(16) | 3 | 4 | 21 | 9 | .216 |
| t(8;21) | 0 | 0 | 26 | 12 | .001* |
| Miscellaneous | 21 | 30 | 48 | 21 | .198 |
Significant P values.
Prognostic impact of NPMc+ and FLT3/ITD
The complete remission (CR) rate was determined for patients with and without NPM mutations at the end of one course of therapy. CR data were available for 293 of the 295 patients with known NPM1 status. Eighteen (78%) of the 23 patients with NPMc+ achieved a CR compared with 230 (85%) of 270 of patients without NPM mutation (P = .369). Thus, NPM1 mutation status did not significantly affect induction CR rate.
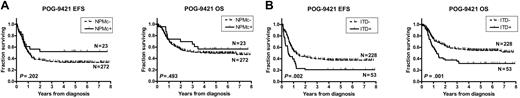
Survival data were examined for the 295 patients for whom the NPM1 mutation status was known (Figure 3A; Table 4). There was a trend toward superior event-free survival (EFS) from diagnosis for the NPMc+ patients (N = 23, 5-year EFS = 50% ± 11%) compared with the NPMc− patients (N = 272, 5-year EFS = 33% ± 3%), but this difference was not statistically significant (hazard ratio, 0.67; 95% confidence interval, 0.43-1.20; P = .202). Overall survival (OS) from diagnosis for the NPMc+ patients was similar, with 5-year OS = 55% ± 11% compared with 49% ± 3% for the NPMc− patients (hazard ratio, 0.8 [0.45-1.46]; P = .493).
Impact of NPMc+ and FLT3/ITD on survival. Kaplan-Meier estimates of event-free survival (EFS) and overall survival (OS) according to (A) NPMc+ and (B) FLT3/ITD. Log-rank P values are shown.
Impact of NPMc+ and FLT3/ITD on survival. Kaplan-Meier estimates of event-free survival (EFS) and overall survival (OS) according to (A) NPMc+ and (B) FLT3/ITD. Log-rank P values are shown.
Survival according to NPM1 mutation status
| . | NPM1−, % ± SEM N = 272 . | NPM1+, % ± SEM N = 23 . | Hazard ratio (95% CI) . | P . |
|---|---|---|---|---|
| 5-year EFS | 33 ± 3 | 50 ± 11 | 0.67 (0.43-1.20) | .202 |
| 5-year OS | 49 ± 3 | 55 ± 11 | 0.8 (0.45-1.46) | .493 |
| . | NPM1−, % ± SEM N = 272 . | NPM1+, % ± SEM N = 23 . | Hazard ratio (95% CI) . | P . |
|---|---|---|---|---|
| 5-year EFS | 33 ± 3 | 50 ± 11 | 0.67 (0.43-1.20) | .202 |
| 5-year OS | 49 ± 3 | 55 ± 11 | 0.8 (0.45-1.46) | .493 |
NPM1 indicates NPM1mutation status (+, NPMc+); EFS, event-free survival; OS, overall survival; SEM, standard error of measurement; and CI, confidence interval.
Survival data were examined for the 281 patients for whom the FLT3/ITD mutation status was known (Figure 3B; Table 5). The presence of a FLT3/ITD mutation was significantly associated with inferior EFS (5-year EFS = 21% ± 6% vs 37% ± 3% for FLT3/ITD negative; hazard ratio, 1.74 [1.29-3.02], P = .002) and OS (5-year OS = 32% ± 6% vs 54% ± 3% for FLT3/ITD negative; hazard ratio, 1.86 [1.35-3.37], P = .001).
Survival according to FLT3/ITD mutation status
| . | ITD−, % ± SEM N = 228 . | ITD+, % ± SEM N = 53 . | Hazard ratio (95% CI) . | P . |
|---|---|---|---|---|
| 5-year EFS | 37 ± 3 | 21 ± 6 | 1.74 (1.29-3.02) | .002 |
| 5-year OS | 54 ± 3 | 32 ± 6 | 1.86 (1.35-3.37) | .001 |
| . | ITD−, % ± SEM N = 228 . | ITD+, % ± SEM N = 53 . | Hazard ratio (95% CI) . | P . |
|---|---|---|---|---|
| 5-year EFS | 37 ± 3 | 21 ± 6 | 1.74 (1.29-3.02) | .002 |
| 5-year OS | 54 ± 3 | 32 ± 6 | 1.86 (1.35-3.37) | .001 |
ITD indicates FLT3/ITD status.
Bone marrow transplantation (BMT) was performed in 45 (15%) of the 295 patients for whom NPM1 mutation status was known, and in 41 (15%) of the 270 patients for whom both NPM1 and FLT3/ITD mutation status were known. There was no difference in the proportion of patients undergoing BMT between subgroups (P = .577). Neither censoring at the time of BMT nor excluding patients undergoing BMT had a significant impact on the survival analysis (data not shown).
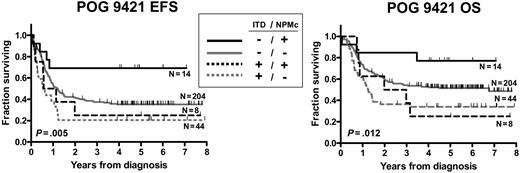
Since there is significant overlap and potential interaction between NPMc+ and FLT3/ITD, subgroup analysis was performed to assess the relative contributions of NPMc+ and FLT3/ITD to prognosis (Figure 4; Tables 6–7). Survival data were examined for the 270 patients for whom both NPM1 and FLT3/ITD mutation status were known. Within the FLT3/ITD-negative subgroup (N = 217), there was a trend toward improved EFS for the NPMc+ patients (N = 13, 5-year EFS = 69% ± 13%) compared with the NPMc− patients (N = 204, 5-year EFS = 35% ± 3%; hazard ratio, 0.39 [0.27-1.00], P = .051). There was also a trend toward improved OS for the NPMc+ patients within the FLT3/ITD-negative subgroup (5-year OS = 77% ± 12% versus 51% ± 4%; hazard ratio, 0.4 [0.25-1.14], P = .106). Neither the EFS nor the OS difference reached the level of statistical significance. A multivariate analysis adjusting for initial WBC and karyotype yielded similar hazard ratios for NPMc+. In contrast, in FLT3/ITD-positive patients, presence of NPMc+ did not impact EFS or OS (P = .568 and 0.824, respectively). The presence of FLT3/ITD was a significant negative prognostic factor within both the NPMc+ and NPMc− subgroups (OS hazard ratio for FLT3/ITD+ was 1.71 [P = .01] within the NPMc− group, and 4.23 [P = .016] within NPMc+ group).
NPMc+ has prognostic value only in FLT3/ITD-negative subgroup. Kaplan-Meier estimates of event-free survival (EFS) and overall survival (OS) according to combination of NPMc+ and FLT3/ITD. Log-rank P values are shown.
NPMc+ has prognostic value only in FLT3/ITD-negative subgroup. Kaplan-Meier estimates of event-free survival (EFS) and overall survival (OS) according to combination of NPMc+ and FLT3/ITD. Log-rank P values are shown.
Survival according to both NPMc and FLT3/ITD status
| . | ITD−/NPMc− . | ITD−/NPMc+ . | ITD+/NPMc− . | ITD+/NPMc+ . |
|---|---|---|---|---|
| N | 204 | 14 | 44 | 8 |
| 5-y EFS, % ± SEM | 35 ± 3 | 69 ± 13 | 21 ± 7 | 25 ±15 |
| 5-y OS, % ± SEM | 51 ± 4 | 77 ± 12 | 34 ± 7 | 25 ± 15 |
| . | ITD−/NPMc− . | ITD−/NPMc+ . | ITD+/NPMc− . | ITD+/NPMc+ . |
|---|---|---|---|---|
| N | 204 | 14 | 44 | 8 |
| 5-y EFS, % ± SEM | 35 ± 3 | 69 ± 13 | 21 ± 7 | 25 ±15 |
| 5-y OS, % ± SEM | 51 ± 4 | 77 ± 12 | 34 ± 7 | 25 ± 15 |
EFS indicates event-free survival; OS, overall survival; and SEM, standard error of measurement.
Statistical comparison of survival data from Table 6
| Comparison . | EFS hazard ratio(95% CI) . | EFS P . | CS hazard ratio(95% CI) . | OS P . |
|---|---|---|---|---|
| ITD−; NPMc− vs NPMc+ | 0.39 (0.27-1.00) | .051 | 0.4 (0.25-1.14) | .106 |
| ITD+; NPMc− vs NPMc+ | 0.78 (0.35-1.77) | .568 | 0.9 (0.39-2.13) | .824 |
| NPMc−; ITD− vs ITD+ | 1.72 (1.22-3.10) | .005 | 1.71 (1.17-3.12) | .01 |
| NPMc+; ITD− vs ITD+ | 3.1 (0.93-13.3) | .063 | 4.23 (1.24-21.4) | .016 |
| Comparison . | EFS hazard ratio(95% CI) . | EFS P . | CS hazard ratio(95% CI) . | OS P . |
|---|---|---|---|---|
| ITD−; NPMc− vs NPMc+ | 0.39 (0.27-1.00) | .051 | 0.4 (0.25-1.14) | .106 |
| ITD+; NPMc− vs NPMc+ | 0.78 (0.35-1.77) | .568 | 0.9 (0.39-2.13) | .824 |
| NPMc−; ITD− vs ITD+ | 1.72 (1.22-3.10) | .005 | 1.71 (1.17-3.12) | .01 |
| NPMc+; ITD− vs ITD+ | 3.1 (0.93-13.3) | .063 | 4.23 (1.24-21.4) | .016 |
EFS indicates event-free survival; OS, overall survival; and CI, confidence interval.
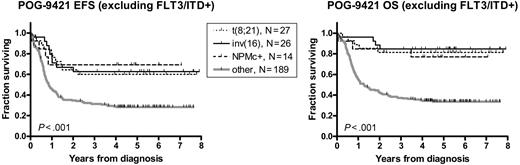
Given the trend of a favorable prognostic influence of NPMc+ in patients who lack FLT3/ITD mutations, we compared NPMc+ to the established favorable prognostic factors in AML, t(8;21), and inv(16), within this FLT3/ITD-negative subset (Figure 5; Table 8). The 5-year EFS and OS for the NPMc+ subgroup (N = 13) were 69% ± 13% and 77% ± 10%, respectively, and were not significantly different from the t(8;21) (N = 27; EFS = 60% ± 10% and OS = 81% ± 8%, P = .729 and P = .766) and inv(16) subgroups and (N = 26, EFS = 63% ± 10% and OS = 77% ± 10%, P = .868 and P = .565). The remainder of the FLT3/ITD-negative patients (N = 189) demonstrated inferior survival (EFS = 28% ± 3% and OS = 34% ± 4%). We separately compared this group with each of the 3 favorable subgroups (NPMc+, t(8;21), and inv(16)). The differences in survival were significant for all 3 subgroups (for EFS: hazard ratio range, 0.32 to 0.40 and P value range, .002 to .017; for OS: hazard ratio range; 0.16-0.25 and P value range, < .001 to .009).
NPMc+ is similar to t(8;21) and inv(16) in predicting favorable outcome in FLT3/ITD negative patients. Kaplan-Meier estimates of event-free survival (EFS) and overall survival (OS) in patients lacking FLT3/ITD mutations. Log-rank P values are shown.
NPMc+ is similar to t(8;21) and inv(16) in predicting favorable outcome in FLT3/ITD negative patients. Kaplan-Meier estimates of event-free survival (EFS) and overall survival (OS) in patients lacking FLT3/ITD mutations. Log-rank P values are shown.
Survival of FLT3/ITD-negative patients according to presence of favorable prognostic factors
| Favorable feature . | No. . | 5-year EFS, ± SEM, % . | Compared with “None” . | 5-year OS, ± SEM, % . | Compared with “None” . | ||
|---|---|---|---|---|---|---|---|
| Hazard ratio (95% CI) . | P . | Hazard ratio (95% CI) . | P . | ||||
| t(8;21) | 27 | 60 ± 10 | 0.40 (0.33-0.80) | .004 | 81 ± 8 | 0.20 (0.25-0.63) | < .001 |
| inv(16) | 26 | 63 ± 10 | 0.36 (0.31-0.77) | .002 | 85 ± 7 | 0.16 (0.23-0.58) | < .001 |
| NPMc+ | 14 | 69 ± 13 | 0.32 (0.26-0.88) | .017 | 77 ± 10 | 0.25 (0.24-0.82) | .009 |
| None | 189 | 28 ± 3 | — | — | 34 ± 4 | — | — |
| Favorable feature . | No. . | 5-year EFS, ± SEM, % . | Compared with “None” . | 5-year OS, ± SEM, % . | Compared with “None” . | ||
|---|---|---|---|---|---|---|---|
| Hazard ratio (95% CI) . | P . | Hazard ratio (95% CI) . | P . | ||||
| t(8;21) | 27 | 60 ± 10 | 0.40 (0.33-0.80) | .004 | 81 ± 8 | 0.20 (0.25-0.63) | < .001 |
| inv(16) | 26 | 63 ± 10 | 0.36 (0.31-0.77) | .002 | 85 ± 7 | 0.16 (0.23-0.58) | < .001 |
| NPMc+ | 14 | 69 ± 13 | 0.32 (0.26-0.88) | .017 | 77 ± 10 | 0.25 (0.24-0.82) | .009 |
| None | 189 | 28 ± 3 | — | — | 34 ± 4 | — | — |
Discussion
Our study demonstrates in a large cohort of children that NPMc+ is significantly less common in childhood than adult AML (8% vs 25%-35%). There is a striking relationship between NPMc+ and age in children (Figure 2), such that NPMc+ appears not to occur in children younger than 3 years (0 of 71 patients). In patients older than 3 years, the incidence of NPMc+ appears to be fairly constant at approximately 10%. FLT3/ITD and t(8;21) are also significantly less common in the youngest age group, while rearrangements of MLL at 11q23 are significantly more common in the youngest patients (Table 3). Beyond the age of 21 years, the incidence of NPMc+ appears to increase in an age-related fashion, such that patients between the ages of 21 and 35 years have an incidence of approximately 15%, patients between 35 and 60 years have an incidence of approximately 40%, and patients older than 60 years have an incidence of approximately 50%.16 Similar age-dependent increases in incidence have been noted for other molecular abnormalities. FLT3/ITD mutations, for example, have an incidence in children of approximately 12% to 18% (vs 25%-35% in adults), are quite uncommon in infants (incidence of approximately 1%), and are more common in elderly adults versus younger adults.2,4,32
These epidemiologic data suggest that the risk of acquiring a mutation such as NPMc+ or FLT3/ITD in a myeloid stem/progenitor cell is cumulative. The rarity of NPMc+ and FLT3/ITD in the youngest children is interesting, and suggests that the latency between the acquisition of NPMc+ or FLT3/ITD and the acquisition of the cooperating mutation(s) required for development of AML may be on the order of years. An alternative explanation is that the myeloid stem/progenitor cells in young children are relatively resistant to the acquisition of NPMc+ or FLT3/ITD mutations. It may be, for example, that these mutations are secondary events that occur only after the acquisition of a common underlying mutation that predisposes a myeloid stem/progenitor cell to errors in DNA replication. This possibility is supported by the data that NPMc+ and FLT3/ITD occur together in many cases, and by the fact that the most common variant (“A” type) of NPM1 mutation is a 4–base pair internal tandem duplication (ITD), the same type of mutation that occurs in FLT3 to cause the FLT3/ITD.
Similar to findings in adult AML,8,16-22 our study shows that in childhood AML, NPMc+ is significantly associated with normal karyotype (OR = 9.8, P < .001) and with FLT3/ITD mutations (OR = 2.6, P = .46). A stronger association between NPMc+ and female sex (OR = 2.9, P = .029) was noted in this study than in most studies of adult AML, where the relationship between sex and NPMc+ has been inconsistent, with no difference noted in some studies,16,19,21 and a slight to significant female predominance noted in others.17,20,22
An interesting difference between NPMc+ in childhood versus adult AML is seen in the distribution of specific mutation variants (Table 2). In adult AML, the “A” variant mutation (tandem duplication of TCTG) accounts for approximately 80% of cases. In our series of children, only 43% were “A” mutants, and 35% of the cases represent novel mutations that have not been reported to occur in adults. One of these novel mutations is of particular interest, since instead of involving a 4–base pair insertion, it involves multiple base pair substitutions, one of which is predicted to cause a change of the tryptophan at position 290 to glycine. To our knowledge, this is the only NPM1 mutation of this type yet reported. The tryptophan 290 residue has been shown to be critical to the nucleolar localization of the NPM1 protein,33,34 so it is likely that this novel mutation, like the more typical 4–base pair insertions, results in abnormal cytoplasmic localization of the NPM1 protein. Further studies will be needed to confirm this.
The different mutation variant distribution in children versus adults raises the possibility that there are significant differences in the mechanisms responsible for the introduction of NPM1 mutations in the 2 populations. Interestingly, there is a fairly extensive literature describing the ontogeny of various DNA repair enzymes and pathways during mammalian development (reviewed in Vinson and Hales35 ), and describing how these developmental patterns may relate to the development of childhood cancer (reviewed in Anderson36 ). It seems reasonable to speculate that such variation may be responsible for our finding that the distribution of specific NPM1 mutations differs in children versus adults.
While the epidemiology of NPMc+ in children differs from that in adults in some respects, there appears to be concordance in the prognostic implications of NPMc+ in the 2 groups of patients. The conclusion of the largest prognostic studies in adult AML is that NPMc+ is a favorable prognostic factor in patients who lack FLT3/ITD mutations.16,17,20,22 Our study suggests that this is true in children as well (Figure 4; Tables 6–7). Since NPMc+ is relatively rare in children, we must be somewhat guarded in our conclusions due to the small numbers of patients in the NPMc+/ITD− subgroup. Nevertheless, it is notable that within the FLT3/ITD− subgroup, the presence of NPMc+ is associated with a near doubling of 5-year EFS (from 35% to 69%, with a P value of .051), and a greater than 50% increase in 5-year OS (from 51% to 77%, with P value of .106).
Our study suggests that the NPMc+, FLT3/ITD-negative subset of childhood AML may be prospectively identifiable as a favorable risk group, similar to patients with favorable risk cytogenetics (t(8;21) and inv(16)). Approximately 60% of children with AML lack FLT3/ITD mutations and have standard risk cytogenetics (ie, lack either favorable or poor risk cytogenetics). For this large group of children with AML, there are currently no reliable prognostic markers. Our study shows that a significant proportion of these patients will have NPMc+, and may therefore join patients with t(8;21) and inv(16) in the favorable risk group (Figure 5). As such, NPMc+ may contribute significantly to risk stratification in childhood AML. One possibility is that NPMc+/ITD− patients may, like their counterparts with t(8;21) or inv(16), be curable in a high percentage of cases without requiring hematopoietic stem cell transplantation (HSCT). This would represent a significant improvement in the risk-based treatment strategy currently used in childhood AML.
Considering the importance of FLT3/ITD and NPM1 mutational status in the prognosis of childhood AML (not to mention their potential roles in minimal residual disease monitoring), it seems appropriate to consider prospective testing for these molecular abnormalities to be standard practice. This is perhaps more clearly the case with FLT3/ITD, where small molecule inhibitors of FLT3 signaling are beginning to be tested in clinical trials for children with AML. We are hopeful that as more is learned about the role of NPM1 mutations in myeloid leukemogenesis, treatments that target NPM1 can also be developed, so that NPM1 mutations will prove to represent not only an important element of risk stratification, but also an important molecular target that can be exploited to improve the outcome for children with AML.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the NCI (K23 CA111728, P.B.), Damon Runyon-Lilly Clinical Investigator Award (P.B.), and Children's Cancer Foundation (P.B.).
National Institutes of Health
Authorship
Contribution: P.B. designed and performed research, analyzed data, and wrote the paper; E.M. performed research and analyzed data; R.R. performed research and wrote the paper; S.M. designed and performed research, analyzed data, and wrote the paper; N.L. designed research and analyzed data; G.D. chaired the clinical trial that made this study possible (POG-9421); T.A.A. designed research, analyzed data, and wrote the paper; M.C. designed research, analyzed data, and wrote the paper; R.J.A. designed research, analyzed data, and wrote the paper; and D.S. designed research and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Patrick Brown, 1650 Orleans St, Rm CRB 2M49, Baltimore, MD, 21231; e-mail: pbrown2@jhmi.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal