The relative importance of various human leukocyte antigen (HLA) loci and the resolution level at which they are matched has not been fully defined for unrelated donor transplantation. To address this question, National Marrow Donor Program data from 3857 transplantations performed from 1988 to 2003 in the United States were analyzed. Patient-donor pairs were fully typed for HLA-A, -B, -C, -DRB1, -DQB1, -DQA1, -DPB1, and -DPA1 alleles. High-resolution DNA matching for HLA-A, -B, -C, and -DRB1 (8/8 match) was the minimum level of matching associated with the highest survival. A single mismatch detected by low- or high-resolution DNA testing at HLA-A, -B, -C or -DRB1 (7/8 match) was associated with higher mortality (relative risk, 1.25; 95% CI, 1.13-1.38; P < .001) and 1-year survival of 43% compared with 52% for 8/8 matched pairs. Single mismatches at HLA-B or HLA-C appear better tolerated than mismatches at HLA-A or HLA-DRB1. Mismatching at 2 or more loci compounded the risk. Mismatching at HLA-DP or -DQ loci and donor factors other than HLA type were not associated with survival. In multivariate modeling, patient age, race, disease stage, and cytomegalovirus status were as predictive of survival as donor HLA matching. High-resolution DNA matching for HLA-A, -B, -C, and -DRB1 alleles is associated with higher rates of survival.
Introduction
Hematopoietic stem cell transplantation (HCT) from volunteer unrelated donors (URDs) can cure patients with malignant and nonmalignant hematologic diseases who lack a suitable family member donor. Although donor human leukocyte antigen (HLA) matching is associated with better outcomes, many are not able to identify an HLA-A, -B, -C, and -DRB1 sequence-matched URD and are faced with choosing the closest match among the available donors. The National Marrow Donor Program (NMDP) has now completed retrospective high-resolution HLA typing on sufficient patient-donor pairs to analyze whether high- or low-resolution mismatches at specific loci are associated with higher mortality. These data are critical for selecting the best available partially HLA-matched donor for patients undergoing transplantation and determining the risk-benefit ratio of URD HCT, including HLA-mismatched URD HCT, compared with other available therapies.
The present analysis focused on 3 questions: First, how different are the outcomes if a mismatched URD is used instead of a fully matched URD? Second, which mismatched donors should be chosen? Third, what is the importance of HLA matching relative to other patient and donor clinical characteristics for the success of URD HCT?
Patients and methods
All research was conducted under the approval of the NMDP Institutional Review Board.
Patients
Patients reported to the NMDP who underwent transplantation between 1988 and 2003 and were fully HLA-typed through the NMDP's ongoing retrospective high-resolution typing project were included in this analysis. Eligible diagnoses included acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic myeloid leukemia (CML), and myelodysplastic syndrome (MDS). Early-stage disease was defined as AML and ALL in first complete remission, CML in first chronic phase, and MDS subtype refractory anemia. Intermediate-stage disease was AML or ALL in second or subsequent complete remission or in first relapse, and CML in accelerated phase or second chronic phase. Advanced-phase disease was AML in second or higher relapse or primary induction failure, CML in blast phase, MDS subtype refractory anemia with excess blasts or in transformation, or MDS, not otherwise classified. All patients received myeloablative conditioning regimens defined as “traditional” if single-dose total body irradiation (TBI) greater than 500 cGy or more than 800 cGy total in fractionated doses (with or without cyclophosphamide) or cyclophosphamide with at least 9.5 mg/kg busulfan or “nontraditional” including at least 9.5 mg/kg busulfan without cyclophosphamide or melphalan with a dose greater than 150 mg/m2.
Patients undergoing conditioning regimens of lower intensity, second or subsequent transplantation, or surviving patients who did not provide signed, informed consent to allow analysis of their clinical data or HLA typing of stored NMDP Research Repository samples were excluded. All surviving recipients included in this analysis were retrospectively contacted and provided informed consent for participation in the NMDP research program. Informed consent was obtained in accordance with the Declaration of Helsinki. Informed consent was waived by the NMDP Institutional Review Board for all deceased recipients. Approximately 4% of surviving patients would not provide consent for use of research data. To adjust for the potential bias introduced by exclusion of nonconsenting surviving patients, a corrective action plan (CAP) modeling process randomly excluded appropriately the same percentage of deceased patients (n = 392) using a biased coin randomization with exclusion probabilities based on characteristics associated with not providing consent for use of the data in survivors. One third (34%) of patients in this study were included in a previous NMDP analysis evaluating the association between HLA matching and outcomes.1 Using the described multivariate model adjustment for multiple mismatches,1 exploratory analyses found similar risk estimates of mortality associated with disparity at each HLA locus in the series of previously analyzed and more recent patients (data not shown). Therefore, the 2 groups were combined in the present analysis of isolated locus-specific effects.
HLA typing
High-resolution typing was performed for HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, -DPA1, and -DPB1, as described.1 Low-resolution (serologic or antigen level) disparities involved conversion of the DNA-based typing to its lower-level serologic equivalent, usually by collapsing the 4-digit typing result back to its first 2 digits, with the exception of a few HLA-B alleles that were mapped to their corresponding serologic specificities.1 Mismatches at HLA-DQ (or HLA-DP) were scored if there was disparity for either the DQA1 (DPA1) or the DQB1 (DPB1) sequence, since both DQA1 (DPA1) and DQB1 (DPB1) genes contribute to the expression of a single heterodimeric HLA-DQ (DP) protein. HLA-DQA1 was not considered for determination of antigen matching. Directional mismatches were considered in the analysis of graft-versus-host disease (GVHD) and engraftment, as described.2 Mismatches at homozygous alleles were considered single mismatches. Only subgroups of single mismatches or multiple mismatches with more than 20 evaluable pairs were included in the subset analyses because fewer pairs would not allow stable estimates of risk. High-resolution HLA matching at HLA-A, -B, -C, and -DRB1 was considered an “8/8 fully-matched pair.” HLA-DQ and HLA-DP match status were considered in dedicated secondary analyses.
Definitions of outcomes
The prespecified primary outcome of the analysis was overall survival, defined as time from graft infusion (day 0) to death from any cause. A number of secondary end points were also analyzed. Failure to engraft (primary graft failure) was defined as failure to achieve an absolute neutrophil count greater than 500 × 106/L by day 28 that was maintained for 3 consecutive measurements. Data about secondary graft failure (decline in absolute neutrophil count less than 100 × 106/L after initial engraftment or loss of donor T-cell chimerism) were not available. Acute GVHD grades III and IV was defined by the Glucksberg scale.3 Extensive chronic GVHD was defined according to the Seattle criteria.4 Clinical relapse of the primary disease was defined by the Center for International Blood and Marrow Transplant Research (CIBMTR) criteria.1 Disease-free survival (DFS) is survival without recurrence of the primary disease. For this end point, either death or relapse was considered an event. Treatment-related mortality (TRM) is death in continuous complete remission of the primary disease.
Biostatistical methods
Medians and ranges are reported for continuous variables and percentages for categoric variables. Probabilities for overall survival and DFS were calculated using the Kaplan-Meier estimator. Survival curves were compared using the log-rank test. Neutrophil engraftment was considered a dichotomous outcome and analyzed by logistic regression. Values for other outcomes were estimated using the cumulative incidence function.5,6 Death was considered a competing risk for all of the end points except overall survival and DFS. Relapse was also considered a competing event for treatment-related mortality. Patients were censored when they underwent a second HCT procedure or if alive at last follow-up.
To analyze the association between number and type of HLA mismatches and clinical outcomes, multivariate proportional hazards models were created that allowed pairs mismatched at specific loci to be compared with HLA-matched pairs. This biostatistical approach allowed precise estimates of the association between number and type of HLA mismatches (for example, 1 or 2, high or low resolution) or locus-specific mismatches (for example, HLA-A, -B, -C, -DR, -DQ, or -DP) and outcomes without confounding by any additional HLA mismatches present. This differs from some previous reports that either used multivariable models to adjust statistically for multiple mismatches1 or grouped different mismatches together before performing subgroup analyses.7 Because of multiple testing, a significant P value was considered less than .01.
All models were tested for significant clinical covariates including disease, disease stage, Karnofsky performance status, donor-patient cytomegalovirus (CMV) serology, patient race, patient age, T-cell depletion, use of TBI, graft source (peripheral blood or bone marrow), donor age, patient-donor sex match, and year of transplantation. Models included any clinical factors that were related to a given outcome at P less than .05. All variables were tested for affirmation of the proportional hazards assumption and to look for interactions with HLA matching. No significant interactions were identified. All variables satisfied the proportional hazards assumption except Karnofsky performance so the analyses were stratified for this variable. Center effect was tested and was not present.
Initial analyses suggested that HLA-DQ mismatching was not associated with worse outcomes if patients were otherwise matched or single locus mismatched, so the HLA-DQ results are presented separately where HLA-A, -B, -C, -DRB1, -DQ (10/10) matched pairs were considered the baseline. Analysis of the effects of HLA-DP mismatching required a separate approach since 86% of otherwise 8/8 matched pairs were mismatched for at least one HLA-DP allele. A separate model was created to compare 266 pairs completely matched at all HLA-A, -B, -C, -DRB1, and -DP loci with pairs mismatched only at one (n = 1029) or both (n = 545) DP loci.
To test the relative contribution of patient and donor clinical factors to predicting survival after URD HCT, a multivariable proportional hazards model was created including HLA matching, significant patient characteristics, and candidate donor variables determined from the literature. Specifically, patient age, patient-donor sex combinations, female parity, and patient-donor CMV serostatus combinations were included in the model.
Results
Patient characteristics
Table 1 shows the patient and transplantation characteristics of the study population (N = 3857). Patients underwent transplantation between 1988 and 2003 using myeloablative conditioning regimens. Seventy-eight percent received T-cell replete grafts and calcineurin inhibitor-based GVHD prophylaxis. Almost all (94%) received bone marrow. The median follow-up was 6 years.
Patient characteristics according to number of matching alleles
| Variable . | 8 of 8 alleles matched, no. (%) [no. eval] . | 7 of 8 alleles matched, no. (%) [no. eval] . | 6 of 8 alleles matched, no. (%) [no. eval] . | 5 or fewer of 8 alleles matched, no. (%) [no. eval] . |
|---|---|---|---|---|
| No. patients | 1840 | 985 | 633 | 399 |
| No. centers | 105 | 100 | 94 | 75 |
| Median age, y (range) | 35 (<1-65) [1839] | 31 (<1-65) [985] | 28 (<1-59) [633] | 28 (<1-55) [399] |
| Age at transplantation, y | [1839] | [985] | [633] | [399] |
| Younger than 11 | 167 (9) | 118 (12) | 79 (12) | 67 (17) |
| 11 to 20 | 194 (11) | 153 (16) | 135 (21) | 66 (17) |
| 21 to 30 | 328 (18) | 175 (18) | 117 (18) | 78 (20) |
| 31 to 40 | 452 (25) | 222 (23) | 142 (22) | 103 (26) |
| 41 to 50 | 479 (26) | 229 (23) | 127 (20) | 67 (17) |
| Older than 50 | 219 (12) | 88 (9) | 33 (5) | 18 (5) |
| Race | [1840] | [985] | [632] | [399] |
| White | 1715 (93) | 858 (87) | 504 (80) | 264 (66) |
| Black | 44 (2) | 50 (5) | 38 (6) | 57 (14) |
| Hispanic | 54 (3) | 53 (5) | 63 (10) | 51 (13) |
| Other | 27 (2) | 24 (3) | 27 (4) | 27 (7) |
| Recipient sex, male | 1041 (57) [1840] | 537 (55) [985] | 361 (57) [633] | 244 (61) [399] |
| Karnofsky prior to TX, 90 or higher | 1328 (74) [1784] | 713 (74) [962] | 453 (73) [622] | 298 (76) [393] |
| HLA-A, -B, -C, -DRB1 matching | [1840] | [985] | [633] | [399] |
| Allele matched | 1840 (100) | 0 | 0 | 0 |
| Single allele mismatch (MM) | 0 | 412 (42) | 0 | 0 |
| Two allele MM | 0 | 0 | 105 (16) | 0 |
| Three or more allele MM | 0 | 0 | 0 | 10 (3) |
| Single antigen MM | 0 | 573 (58) | 0 | 0 |
| Two antigen MM | 0 | 0 | 239 (38) | 0 |
| Three or more antigen MM | 0 | 0 | 0 | 36 (9) |
| One or more allele MM and 1 or more antigen MM | 0 | 0 | 289 (46) | 353 (88) |
| Disease at transplantation | [1840] | [985] | [633] | [399] |
| AML | 496 (27) | 294 (30) | 179 (28) | 86 (21) |
| ALL | 410 (22) | 253 (26) | 171 (27) | 118 (30) |
| CML | 759 (41) | 366 (37) | 242 (38) | 175 (44) |
| MDS | 175 (10) | 72 (7) | 41 (7) | 20 (5) |
| Disease status at transplantation | [1840] | [985] | [633] | [399] |
| Early | 835 (45) | 378 (38) | 241 (38) | 149 (37) |
| Intermediate | 674 (37) | 410 (42) | 268 (42) | 180 (45) |
| Advanced | 327 (18) | 195 (20) | 123 (19) | 69 (17) |
| Unknown | 4 (<1) | 2 (<1) | 1 (<1) | 1 (<1) |
| Conditioning regimen, TBI based | 1492 (81) [1840] | 807 (82) [985] | 553 (87) [633] | 357 (89) [399] |
| GVHD prophylaxis, T-cell depleted | 307 (17) [1840] | 241 (24) [985] | 167 (26) [633] | 115 (29) [399] |
| Graft type | [1840] | [985] | [633] | [399] |
| Bone marrow | 1698 (92) | 916 (93) | 604 (95) | 397 (99) |
| PBSC | 142 (8) | 69 (7) | 29 (5) | 2 (1) |
| Donor/recipient sex match | [1840] | [985] | [633] | [399] |
| Male/male | 717 (39) | 331 (34) | 198 (31) | 135 (34) |
| Male/female | 446 (24) | 237 (24) | 141 (22) | 74 (19) |
| Female/male | 324 (18) | 206 (21) | 163 (26) | 109 (27) |
| Female/female | 353 (19) | 211 (21) | 131 (21) | 81 (20) |
| Donor/recipient CMV match | [1840] | [985] | [633] | [399] |
| Negative/negative | 667 (36) | 341 (35) | 201 (32) | 104 (26) |
| Negative/positive | 516 (28) | 266 (27) | 187 (30) | 118 (30) |
| Positive/negative | 298 (16) | 160 (16) | 97 (15) | 77 (19) |
| Positive/positive | 303 (17) | 192 (19) | 128 (20) | 90 (23) |
| Unknown | 56 (3) | 26 (3) | 20 (3) | 10 (2) |
| Donor age, median (range), y | 36 (18-60) [1840] | 36 (19-59) [985] | 36 (18-59) [633] | 36 (19-60) [399] |
| Donor age, y | [1840] | [985] | [633] | [399] |
| 18 to 19 | 15 (1) | 6 (1) | 5 (1) | 3 (1) |
| 20 to 29 | 463 (25) | 237 (24) | 161 (25) | 102 (25) |
| 30 to 39 | 726 (39) | 376 (38) | 238 (38) | 146 (37) |
| 40 to 49 | 511 (28) | 284 (29) | 186 (29) | 119 (30) |
| 50 and older | 125 (7) | 82 (8) | 43 (7) | 29 (7) |
| Year of transplantation | [1840] | [985] | [633] | [399] |
| 1988 to 1989 | 44 (2) | 19 (2) | 17 (3) | 18 (5) |
| 1990 to 1994 | 432 (24) | 224 (23) | 182 (29) | 140 (35) |
| 1995 to 1999 | 787 (43) | 441 (45) | 292 (46) | 176 (44) |
| 2000 to 2003 | 577 (31) | 301 (30) | 142 (22) | 65 (16) |
| Median follow-up of survivors, mo | 73 (3-194) [671] | 63 (6-191) [297] | 84 (11-192) [134] | 96 (4-183) [75] |
| Variable . | 8 of 8 alleles matched, no. (%) [no. eval] . | 7 of 8 alleles matched, no. (%) [no. eval] . | 6 of 8 alleles matched, no. (%) [no. eval] . | 5 or fewer of 8 alleles matched, no. (%) [no. eval] . |
|---|---|---|---|---|
| No. patients | 1840 | 985 | 633 | 399 |
| No. centers | 105 | 100 | 94 | 75 |
| Median age, y (range) | 35 (<1-65) [1839] | 31 (<1-65) [985] | 28 (<1-59) [633] | 28 (<1-55) [399] |
| Age at transplantation, y | [1839] | [985] | [633] | [399] |
| Younger than 11 | 167 (9) | 118 (12) | 79 (12) | 67 (17) |
| 11 to 20 | 194 (11) | 153 (16) | 135 (21) | 66 (17) |
| 21 to 30 | 328 (18) | 175 (18) | 117 (18) | 78 (20) |
| 31 to 40 | 452 (25) | 222 (23) | 142 (22) | 103 (26) |
| 41 to 50 | 479 (26) | 229 (23) | 127 (20) | 67 (17) |
| Older than 50 | 219 (12) | 88 (9) | 33 (5) | 18 (5) |
| Race | [1840] | [985] | [632] | [399] |
| White | 1715 (93) | 858 (87) | 504 (80) | 264 (66) |
| Black | 44 (2) | 50 (5) | 38 (6) | 57 (14) |
| Hispanic | 54 (3) | 53 (5) | 63 (10) | 51 (13) |
| Other | 27 (2) | 24 (3) | 27 (4) | 27 (7) |
| Recipient sex, male | 1041 (57) [1840] | 537 (55) [985] | 361 (57) [633] | 244 (61) [399] |
| Karnofsky prior to TX, 90 or higher | 1328 (74) [1784] | 713 (74) [962] | 453 (73) [622] | 298 (76) [393] |
| HLA-A, -B, -C, -DRB1 matching | [1840] | [985] | [633] | [399] |
| Allele matched | 1840 (100) | 0 | 0 | 0 |
| Single allele mismatch (MM) | 0 | 412 (42) | 0 | 0 |
| Two allele MM | 0 | 0 | 105 (16) | 0 |
| Three or more allele MM | 0 | 0 | 0 | 10 (3) |
| Single antigen MM | 0 | 573 (58) | 0 | 0 |
| Two antigen MM | 0 | 0 | 239 (38) | 0 |
| Three or more antigen MM | 0 | 0 | 0 | 36 (9) |
| One or more allele MM and 1 or more antigen MM | 0 | 0 | 289 (46) | 353 (88) |
| Disease at transplantation | [1840] | [985] | [633] | [399] |
| AML | 496 (27) | 294 (30) | 179 (28) | 86 (21) |
| ALL | 410 (22) | 253 (26) | 171 (27) | 118 (30) |
| CML | 759 (41) | 366 (37) | 242 (38) | 175 (44) |
| MDS | 175 (10) | 72 (7) | 41 (7) | 20 (5) |
| Disease status at transplantation | [1840] | [985] | [633] | [399] |
| Early | 835 (45) | 378 (38) | 241 (38) | 149 (37) |
| Intermediate | 674 (37) | 410 (42) | 268 (42) | 180 (45) |
| Advanced | 327 (18) | 195 (20) | 123 (19) | 69 (17) |
| Unknown | 4 (<1) | 2 (<1) | 1 (<1) | 1 (<1) |
| Conditioning regimen, TBI based | 1492 (81) [1840] | 807 (82) [985] | 553 (87) [633] | 357 (89) [399] |
| GVHD prophylaxis, T-cell depleted | 307 (17) [1840] | 241 (24) [985] | 167 (26) [633] | 115 (29) [399] |
| Graft type | [1840] | [985] | [633] | [399] |
| Bone marrow | 1698 (92) | 916 (93) | 604 (95) | 397 (99) |
| PBSC | 142 (8) | 69 (7) | 29 (5) | 2 (1) |
| Donor/recipient sex match | [1840] | [985] | [633] | [399] |
| Male/male | 717 (39) | 331 (34) | 198 (31) | 135 (34) |
| Male/female | 446 (24) | 237 (24) | 141 (22) | 74 (19) |
| Female/male | 324 (18) | 206 (21) | 163 (26) | 109 (27) |
| Female/female | 353 (19) | 211 (21) | 131 (21) | 81 (20) |
| Donor/recipient CMV match | [1840] | [985] | [633] | [399] |
| Negative/negative | 667 (36) | 341 (35) | 201 (32) | 104 (26) |
| Negative/positive | 516 (28) | 266 (27) | 187 (30) | 118 (30) |
| Positive/negative | 298 (16) | 160 (16) | 97 (15) | 77 (19) |
| Positive/positive | 303 (17) | 192 (19) | 128 (20) | 90 (23) |
| Unknown | 56 (3) | 26 (3) | 20 (3) | 10 (2) |
| Donor age, median (range), y | 36 (18-60) [1840] | 36 (19-59) [985] | 36 (18-59) [633] | 36 (19-60) [399] |
| Donor age, y | [1840] | [985] | [633] | [399] |
| 18 to 19 | 15 (1) | 6 (1) | 5 (1) | 3 (1) |
| 20 to 29 | 463 (25) | 237 (24) | 161 (25) | 102 (25) |
| 30 to 39 | 726 (39) | 376 (38) | 238 (38) | 146 (37) |
| 40 to 49 | 511 (28) | 284 (29) | 186 (29) | 119 (30) |
| 50 and older | 125 (7) | 82 (8) | 43 (7) | 29 (7) |
| Year of transplantation | [1840] | [985] | [633] | [399] |
| 1988 to 1989 | 44 (2) | 19 (2) | 17 (3) | 18 (5) |
| 1990 to 1994 | 432 (24) | 224 (23) | 182 (29) | 140 (35) |
| 1995 to 1999 | 787 (43) | 441 (45) | 292 (46) | 176 (44) |
| 2000 to 2003 | 577 (31) | 301 (30) | 142 (22) | 65 (16) |
| Median follow-up of survivors, mo | 73 (3-194) [671] | 63 (6-191) [297] | 84 (11-192) [134] | 96 (4-183) [75] |
No. eval indicates the number evaluable for each characteristic; TX, transplantation; and PBSC, peripheral blood stem cell.
Single locus mismatches
Mismatching at a single HLA-A, -B, -C, or -DRB1 locus (7/8) was associated with lower survival and DFS, and higher treatment-related mortality and more acute GVHD compared with 8/8 HLA-matched pairs (Table 2). There were no statistically significant differences in risk associated with single high-resolution (allele) versus single low-resolution (antigen) mismatches, so all subsequent analyses considered allele and antigen mismatches equivalent. Examination of the relative risk point estimates of mismatches at specific loci for survival and the secondary outcomes supports this approach for all loci except for HLA-C, where allele mismatches were not associated with worse outcomes (Table 3). There was no statistically significant difference between 7/8 (either allele or antigen) and 8/8 matched pairs for relapse, chronic GVHD, and engraftment. One-year survival was 43% for 7/8 compared with 52% for 8/8 matched pairs (Table 4).
Single locus mismatches at HLA-A, -B, -C, and -DRB1
| Factor . | Survival . | Disease-free survival . | Treatment-related mortality . | Acute graft-versus-host disease . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | |
| Any single locus (n = 985) vs matched (n=1840) | 1.25 | 1.13-1.38 | <.001 | 1.23 | 1.12-1.36 | <.001 | 1.40 | 1.25-1.56 | <.001 | 1.48 | 1.29-1.68 | <.001 |
| Any single allele (n=412) vs matched | 1.30 | 1.14-1.46 | <.001 | 1.28 | 1.13-1.46 | .002 | 1.40 | 1.20-1.63 | <.001 | 1.34 | 1.12-1.61 | .002 |
| Any single antigen (n=573) vs matched | 1.22 | 1.08-1.37 | .001 | 1.20 | 1.07-1.35 | .002 | 1.40 | 1.22-1.60 | <.001 | 1.59 | 1.35-1.86 | <.001 |
| Any single allele vs any single antigen | 1.07 | 0.92-1.24 | .40 | 1.07 | 0.92-1.24 | .39 | 1.00 | 0.83-1.19 | .98 | 0.85 | 0.68-1.04 | .12 |
| Factor . | Survival . | Disease-free survival . | Treatment-related mortality . | Acute graft-versus-host disease . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | |
| Any single locus (n = 985) vs matched (n=1840) | 1.25 | 1.13-1.38 | <.001 | 1.23 | 1.12-1.36 | <.001 | 1.40 | 1.25-1.56 | <.001 | 1.48 | 1.29-1.68 | <.001 |
| Any single allele (n=412) vs matched | 1.30 | 1.14-1.46 | <.001 | 1.28 | 1.13-1.46 | .002 | 1.40 | 1.20-1.63 | <.001 | 1.34 | 1.12-1.61 | .002 |
| Any single antigen (n=573) vs matched | 1.22 | 1.08-1.37 | .001 | 1.20 | 1.07-1.35 | .002 | 1.40 | 1.22-1.60 | <.001 | 1.59 | 1.35-1.86 | <.001 |
| Any single allele vs any single antigen | 1.07 | 0.92-1.24 | .40 | 1.07 | 0.92-1.24 | .39 | 1.00 | 0.83-1.19 | .98 | 0.85 | 0.68-1.04 | .12 |
Numbers in each group are for the survival model. Other models may have had fewer evaluable patients, as described in “Biostatistical methods.”
RR indicates relative risk; and CI, confidence interval.
Single allele or antigen mismatches at HLA-A, -B, -C, and -DRB1
| . | n* . | Survival . | Disease-free survival . | Treatment-related mortality . | Acute graft-versus-host disease . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | ||
| Matched | 1840 | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — |
| HLA-A | |||||||||||||
| Allele | 113 | 1.50 | 1.20-1.88 | <.001 | 1.46 | 1.71-1.82 | .001 | 1.65 | 1.24-2.10 | <.001 | 1.62 | 1.19-2.20 | .002 |
| Antigen | 161 | 1.24 | 1.02-1.52 | .03 | 1.27 | 1.05-1.55 | .02 | 1.39 | 1.10-1.77 | .006 | 1.54 | 1.18-2.03 | .002 |
| Allele vs antigen | — | 0.83 | 0.62-1.10 | .19 | 0.87 | 0.65-1.15 | .34 | 0.86 | 0.62-1.21 | .39 | 0.95 | 0.64-1.41 | .81 |
| HLA-B | |||||||||||||
| Allele | 99 | 1.25 | 0.97-1.60 | .09 | 1.18 | 0.92-1.51 | .20 | 1.41 | 1.06-1.87 | .02 | 1.63 | 1.19-2.23 | .002 |
| Antigen | 17 | 0.78 | 0.42-1.45 | .43 | 0.72 | 0.38-1.34 | .29 | 1.01 | 0.52-1.96 | .97 | 1.60 | 0.79-3.21 | .19 |
| Allele vs antigen | — | 0.62 | 0.32-1.21 | .17 | 0.61 | 0.31-1.18 | .14 | 0.72 | 0.35-1.46 | .36 | 0.98 | 0.46-2.08 | .96 |
| HLA-C | |||||||||||||
| Allele | 96 | 1.03 | 0.79-1.34 | .84 | 1.11 | 0.86-1.42 | .43 | 1.05 | 0.77-1.44 | .76 | 0.98 | 0.68-1.40 | .90 |
| Antigen | 382 | 1.22 | 1.06-1.39 | .004 | 1.19 | 1.04-1.36 | .009 | 1.40 | 1.20-1.64 | <.001 | 1.60 | 1.33-1.93 | <.001 |
| Allele vs antigen | — | 1.18 | 0.89-1.57 | .24 | 1.08 | 0.82-1.41 | .59 | 1.33 | 0.95-1.87 | .09 | 1.63 | 1.11-2.42 | .01 |
| HLA-DRB1 | |||||||||||||
| Allele | 104 | 1.42 | 1.13-1.80 | .003 | 1.39 | 1.10-1.75 | .005 | 1.52 | 1.16-4.48 | .002 | 1.20 | 0.83-1.73 | .32 |
| Antigen | 13 | 1.81 | 0.96-3.41 | .07 | 1.65 | 0.85-3.10 | .12 | 2.29 | 1.17-4.48 | .02 | 1.77 | 0.83-3.78 | .14 |
| Allele vs antigen | — | 1.27 | 0.64-2.48 | .49 | 1.19 | 0.61-2.31 | .62 | 1.50 | 0.72-3.08 | .26 | 1.46 | 0.64-3.37 | .36 |
| . | n* . | Survival . | Disease-free survival . | Treatment-related mortality . | Acute graft-versus-host disease . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | ||
| Matched | 1840 | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — |
| HLA-A | |||||||||||||
| Allele | 113 | 1.50 | 1.20-1.88 | <.001 | 1.46 | 1.71-1.82 | .001 | 1.65 | 1.24-2.10 | <.001 | 1.62 | 1.19-2.20 | .002 |
| Antigen | 161 | 1.24 | 1.02-1.52 | .03 | 1.27 | 1.05-1.55 | .02 | 1.39 | 1.10-1.77 | .006 | 1.54 | 1.18-2.03 | .002 |
| Allele vs antigen | — | 0.83 | 0.62-1.10 | .19 | 0.87 | 0.65-1.15 | .34 | 0.86 | 0.62-1.21 | .39 | 0.95 | 0.64-1.41 | .81 |
| HLA-B | |||||||||||||
| Allele | 99 | 1.25 | 0.97-1.60 | .09 | 1.18 | 0.92-1.51 | .20 | 1.41 | 1.06-1.87 | .02 | 1.63 | 1.19-2.23 | .002 |
| Antigen | 17 | 0.78 | 0.42-1.45 | .43 | 0.72 | 0.38-1.34 | .29 | 1.01 | 0.52-1.96 | .97 | 1.60 | 0.79-3.21 | .19 |
| Allele vs antigen | — | 0.62 | 0.32-1.21 | .17 | 0.61 | 0.31-1.18 | .14 | 0.72 | 0.35-1.46 | .36 | 0.98 | 0.46-2.08 | .96 |
| HLA-C | |||||||||||||
| Allele | 96 | 1.03 | 0.79-1.34 | .84 | 1.11 | 0.86-1.42 | .43 | 1.05 | 0.77-1.44 | .76 | 0.98 | 0.68-1.40 | .90 |
| Antigen | 382 | 1.22 | 1.06-1.39 | .004 | 1.19 | 1.04-1.36 | .009 | 1.40 | 1.20-1.64 | <.001 | 1.60 | 1.33-1.93 | <.001 |
| Allele vs antigen | — | 1.18 | 0.89-1.57 | .24 | 1.08 | 0.82-1.41 | .59 | 1.33 | 0.95-1.87 | .09 | 1.63 | 1.11-2.42 | .01 |
| HLA-DRB1 | |||||||||||||
| Allele | 104 | 1.42 | 1.13-1.80 | .003 | 1.39 | 1.10-1.75 | .005 | 1.52 | 1.16-4.48 | .002 | 1.20 | 0.83-1.73 | .32 |
| Antigen | 13 | 1.81 | 0.96-3.41 | .07 | 1.65 | 0.85-3.10 | .12 | 2.29 | 1.17-4.48 | .02 | 1.77 | 0.83-3.78 | .14 |
| Allele vs antigen | — | 1.27 | 0.64-2.48 | .49 | 1.19 | 0.61-2.31 | .62 | 1.50 | 0.72-3.08 | .26 | 1.46 | 0.64-3.37 | .36 |
is the number for survival model. Other models may have had fewer evaluable patients, as described in “Biostatistical methods.”
—, indicates not applicable.
Unadjusted clinical outcomes by degree of HLA-A, -B, -C, -DRB1 match
| . | 8/8 . | 7/8 . | 6/8 . |
|---|---|---|---|
| No. patients | 1840 | 985 | 633 |
| Overall survival at 5 y | 37 (35-40) | 29 (26-32) | 22 (19-26) |
| Overall survival at 1 y | 52 (50-54) | 43 (40-46) | 33 (30-37) |
| Disease-free survival at 1 y | 47 (44-49) | 38 (35-42) | 29 (26-33) |
| Treatment-related mortality at 1 y | 36 (34-38) | 45 (42-49) | 55 (51-59) |
| Relapse at 1 y | 18 (16-19) | 16 (14-18) | 15 (13-18) |
| Chronic GVHD at 1 y | 44 (41-46) | 36 (33-39) | 32 (29-36) |
| Acute GVHD grade III-IV at 100 days | 28 (26-30) | 37 (34-40) | 44 (40-48) |
| Failure to engraft at 28 days | 10 (9-11) | 13 (10-15) | 17 (14-20) |
| . | 8/8 . | 7/8 . | 6/8 . |
|---|---|---|---|
| No. patients | 1840 | 985 | 633 |
| Overall survival at 5 y | 37 (35-40) | 29 (26-32) | 22 (19-26) |
| Overall survival at 1 y | 52 (50-54) | 43 (40-46) | 33 (30-37) |
| Disease-free survival at 1 y | 47 (44-49) | 38 (35-42) | 29 (26-33) |
| Treatment-related mortality at 1 y | 36 (34-38) | 45 (42-49) | 55 (51-59) |
| Relapse at 1 y | 18 (16-19) | 16 (14-18) | 15 (13-18) |
| Chronic GVHD at 1 y | 44 (41-46) | 36 (33-39) | 32 (29-36) |
| Acute GVHD grade III-IV at 100 days | 28 (26-30) | 37 (34-40) | 44 (40-48) |
| Failure to engraft at 28 days | 10 (9-11) | 13 (10-15) | 17 (14-20) |
Data are given as percent (95% CI).
Table 5 shows the association between specific HLA locus mismatches and survival. A single mismatch at HLA-A, -C, or -DRB1 was statistically associated with lower survival compared with fully matched pairs. Survival, however, was not statistically worse with single mismatches at HLA-B compared with 8/8 matches. Examination of the relative risks for each locus suggests that single mismatches at HLA-A (n = 274; RR, 1.36; 95% CI, 1.17-1.59) or HLA-DRB1 (n = 117; RR, 1.48; 95% CI, 1.19-1.85) are more poorly tolerated than single mismatches at HLA-B (n = 116; RR, 1.16; 95% CI, 0.92-1.47) or HLA-C (n = 478; RR, 1.19; 95% CI, 1.05-1.35), although none was significantly different in pairwise comparisons (data not shown). When single mismatches at either HLA-A or HLA-DRB1 were compared with single mismatches at either HLA-B or HLA-C, the relative risk was 1.18 (95% CI, 1.01-1.37; P = .04).
Effect of HLA mismatching on survival
| . | No. . | RR (95% CI) . | P . |
|---|---|---|---|
| Fully matched (8/8)* | 1840 | 1.00 | — |
| Single mismatch (7/8) | 985 | 1.26 (1.15-1.39) | <.001 |
| A | 274 | 1.36 (1.17-1.58) | <.001 |
| B | 116 | 1.16 (0.92-1.47) | .20 |
| C | 478 | 1.19 (1.05-1.35) | .006 |
| DRB1 | 117 | 1.48 (1.19-1.85) | .001 |
| Double mismatch (6/8) | 633 | 1.66 (1.48-1.85) | <.001 |
| A+B | 41 | 1.13 (0.77-1.65) | .53 |
| A+C | 130 | 1.68 (1.37-2.07) | <.001 |
| A+DRB1 | 20 | 1.96 (1.19-3.23) | .008 |
| B+DRB1 | 29 | 1.51 (1.00-2.27) | .05 |
| B+C | 284 | 1.87 (1.62-2.16) | <.001 |
| C+C | 36 | 1.73 (1.18-2.54) | .005 |
| C+DRB1 | 72 | 1.27 (0.96-1.67) | .09 |
| Others | 241 | — | — |
| Triple mismatch (5/8) | 275 | 1.64 (1.42-1.91) | <.001 |
| A+B+C | 97 | 1.77 (1.40-2.24) | <.001 |
| A+C+DRB1 | 20 | 1.82 (1.11-2.99) | .02 |
| B+C+C | 41 | 1.96 (1.40-2.75) | <.001 |
| B+C+DRB1 | 48 | 1.64 (1.19-2.25) | .002 |
| B+B+C | 25 | 0.93 (0.56-1.56) | .79 |
| Others | 44 | — | — |
| Quadruple mismatch (4/8) | 91 | 2.05 (1.61-2.60) | <.001 |
| A+B+C+DRB1 | 23 | 2.39 (1.51-3.77) | <.001 |
| Others | 68 | — | — |
| . | No. . | RR (95% CI) . | P . |
|---|---|---|---|
| Fully matched (8/8)* | 1840 | 1.00 | — |
| Single mismatch (7/8) | 985 | 1.26 (1.15-1.39) | <.001 |
| A | 274 | 1.36 (1.17-1.58) | <.001 |
| B | 116 | 1.16 (0.92-1.47) | .20 |
| C | 478 | 1.19 (1.05-1.35) | .006 |
| DRB1 | 117 | 1.48 (1.19-1.85) | .001 |
| Double mismatch (6/8) | 633 | 1.66 (1.48-1.85) | <.001 |
| A+B | 41 | 1.13 (0.77-1.65) | .53 |
| A+C | 130 | 1.68 (1.37-2.07) | <.001 |
| A+DRB1 | 20 | 1.96 (1.19-3.23) | .008 |
| B+DRB1 | 29 | 1.51 (1.00-2.27) | .05 |
| B+C | 284 | 1.87 (1.62-2.16) | <.001 |
| C+C | 36 | 1.73 (1.18-2.54) | .005 |
| C+DRB1 | 72 | 1.27 (0.96-1.67) | .09 |
| Others | 241 | — | — |
| Triple mismatch (5/8) | 275 | 1.64 (1.42-1.91) | <.001 |
| A+B+C | 97 | 1.77 (1.40-2.24) | <.001 |
| A+C+DRB1 | 20 | 1.82 (1.11-2.99) | .02 |
| B+C+C | 41 | 1.96 (1.40-2.75) | <.001 |
| B+C+DRB1 | 48 | 1.64 (1.19-2.25) | .002 |
| B+B+C | 25 | 0.93 (0.56-1.56) | .79 |
| Others | 44 | — | — |
| Quadruple mismatch (4/8) | 91 | 2.05 (1.61-2.60) | <.001 |
| A+B+C+DRB1 | 23 | 2.39 (1.51-3.77) | <.001 |
| Others | 68 | — | — |
— indicates not calculated.
Matched at HLA-A, -B, -C, -DRB1.
Multiple loci mismatches
Subsets with greater than 20 pairs were analyzed. The most common double mismatches were HLA-B + C and HLA-A + C. The most common triple mismatch was HLA-A + B + C. These as well as other multiple mismatches were associated with lower survival compared with 8/8 matched pairs (Table 5).
DQ and DP mismatches
A single HLA-DQ mismatch was not associated with any adverse outcomes. Within 7/8 matched pairs, the addition of an HLA-DQ mismatch was associated with a mortality risk of 1.15 (95% CI, 0.95-1.39; P = .14). Among 6/8 matched pairs, the mortality risk associated with an additional HLA-DQ mismatch was 1.20 (95% CI, 0.98-1.46; P = .08). Thus, addition of HLA-DQ mismatching to one or more mismatches at other loci was associated with a small, but not statistically significant, adverse effect on survival.
HLA-DP locus mismatches were common in otherwise well-matched pairs. Specifically, in a cohort of 8/8 matched pairs, 14% were matched for both HLA-DP loci, 56% had 1 mismatch, and 30% contained 2 mismatches. There was no association of HLA-DP mismatching with survival (RR, 1.07; 95% CI, 0.90-1.26; P = .46) or DFS (RR, 1.02; 95% CI, 0.86-1.20; P = .86) in the multivariate analysis. There was an association with increased acute GVHD (RR, 1.43; 95% CI, 1.16-1.76; P = .001). There was a suggestion of an increased risk of TRM (RR, 1.22; 95% CI, 0.99-1.52; P = .06) and decreased risk of relapse (RR, 0.74; 95% CI, 0.57-0.96; P = .02), although these were not significant.
Number of relevant mismatches
Increasing the number of HLA mismatches was associated with clinically important and statistically significantly worse outcomes (Table 5). In particular, comparison of 8/8, 7/8, and 6/8 HLA-matched cohorts showed a 1-year survival of 52%, 43%, and 33%, respectively, suggesting that each additional HLA mismatch is associated with absolute unadjusted survival differences of 9% to 10% (Table 3). This difference was highly statistically significant for 7/8 versus 8/8 (RR, 1.25; 95% CI, 1.13-1.37; P < .001) and for 6/8 versus 7/8 (RR, 1.31; 95% CI, 1.17-1.47; P < .001).
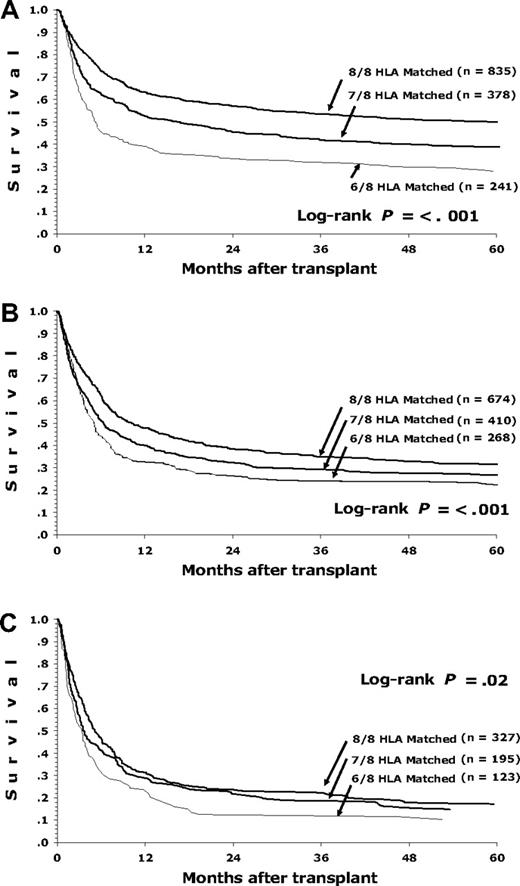
Figure 1 shows the association of the number of HLA mismatches with survival for patients with early, intermediate, or advanced disease. More advanced disease before HCT is associated with a greater absolute impact on survival compared with increasing HLA mismatching. For example, 1-year survival for a patient with early-stage disease is 63% for an 8/8 matched donor and 52% for a 7/8 matched donor. However, for a patient with intermediate-risk disease, the survival with an 8/8 matched donor is 48% compared with 40% with a 7/8 donor.
Survival of patients with early, intermediate, and advanced disease depending on degree of HLA matching (8/8, 7/8, and 6/8) for HLA-A, -B, -C, and -DRB1. (A) Early-stage disease for 8/8, 7/8, and 6/8, respectively: 1-year survival 63%, 52%, and 39%; 5-year survival 50%, 39%, and 28%. (B) Intermediate-stage disease for 8/8, 7/8, and 6/8, respectively: 1-year survival 48%, 40%, and 32%; 5-year survival 32%, 27%, and 22%. (C) Advanced-stage disease for 8/8, 7/8, and 6/8, respectively: 1-year survival 31%, 29%, and 24%; 5-year survival 17%, 15%, and 10%.
Survival of patients with early, intermediate, and advanced disease depending on degree of HLA matching (8/8, 7/8, and 6/8) for HLA-A, -B, -C, and -DRB1. (A) Early-stage disease for 8/8, 7/8, and 6/8, respectively: 1-year survival 63%, 52%, and 39%; 5-year survival 50%, 39%, and 28%. (B) Intermediate-stage disease for 8/8, 7/8, and 6/8, respectively: 1-year survival 48%, 40%, and 32%; 5-year survival 32%, 27%, and 22%. (C) Advanced-stage disease for 8/8, 7/8, and 6/8, respectively: 1-year survival 31%, 29%, and 24%; 5-year survival 17%, 15%, and 10%.
Younger patients (< 40 years) with early-stage disease and 8/8 donors had the best survival. For this group, 1-year survival was 67% (95% CI, 62%-71%) and 5-year survival was 54% (95% CI, 49%-58%).
Patient and donor characteristics versus HLA matching
Results of a multivariate model including 8/8, 7/8, and 6/8 mismatched pairs are shown in Table 6. The relative risk for a single HLA mismatch was 1.25 (25% higher risk for death with a single mismatch), similar to the relative risk associated with unmodifiable patient characteristics such as patient age, patient race, and patient CMV status. The sole potentially modifiable patient characteristic is disease status, in that physicians may be able to perform transplantation in patients before disease progression to intermediate or advanced stages. Intermediate- rather than early-stage disease was associated with a 38% greater risk of mortality, while advanced-stage patients have approximately double the mortality risk as patients with early-stage disease. No significant interactions between variables were identified, suggesting that HLA mismatching exerts a similar effect regardless of patient age, disease status, use of T-cell depletion, or other factors. There was no association between donor age, donor parity, donor CMV status, and donor sex with patient survival.
Association between patient and donor characteristics and survival
| Variable/category . | No. . | Hazard ratio . | 95% CI . | P . |
|---|---|---|---|---|
| HLA match | ||||
| 8/8 | 1840 | 1.00 | — | — |
| 7/8 | 985 | 1.25 | 1.13-1.37 | <.001 |
| 6/8 | 633 | 1.65 | 1.48-1.84 | <.001 |
| Disease diagnosis | ||||
| AML | 969 | 1.00 | — | — |
| ALL | 834 | 1.07 | 0.95-1.20 | .25 |
| CML | 1367 | 0.78 | 0.69-0.87 | <.001 |
| MDS | 288 | 0.73 | 0.62-0.86 | <.001 |
| Disease status | ||||
| Early | 1454 | 1.00 | — | — |
| Intermediate | 1352 | 1.38 | 1.25-1.53 | <.001 |
| Late | 645 | 1.90 | 1.67-2.16 | <.001 |
| Patient age, y | ||||
| Younger than 31 | 1467 | 1.00 | — | — |
| 31 to 45 | 1263 | 1.51 | 1.36-1.67 | <.001 |
| Older than 45 | 728 | 1.79 | 1.59-2.02 | <.001 |
| Patient race | ||||
| White | 3077 | 1.00 | — | — |
| Black | 132 | 1.53 | 1.26-1.87 | <.001 |
| Hispanic | 170 | 1.05 | 0.87-1.27 | .62 |
| Other | 78 | 0.68 | 0.51-0.92 | .012 |
| Donor/recipient, CMV | ||||
| −/− | 1209 | 1.00 | — | — |
| −/+ | 969 | 1.31 | 1.18-1.45 | <.001 |
| +/− | 555 | 1.08 | 0.95-1.23 | .23 |
| +/+ | 623 | 1.36 | 1.20-1.54 | <.001 |
| Unknown | 102 | 1.34 | 1.06-1.71 | .016 |
| Donor/recipient, sex match | ||||
| M/M | 1246 | 1.00 | — | — |
| M/F | 824 | 1.00 | 0.90-1.11 | .99 |
| F/M | 693 | 0.99 | 0.86-1.15 | .89 |
| F/F | 695 | 1.03 | 0.90-1.19 | .65 |
| Donor parity | ||||
| Male or not parous | 2615 | 1.00 | — | — |
| Parous | 814 | 1.10 | 0.96-1.27 | .17 |
| Donor age, y | ||||
| Younger than 31 | 887 | 1.00 | — | — |
| 31 to 45 | 1929 | 1.05 | 0.95-1.16 | .38 |
| Older than 45 | 642 | 1.06 | 0.93-1.20 | .42 |
| Variable/category . | No. . | Hazard ratio . | 95% CI . | P . |
|---|---|---|---|---|
| HLA match | ||||
| 8/8 | 1840 | 1.00 | — | — |
| 7/8 | 985 | 1.25 | 1.13-1.37 | <.001 |
| 6/8 | 633 | 1.65 | 1.48-1.84 | <.001 |
| Disease diagnosis | ||||
| AML | 969 | 1.00 | — | — |
| ALL | 834 | 1.07 | 0.95-1.20 | .25 |
| CML | 1367 | 0.78 | 0.69-0.87 | <.001 |
| MDS | 288 | 0.73 | 0.62-0.86 | <.001 |
| Disease status | ||||
| Early | 1454 | 1.00 | — | — |
| Intermediate | 1352 | 1.38 | 1.25-1.53 | <.001 |
| Late | 645 | 1.90 | 1.67-2.16 | <.001 |
| Patient age, y | ||||
| Younger than 31 | 1467 | 1.00 | — | — |
| 31 to 45 | 1263 | 1.51 | 1.36-1.67 | <.001 |
| Older than 45 | 728 | 1.79 | 1.59-2.02 | <.001 |
| Patient race | ||||
| White | 3077 | 1.00 | — | — |
| Black | 132 | 1.53 | 1.26-1.87 | <.001 |
| Hispanic | 170 | 1.05 | 0.87-1.27 | .62 |
| Other | 78 | 0.68 | 0.51-0.92 | .012 |
| Donor/recipient, CMV | ||||
| −/− | 1209 | 1.00 | — | — |
| −/+ | 969 | 1.31 | 1.18-1.45 | <.001 |
| +/− | 555 | 1.08 | 0.95-1.23 | .23 |
| +/+ | 623 | 1.36 | 1.20-1.54 | <.001 |
| Unknown | 102 | 1.34 | 1.06-1.71 | .016 |
| Donor/recipient, sex match | ||||
| M/M | 1246 | 1.00 | — | — |
| M/F | 824 | 1.00 | 0.90-1.11 | .99 |
| F/M | 693 | 0.99 | 0.86-1.15 | .89 |
| F/F | 695 | 1.03 | 0.90-1.19 | .65 |
| Donor parity | ||||
| Male or not parous | 2615 | 1.00 | — | — |
| Parous | 814 | 1.10 | 0.96-1.27 | .17 |
| Donor age, y | ||||
| Younger than 31 | 887 | 1.00 | — | — |
| 31 to 45 | 1929 | 1.05 | 0.95-1.16 | .38 |
| Older than 45 | 642 | 1.06 | 0.93-1.20 | .42 |
M indicates male; F, female; and —, not applicable.
Discussion
High-resolution matching of HLA-A, -B, -C, and -DRB1 between volunteer hematopoietic stem cell donors and recipients is associated with the best survival. Thus, low- to intermediate-resolution HLA typing technologies do not provide adequate information for the selection of an unrelated donor. If a fully allele-matched donor at HLA-A, -B, -C, and -DRB1 is not available, then single mismatches at HLA-B or HLA-C appear to be better tolerated than single HLA-A or -DRB1 mismatches. Each additional mismatch is associated with a 9% to 10% absolute decrease in survival. Mismatching at HLA-DQ or HLA-DP alleles is not associated with survival. We found no differences in the adverse effects of allele and antigen mismatches, contradicting the common perception that allele mismatches are more tolerable than antigen mismatches. Examination of the risks associated with allele or antigen mismatching at individual loci supported the aggregate conclusion, except perhaps for HLA-C where only antigen mismatches were associated with worse outcomes. Separate studies are evaluating whether these results may be attributable to the poor immunogenicity of common HLA-C allelic products or the presence of killer immunoglobulin-like receptor (KIR)–binding HLA-C epitopes that are more common in antigen than allele mismatches. Additional studies are investigating whether clinical outcomes are associated with physical linkage on HLA haplotypes8 or specific combinations of donor-patient mismatches within HLA loci.9
Our analysis suggests that matching for HLA-DQ does not have a significant impact on survival after URD HCT, raising the issue of whether typing for HLA-DQ and selection of donors on this basis is warranted. If a pair is otherwise matched at HLA-A, -B, -C, and -DRB1, then we found no evidence that additional testing for HLA-DQ is helpful since single HLA-DQ mismatches were not associated with increased mortality. However, if 1 or 2 HLA-A, -B, -C, or -DRB1 mismatches are present, there was a small, albeit statistically insignificant, adverse effect of an additional HLA-DQ mismatch in combination with other mismatches, and therefore HLA-DQ–matched donors should be favored given the same degree of matching at other loci.
Although analysis of HLA-DP was not the primary focus of the paper, available data were analyzed to the extent possible. HLA-DP mismatching was not associated with overall or DFS, although acute GVHD was higher consistent with prior reports.10,–12
Three groups have used high-resolution matching for HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, -DPA1, and -DPB1 in URD transplant pairs from series larger than 300. The Japan Marrow Donor Program (JMDP) first concluded that high-resolution mismatching at HLA-A and/or HLA-B, but not at HLA-C or -DRB1, was associated with higher mortality.7,13 We confirm in the present study that high-resolution mismatching at HLA-A and -B is associated with an increased risk compared with full match, and show in addition that such a risk is similar to low-resolution mismatches. Like the JMDP, we also failed to appreciate increased mortality with high-resolution mismatching at HLA-C, whereas low-resolution mismatch at HLA-C contributed significant risk (Table 3). Unlike the JMDP, we found increased mortality among 104 transplantations with a single DRB1 high-resolution mismatch, while the JMDP did not find increased risk among the 17 pairs with a single DRB1 mismatch available in their dataset.13 In a larger series reported by the JMDP, DRB1 mismatching was associated with increased risk of acute GVHD but was not associated with survival.7 We hypothesize the apparent discrepancy between our results and the JMDP studies is related to study power rather than the biology of HLA-DRB1 in the 2 ethnic groups. Petersdorf et al found that a single disparity at HLA-C (n = 24) was associated with increased mortality in patients with early- but not more advanced-stage disease, whereas no statistically significant association with survival was seen in pairs mismatched at HLA-A (n = 26), HLA-B (n = 9), or HLA-DRB1 (n = 7).14 The large confidence intervals of the Petersdorf et al study may explain the apparent discrepancy with the results of the present analysis, where mismatching at HLA-A, -B, -C, and -DRB1 was associated with significant risk in patients with both early and more advanced disease. Flomenberg et al analyzed data from the NMDP (n = 1874) and reported that mismatches at HLA-A, -B, -C, and -DRB1 were associated with similar decrements in survival regardless of disease stage. Multiple mismatches were included in the analysis by Flomenberg et al, and each locus was compared with locus-matched cases as the baseline risk, with adjustment for additional mismatched loci accomplished through multivariate modeling.1 Since almost all studies, including 2 smaller European reports,15,16 show that multiple mismatches increase mortality, statistical adjustment for additional mismatches may not allow accurate risk estimates for isolated mismatches. In contrast, the present study compared outcomes between fully matched patients and defined subsets of patients with specific mismatches, providing independent risk estimates according to locus and resolution mismatch.
This report includes a large number of fully typed patients, but there are a number of limitations. First, the power to detect statistically significant differences remains limited by the low number of observations in some subgroups. For example, there were only 17 single HLA-B antigen mismatches and 13 single DRB1 antigen mismatches in the dataset. Single mismatches at these loci are rarely observed because they are commonly linked with mismatches at other loci.17 Known DRB1-mismatched donors may be underrepresented because of prior studies suggesting worse survival18,19 and the earlier availability of high-resolution DNA typing methodology for DRB1 compared with HLA class I. Second, while there are specific strengths to subset comparisons since they avoid confounding by mismatches at other loci, a functional role of mismatching for HLA-A, -B, -C, or -DR operating only in combination with one or more mismatches may be overlooked by this analytic method. Although some groups of double and triple mismatches were large enough to analyze, many potential combinations were too rare to allow comment using this statistical method. Third, our population all received myeloablative conditioning regimens, and 94% underwent bone marrow transplantation for acute leukemia, CML, and MDS. The most recent statistics from the NMDP show that 36% of HCT procedures were nonmyeloablative, 64% used peripheral blood, and 9% used cord blood, while the spectrum of diseases treated by transplantation is shifting. Although we did not find an interaction between HLA mismatching and underlying disease, conditioning regimens, or graft type, URD HCT technology is rapidly evolving. The NMDP is evaluating the effect of HLA mismatching in myeloablative URD peripheral blood stem cell transplantation. One report in reduced intensity URD HCT suggested that HLA-C mismatches are associated with worse survival.20 Finally, while optimizing donor selection would be easier if all potential donors were fully typed and available, in reality many factors, including the patient's HLA type, linkage disequilibrium, the urgency of the HCT procedure, and resources available for high-resolution HLA typing influence which donors are considered and ultimately selected. While a single mismatch at HLA-B or -C appears better tolerated than mismatch at other loci, finding an isolated mismatch at HLA-B or -C may be difficult. Of approximately 250 searches facilitated by the NMDP where an HLA-A locus–mismatched donor was ultimately selected, about 30% would have had an equivalent HLA-B– or HLA-C–mismatched donor available (M. Setterholm, written communication, March 2007).
In multivariate analysis, the only donor factor associated with patient survival was HLA matching. We did not find an association between donor age and patient survival, in contrast to a previous NMDP study by Kollman et al21 Kollman et al's study included twice as many patients, used serologic typing for class I loci, had a median follow up of only 2 years, and included 17% with different diseases than our population. In that multivariate analysis, each decade of donor age was associated with an RR of 1.10 (95% CI, 1.06-1.14; P < .001) suggesting a relatively small effect despite a highly statistically significant P value.
While HLA matching is important for maximizing the success of URD HCT, patient factors such as patient age, disease diagnosis, disease stage, CMV status, and race are still the most critical predictors of survival. Disease stage is the only patient characteristic that physicians can affect by performing the transplantation earlier in the course of the patient's disease. If no 8/8 HLA-matched donors appear available for a patient needing HCT, further searching to identify a fully matched donor may not significantly improve survival and must be balanced against the risk that the disease will progress while a prolonged search is ongoing. In most instances, the adverse consequences of using an HLA-mismatched donor are less serious than proceeding to HCT with more advanced disease and may still offer better outcomes than other available treatments. Expeditious transplantation with the best available donor, even if mismatched, may offer the best chance for survival.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This project was supported by funding from the NMDP and the Department of the Navy, Office of Naval Research Cooperative Agreement N00014-99-2–0006 and grant N00014-05-1–0859 to the NMDP.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the Office of Naval Research or the NMDP.
Authorship
Contribution: S.J.L., J.K., M.H., L.A.B.-L., D.L.C., M.E., M.F.-V., N.F., M.H., C.K.H., H.N., M.O., E.P., M.S., S.S., D.W., T.M.W., and C.A. designed research; J.K. and M.H. performed statistics; S.J.L., J.K., M.H., L.A.B.-L., D.L.C., M.E., M.F.-V., N.F., M.H., C.K.H., H.N., M.O., E.P., M.S., S.S., D.W., T.M.W., and C.A. analyzed and interpreted data; S.J.L. and C.A. drafted the paper; S.J.L., J.K., M.H., L.A.B.-L., D.L.C., M.E., M.F.-V., N.F., M.H., C.K.H., H.N., M.O., E.P., M.S., S.S., D.W., T.M.W., and C.A. critically revised the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephanie J. Lee, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, D5-290, Seattle, WA 98109; e-mail:sjlee@fhcrc.org.