We have generated a large, unique database that includes morphologic, clinical, cytogenetic, and follow-up data from 2124 patients with myelodysplastic syndromes (MDSs) at 4 institutions in Austria and 4 in Germany. Cytogenetic analyses were successfully performed in 2072 (97.6%) patients, revealing clonal abnormalities in 1084 (52.3%) patients. Numeric and structural chromosomal abnormalities were documented for each patient and subdivided further according to the number of additional abnormalities. Thus, 684 different cytogenetic categories were identified. The impact of the karyotype on the natural course of the disease was studied in 1286 patients treated with supportive care only. Median survival was 53.4 months for patients with normal karyotypes (n = 612) and 8.7 months for those with complex anomalies (n = 166). A total of 13 rare abnormalities were identified with good (+1/+1q, t(1q), t(7q), del(9q), del(12p), chromosome 15 anomalies, t(17q), monosomy 21, trisomy 21, and −X), intermediate (del(11q), chromosome 19 anomalies), or poor (t(5q)) prognostic impact, respectively. The prognostic relevance of additional abnormalities varied considerably depending on the chromosomes affected. For all World Health Organization (WHO) and French-American-British (FAB) classification system subtypes, the karyotype provided additional prognostic information. Our analyses offer new insights into the prognostic significance of rare chromosomal abnormalities and specific karyotypic combinations in MDS.
Introduction
The myelodysplastic syndromes (MDSs) are a heterogeneous group of clonal hematopoietic stem cell disorders that cause one or more peripheral cytopenias due to ineffective hematopoiesis.1,,,,,,–8 The sequelae of MDS result from the underlying cytopenias and include hemorrhage (thrombocytopenia), infections (neutropenia), and, most commonly, anemia. Complications from cytopenias lead to higher morbidity and mortality among patients with MDS compared with an age-matched population.9 The variable risk of progression to acute myeloid leukemia (AML) is of major prognostic significance in early MDS without blast excess.2
A succession of MDS classification systems have been developed to facilitate prediction of the risk of progression to AML and overall survival.10,,,,–15 The first of these was the French-American-British (FAB) system,10 which used cytomorphologic abnormalities and blast percentage as criteria for classification. The World Health Organization (WHO) classification system subsequently improved homogeneity and discrimination between lower-risk MDS categories,11,12,16,17 and was prospectively validated recently on a large number of patients.18 The International Prognostic Scoring System (IPSS), which applies only to de novo MDS, assigns 4 categories of risk for death or transformation to AML (Low, Int-1, Int-2, and High) based on a numeric score that reflects the percentage of bone marrow blasts, number of cytopenias, and presence or absence and type of chromosomal abnormalities.14 Cytogenetic risk groups are defined by the IPSS as good (normal, isolated −Y, del(5q), and del(20q)), poor (complex [≥ 3 abnormalities] and/or any chromosome 7 anomalies), and intermediate (all other abnormalities).14 Due to the growing awareness of shortcomings of established classification and prognostic scoring systems, and the increasing number of emerging therapeutic targets, a consensus statement was published recently revising standards for diagnostic criteria and prognostication.19,20
Cytogenetic abnormalities are major determinants in the pathogenesis, diagnosis, and prognosis, and, increasingly, the basis for selection of drugs in individual patients with MDS.1,17,21,,,,–26 Chromosomal anomalies are detected in approximately 50% of patients with de novo MDS and in up to 80% of patients with MDS secondary to chemotherapy or other toxic agents.8,27,,,–31 Balanced cytogenetic abnormalities, including reciprocal translocations, inversions, and insertions, are prevalent in myeloid leukemias such as AML or chronic myelogenous leukemia but are uncommon in MDS, in which unbalanced chromosomal abnormalities reflecting a gain or loss of chromosomal material are more prevalent.1,8,15,24,32,33 Interstitial deletions of the long arm of chromosome 5 (del(5q) or 5q−) with or without additional karyotypic abnormalities are the most frequent chromosomal abnormalities in de novo MDS.8,15,28,34 The WHO defined the 5q− syndrome as a distinct subtype of isolated del(5q) MDS characterized by the presence of less than 5% blasts in the bone marrow.12,35,36 Although cytogenetic abnormalities in MDS are among the most valuable independent prognostic determinants, analyses of MDS populations have been hampered by small sample size, varying treatment approaches, and profound karyotypic diversity. Therefore, it is not yet possible to determine the impact of rare as well as combined karyotype aberrations on prognosis in MDS. While the IPSS cytogenetic risk stratification system provides adequate prognostic information for common karyotypic abnormalities,14,31 its prognostic value is limited by the inherent cytogenetic diversity of these diseases, which is reflected not only by the numerous rare cytogenetic abnormalities but also by the various combinations of cytogenetic changes that can occur in these patients. Although supportive care has been the mainstay of treatment for MDS, therapies influencing the natural course of the disease can lead to problems in prognostication. Since these treatments are more commonly used, there remains only a limited time for studying the influence of cytogenetic abnormalities on the natural course of MDS.
A large, well-characterized patient cohort is needed to assess prognosis, survival, and response to therapies among patients with infrequent karyotypic lesions and combinations. Toward this goal, we have assembled cytogenetic, morphologic, prognostic, and therapeutic data on 2124 patients with MDS, representing the largest dataset published to date.
Patients and methods
Patients
Clinical data were obtained on 1981 patients with primary MDS and 143 with secondary MDS from 4 institutions in Austria (Hanusch Hospital, Vienna; Elisabethinen Hospital, Linz; University of Vienna; and Innsbruck Medical University) and 4 institutions in Germany (University of Düsseldorf; University of Göttingen; University of Freiburg; and Johannes Hospital, Duisburg). All patients with a confirmed diagnosis of MDS and available cytogenetics were eligible for this retrospective study. Patients were treated with supportive care, chemotherapy (induction type or low-dose) or amifostine as an alternative treatment regimen from 1964 to 2004. Patients treated with lenalidomide were not included in this study. Follow-up was finished by the end of 2005.
Pathology review and bone marrow morphology
Cytogenetic analysis
Cytogenetic analysis was performed at the individual centers, and karyotypes were documented according to the 1995 International System for Human Cytogenetic Nomenclature (ISCN) recommendations.37 Results were reviewed centrally by D.H. and C.S. The mean number of metaphases in the entire cohort was 21.9 plus or minus 10.6, fitting high-quality criteria for cytogenetics in MDS that we demonstrated recently.38 Details of cultivation, banding procedure, and processing have been presented elsewhere, as have procedures of fluorescence in situ hybridization, which was performed in several cases to verify or supplement classic cytogenetics.39,40 Additional abnormalities were defined as those changes accompanying a given abnormality that was used to define a cytogenetic subgroup (eg, del(5q)).
Statistical analysis
Statistical analysis procedures always included all patients of the sample for which the required covariate data were available. Time-to-event variables were analyzed using the Kaplan-Meier method and comparisons of groups were done by means of the log-rank test. Sample subgroups were analyzed based on WHO and FAB classifications as well as the occurrence of chromosomal aberrations. Statistical analysis was performed using the open-source software R version 2.1.1 (Free Software Foundation, Boston, MA; www.r-project.org). Median follow-up was 29.2 months.
Multiple comparisons were made from the sample underlying this study. This should be considered when appraising the result of individual statistical tests. Instead of applying a formal procedure such as Bonferroni adjustment of alpha levels, we have carefully chosen the statistical comparisons made and taken P values less than .01 as an indication of significance.41
Informed consent was obtained in accordance with the modified Declaration of Helsinki.
Results
Patient characteristics
Patient characteristics are listed in Table 1. The median age was 65.7 years and the male-to-female ratio was 1.29, consistent with the well-known male predominance of MDS. Of the 2124 patients, 1286 (60.5%) received supportive care only, 462 (21.8%) were treated with chemotherapy (induction type or low-dose), and 22 (1.0%) received amifostine. Therapeutic data were unavailable for 354 patients (16.7%). Follow-up data were available for 1841 patients (86.7%).
Patient characteristics
| Characteristic . | Quantity . |
|---|---|
| Patients, no. | 2124 |
| Median age, y (range) | 65.7 (0.1-96.1) |
| Sex, male/female | 1197/927 |
| Type of MDS, no. (%) | |
| Primary | 1981 (93.3) |
| Secondary | 143 (6.7) |
| FAB classification (n = 2124), n (%) | |
| RA | 590 (27.8) |
| RARS | 256 (12.1) |
| RAEB | 425 (20.0) |
| RAEB-t | 311 (14.6) |
| CMML | 287 (13.5) |
| MDS-AL | 132 (6.2) |
| Unknown | 123 (5.8) |
| WHO classification (n = 598), no. (%) | |
| 5q− syndrome | 61 (10.2) |
| RA | 56 (9.4) |
| RARS | 26 (4.3) |
| RCMD | 165 (27.6) |
| RSCMD | 77 (12.9) |
| RAEB-I | 90 (15.1) |
| RAEB-II | 123 (20.6) |
| Follow-up | |
| Patients with follow-up data, no. (%) | 1841 (86.7) |
| Mean observation time, mo | 29.2 |
| Therapies, no. (%) | |
| Supportive care only | 1286 (60.5) |
| Chemotherapy (intensive and low-dose) | 462 (21.8) |
| Amifostine | 22 (1.0) |
| No therapeutic data | 354 (16.7) |
| Cytogenetic overview | |
| Mean no. of metaphases analyzed | 21.9 |
| Successful cytogenetic analyses, no. (%) | 2072 (97.6) |
| Clonal cytogenetic abnormalities, no. (%) | 1084 (52.3) |
| Mean number of aberrations per case | 1.52 |
| Clonal abnormalities in primary MDS, no. (%) | 986 (49.8) |
| Clonal abnormalities in secondary MDS, no. (%) | 96 (67.1) |
| IPSS cytogenetic risk group, no. (%) | |
| Good | 1217 (58.7) |
| Intermediate | 401 (19.4) |
| Poor | 454 (21.9) |
| Characteristic . | Quantity . |
|---|---|
| Patients, no. | 2124 |
| Median age, y (range) | 65.7 (0.1-96.1) |
| Sex, male/female | 1197/927 |
| Type of MDS, no. (%) | |
| Primary | 1981 (93.3) |
| Secondary | 143 (6.7) |
| FAB classification (n = 2124), n (%) | |
| RA | 590 (27.8) |
| RARS | 256 (12.1) |
| RAEB | 425 (20.0) |
| RAEB-t | 311 (14.6) |
| CMML | 287 (13.5) |
| MDS-AL | 132 (6.2) |
| Unknown | 123 (5.8) |
| WHO classification (n = 598), no. (%) | |
| 5q− syndrome | 61 (10.2) |
| RA | 56 (9.4) |
| RARS | 26 (4.3) |
| RCMD | 165 (27.6) |
| RSCMD | 77 (12.9) |
| RAEB-I | 90 (15.1) |
| RAEB-II | 123 (20.6) |
| Follow-up | |
| Patients with follow-up data, no. (%) | 1841 (86.7) |
| Mean observation time, mo | 29.2 |
| Therapies, no. (%) | |
| Supportive care only | 1286 (60.5) |
| Chemotherapy (intensive and low-dose) | 462 (21.8) |
| Amifostine | 22 (1.0) |
| No therapeutic data | 354 (16.7) |
| Cytogenetic overview | |
| Mean no. of metaphases analyzed | 21.9 |
| Successful cytogenetic analyses, no. (%) | 2072 (97.6) |
| Clonal cytogenetic abnormalities, no. (%) | 1084 (52.3) |
| Mean number of aberrations per case | 1.52 |
| Clonal abnormalities in primary MDS, no. (%) | 986 (49.8) |
| Clonal abnormalities in secondary MDS, no. (%) | 96 (67.1) |
| IPSS cytogenetic risk group, no. (%) | |
| Good | 1217 (58.7) |
| Intermediate | 401 (19.4) |
| Poor | 454 (21.9) |
Using FAB criteria, the largest number of patients (27.8%) was classified as having refractory anemia (RA). The other FAB subgroups were as follows: RA with ringed sideroblasts (RARS), 12.1%; RA with excess blasts (RAEB), 20.0%; RAEB in transformation (RAEB-t), 14.6%; chronic myelomonocytic leukemia (CMML), 13.5%; acute leukemia following MDS (MDS-AL), 6.2%; and unclassifiable, 5.8%. WHO classification was available for 598 patients, of whom 10.2% had 5q− syndrome, 9.4% had RA, 4.3% had RARS, 27.6% had refractory cytopenia with multilineage dysplasia (RCMD), 12.9% had RCMD with ringed sideroblasts (RSCMD), 15.1% had RAEB-I, and 20.6% had RAEB-II.
Cytogenetic profile of MDS based on analysis of 2072 patients
Successful cytogenetic analyses were available for 2072 patients (1931 with primary MDS and 141 with secondary MDS). Of these, 1217 (58.7%) were classified as having good IPSS cytogenetic risk, 401 (19.4%) as having intermediate risk, and 454 (21.9%) as having poor risk (Table 1). Among the 2072 patients with successful cytogenetic analyses, 988 (48%) had no karyotype anomalies and 1084 (52%) had clonal cytogenetic abnormalities, with 605 (29%) having 1, 180 (9%) having 2, and 299 (14%) having complex (≥ 3) abnormalities. In the latter group, 65 patients (3%) had 3 abnormalities, 39 (2%) had 4 abnormalities, 31 (1%) had 5 abnormalities, 38 (2%) had 6 abnormalities, 27 (1%) had 7 abnormalities, 26 (1%) had 8 abnormalities, and 47 (2%) had more than 8 abnormalities. In 26 patients with complex karyotype, no ISCN code was available.
A systematic documentation of cytogenetic abnormalities (monosomies, trisomies, deletions of short or long arms, trisomies of short or long arms, additional material on short or long arms, translocations involving short or long arms, inversions, isochromosomes, dicentrics, or derivative chromosomes or chromosome arms) was performed for every patient. Furthermore, occurrences as isolated abnormalities, accompanied by 1 additional change, as noncomplex abnormalities (sole and +1 abnormality), and as part of complex abnormalities were recorded (Table S1, available on the Blood website; see the Supplemental Tables link at the top of the online article).
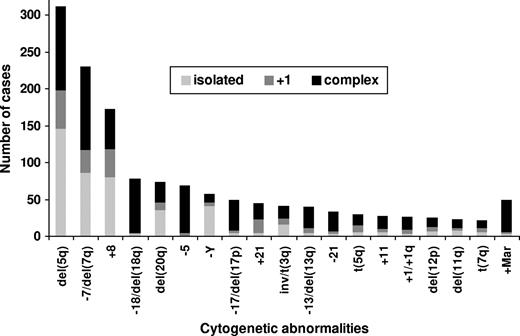
Karyotype abnormalities involving deletions of 5q were the most frequent, occurring in 30% of the 1080 patients with clonal cytogenetic abnormalities (15% of the 2072 patients with successful cytogenetic analyses; Figure 1). Other frequent anomalies were −7/del(7q) (21% of 1080 patients with chromosomal abnormalities), +8 (16%), −18/18q− (7%), 20q− (7%), −5 (6%), −Y (5%), −17/17p− (including isochromosome (17q)) (5%), +Mar (5%), +21 (4%), inv/t(3q) (4%), −13/13q− (4%), +1/+1q (3%), −21 (3%), +11 (3%), 12p− (2%), t(5q) (2%), 11q− (2%), and t(7q) (2%). Isolated del(5q) was seen in 14% of patients with clonal abnormalities, del(5q) with 1 additional anomaly occurred in 5% of patients, and complex anomalies including del(5q) were seen in 11% of patients. The −7/del(7q) anomaly was seen in 8% of patients in isolation, in 3% of patients with 1 additional anomaly, and in 10% of patients as a complex karyotype. Trisomy 8 was seen in 8% of patients in isolation, in 3% of patients with an additional anomaly, and in 5% of patients as a complex anomaly. Loss of the Y chromosome as a sole anomaly was present in 4%, with 1 additional change in less than 1%, and as a complex abnormality in 2%. Isolated 20q− was seen in 3% of patients, 20q− with 1 additional anomaly was seen in less than 1% of patients, and complex anomalies involving 20q− were seen in 4% of patients. Partial or total monosomy 18 occurred nearly exclusively as part of complex anomalies (7% of abnormal patients), except for 3 patients (< 1%) in whom it occurred as isolated anomaliy and 2 (< 1%) with 1 further anomaly each. Cytogenetic anomaly involving +1/+1q, −5, +11, −13/13q−, −17/17p−, −21, and +Mar were also seen predominantly in complex karyotypes.
Frequencies of most common cytogenetic anomalies subdivided into isolated, with 1 additional anomaly, and complex anomalies.
Frequencies of most common cytogenetic anomalies subdivided into isolated, with 1 additional anomaly, and complex anomalies.
Correlation of cytogenetics with FAB and WHO classifications
Of the 2072 patients with successful cytogenetic analyses, FAB classification was available for 1949 patients; of these 1949 patients, WHO classification was available for 595 patients examined by U.G. at the University of Düsseldorf. The distributions of most frequent karyotype abnormalities and IPSS cytogenetic risk categories among the FAB and WHO subgroups are shown in Tables 2 and 3.
Frequency of common karyotypic abnormalities among WHO and FAB subgroups
| Classification . | No. . | Karyotype, no. (%) . | |||||
|---|---|---|---|---|---|---|---|
| Normal . | del(5q) . | −7/del(7q) . | +8 . | −20/del(20q) . | Complex . | ||
| All FAB | 1949 | 942 (48.3) | 295 (15.1) | 209 (10.7) | 162 (8.3) | 86 (4.4) | 282 (14.5) |
| RA | 573 | 267 (46.6) | 139 (24.3) | 30 (5.2) | 37 (6.5) | 31 (5.4) | 47 (8.2) |
| RARS | 252 | 147 (58.3) | 23 (9.1) | 24 (9.5) | 14 (5.6) | 9 (3.6) | 20 (7.9) |
| RAEB | 415 | 179 (43.1) | 71 (17.1) | 60 (23.8) | 39 (9.4) | 21 (5.1) | 98 (23.6) |
| RAEB-t | 305 | 132 (43.3) | 38 (12.5) | 50 (16.4) | 30 (9.8) | 16 (5.2) | 68 (22.3) |
| CMML | 272 | 170 (62.5) | 4 (1.5) | 23 (8.5) | 18 (6.6) | 2 (<1) | 12 (4.4) |
| MDS-AL | 132 | 47 (30.9) | 20 (15.2) | 22 (16.7) | 25 (18.9) | 7 (5.3) | 37 (28.0) |
| All WHO | 595 | 285 (47.8) | 110 (18.5) | 53 (8.9) | 40 (6.7) | 22 (3.7) | 71 (11.9) |
| 5q− syndrome | 61 | 0 (0.0) | 61 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| RA | 56 | 38 (67.9) | 3 (6.5) | 5 (10.9) | 1 (2.2) | 1 (2.2) | 6 (13.0) |
| RARS | 26 | 23 (88.5) | 0 (0.0) | 0 (0.0) | 1 (3.8) | 0 (0.0) | 0 (0.0) |
| RCMD | 164 | 88 (53.7) | 11 (6.7) | 20 (12.2) | 12 (7.3) | 8 (4.8) | 18 (11.0) |
| RSCMD | 77 | 34 (44.2) | 8 (10.4) | 8 (10.4) | 8 (10.4) | 3 (3.9) | 12 (15.6) |
| RAEB-I | 90 | 42 (45.7) | 16 (17.8) | 10 (11.1) | 5 (5.6) | 4 (4.4) | 15 (16.7) |
| RAEB-II | 121 | 60 (49.6) | 11 (9.1) | 8 (6.6) | 13 (10.7) | 5 (4.1) | 19 (15.7) |
| Classification . | No. . | Karyotype, no. (%) . | |||||
|---|---|---|---|---|---|---|---|
| Normal . | del(5q) . | −7/del(7q) . | +8 . | −20/del(20q) . | Complex . | ||
| All FAB | 1949 | 942 (48.3) | 295 (15.1) | 209 (10.7) | 162 (8.3) | 86 (4.4) | 282 (14.5) |
| RA | 573 | 267 (46.6) | 139 (24.3) | 30 (5.2) | 37 (6.5) | 31 (5.4) | 47 (8.2) |
| RARS | 252 | 147 (58.3) | 23 (9.1) | 24 (9.5) | 14 (5.6) | 9 (3.6) | 20 (7.9) |
| RAEB | 415 | 179 (43.1) | 71 (17.1) | 60 (23.8) | 39 (9.4) | 21 (5.1) | 98 (23.6) |
| RAEB-t | 305 | 132 (43.3) | 38 (12.5) | 50 (16.4) | 30 (9.8) | 16 (5.2) | 68 (22.3) |
| CMML | 272 | 170 (62.5) | 4 (1.5) | 23 (8.5) | 18 (6.6) | 2 (<1) | 12 (4.4) |
| MDS-AL | 132 | 47 (30.9) | 20 (15.2) | 22 (16.7) | 25 (18.9) | 7 (5.3) | 37 (28.0) |
| All WHO | 595 | 285 (47.8) | 110 (18.5) | 53 (8.9) | 40 (6.7) | 22 (3.7) | 71 (11.9) |
| 5q− syndrome | 61 | 0 (0.0) | 61 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| RA | 56 | 38 (67.9) | 3 (6.5) | 5 (10.9) | 1 (2.2) | 1 (2.2) | 6 (13.0) |
| RARS | 26 | 23 (88.5) | 0 (0.0) | 0 (0.0) | 1 (3.8) | 0 (0.0) | 0 (0.0) |
| RCMD | 164 | 88 (53.7) | 11 (6.7) | 20 (12.2) | 12 (7.3) | 8 (4.8) | 18 (11.0) |
| RSCMD | 77 | 34 (44.2) | 8 (10.4) | 8 (10.4) | 8 (10.4) | 3 (3.9) | 12 (15.6) |
| RAEB-I | 90 | 42 (45.7) | 16 (17.8) | 10 (11.1) | 5 (5.6) | 4 (4.4) | 15 (16.7) |
| RAEB-II | 121 | 60 (49.6) | 11 (9.1) | 8 (6.6) | 13 (10.7) | 5 (4.1) | 19 (15.7) |
Frequency of poor risk cytogenetics (according to IPSS) in morphologically defined MDS subgroups (FAB and WHO)
| Groups compared . | No. . | Cases with unfavorable karyotype according to IPSS, no. (%) . | P (χ2 value) . |
|---|---|---|---|
| FAB | |||
| RA vs | 573 | 76 (13) | .25 (1.30) |
| RARS | 252 | 41 (16) | |
| RAEB vs | 415 | 133 (32) | .94 (0.00) |
| RAEB-t | 305 | 97 (32) | |
| RA + RARS vs | 825 | 117 (14) | < .001 (69.65) |
| RAEB + RAEB-t | 720 | 230 (32) | |
| RAEB + RAEB-t vs | 720 | 230 (32) | .32 (0.99) |
| AML | 132 | 48 (36) | |
| WHO | |||
| RA vs | 56 | 9 (16) | .03 (4.69) |
| RARS | 26 | 0 (0) | |
| RCMD vs | 164 | 33 (20) | .15 (2.12) |
| RSCMD | 77 | 22 (29) | |
| RAEB-I vs | 90 | 28 (31) | .19 (1.68) |
| RAEB-II | 121 | 28 (23) | |
| RA + RARS vs | 82 | 9 (11) | .03 (4.68) |
| RCMD + RSCMD | 241 | 55 (23) | |
| RA + RARS vs | 82 | 9 (11) | .006 (7.41) |
| RAEB-I + RAEB-II | 211 | 56 (27) | |
| RCMD + RSCMD vs | 241 | 55 (23) | .36 (0.84) |
| RAEB-I + RAEB-II | 211 | 56 (27) |
| Groups compared . | No. . | Cases with unfavorable karyotype according to IPSS, no. (%) . | P (χ2 value) . |
|---|---|---|---|
| FAB | |||
| RA vs | 573 | 76 (13) | .25 (1.30) |
| RARS | 252 | 41 (16) | |
| RAEB vs | 415 | 133 (32) | .94 (0.00) |
| RAEB-t | 305 | 97 (32) | |
| RA + RARS vs | 825 | 117 (14) | < .001 (69.65) |
| RAEB + RAEB-t | 720 | 230 (32) | |
| RAEB + RAEB-t vs | 720 | 230 (32) | .32 (0.99) |
| AML | 132 | 48 (36) | |
| WHO | |||
| RA vs | 56 | 9 (16) | .03 (4.69) |
| RARS | 26 | 0 (0) | |
| RCMD vs | 164 | 33 (20) | .15 (2.12) |
| RSCMD | 77 | 22 (29) | |
| RAEB-I vs | 90 | 28 (31) | .19 (1.68) |
| RAEB-II | 121 | 28 (23) | |
| RA + RARS vs | 82 | 9 (11) | .03 (4.68) |
| RCMD + RSCMD | 241 | 55 (23) | |
| RA + RARS vs | 82 | 9 (11) | .006 (7.41) |
| RAEB-I + RAEB-II | 211 | 56 (27) | |
| RCMD + RSCMD vs | 241 | 55 (23) | .36 (0.84) |
| RAEB-I + RAEB-II | 211 | 56 (27) |
All FAB subgroups contained IPSS good-, intermediate-, and poor-risk cytogenetics. Each subgroup included patients with normal, 5q−, −7/del (7q), +8, −20/20q−, and complex karyotypes. The distribution of normal karyotypes among these subgroups ranged from 30.9% (MDS-AL) to 62.5% (CMML). Del(5q) was slightly more prevalent in RA than in other subtypes and was very rare in CMML. The frequency of complex karyotypes was highest in RAEB, RAEB-t, and MDS-AL. No substantial differences were noted between RA and RARS, except 5q deletions, which were more frequent in RA, or between RAEB and RAEB-t. The proportion of poor cytogenetics was higher in FAB subgroups with more than 5% blasts than in subgroups with less than 5% blasts. MDS-AL had the highest proportion of poor cytogenetics.
In contrast to FAB classification, karyotypic abnormalities were distinctly distributed among the WHO subgroups. Per definition, the WHO 5q− syndrome was entirely made up of IPSS good-risk cytogenetics; RARS comprised only good- and intermediate-risk cytogenetics. Poor-risk cytogenetics were seen in all other WHO subgroups, but RAEB-I contained the highest proportion of poor-risk cytogenetics.
A statistical comparison (χ2 test) of the distribution of the karyotypic findings according to IPSS in the different FAB and WHO subgroups was performed (Table 3). In the FAB subgroups, no significant differences were observed between RA and RARS, RAEB and RAEB-t, and RAEB plus RAEB-t and AML. A highly significant difference (P < .001) became evident comparing cases with fewer than 5% blasts (RA + RARS) with those with at least 5% blasts (RAEB + RAEB-t), with the latter ones showing an accumulation of poor-risk cytogenetics. In WHO subgroups, no significant differences were observed between RCMD and RSCMD, RAEB-I and RAEB-II, and RCMD plus RSCMD and RAEB-I plus RAEB-II. A difference with borderline significance was seen between RA plus RARS and RCMD plus RSCMD, demonstrating a likely association between multilineage dysplasia and poor-risk cytogenetics in cases with low blast counts. This observation is consistent with the lack of statistical differences between early MDS with multilineage dysplasia (RCMD + RSCMD) and advanced MDS (RAEB-I + RAEB-II). As in the FAB system, the comparison of RA plus RARS versus RAEB-I plus RAEB-II again revealed the relevance of the blast count threshold of 5% for the incidence of poor-risk cytogenetics.
Correlation of cytogenetics and survival
Common cytogenetic findings.
Median survival in the cohort of patients with successful cytogenetic analyses (comprising all therapies) was 48 months for patients with good IPSS cytogenetics (n = 1217), 30 months for those with intermediate cytogenetics (n = 401), and 14 months for those with poor cytogenetics (n = 454).
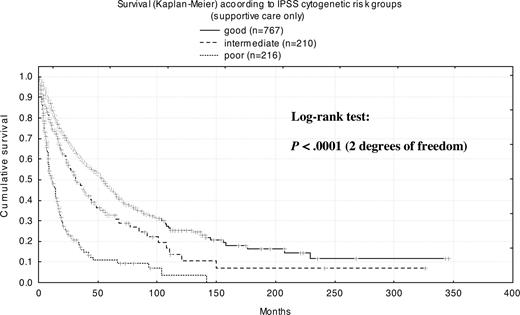
To determine the impact of cytogenetics on the natural course of MDS, we included only the subset of patients (n = 1237) receiving exclusively supportive care for a more detailed analysis (Table S2). Patients with normal karyotype (n = 612) had a median survival time of 53 months, and those with any chromosome abnormality (n = 625) had a median survival time of 24 months. For patients with a mosaic of normal and abnormal metaphases (n = 225), median survival was 27 months, whereas for those with only abnormal metaphases (n = 176), median survival was 18 months (P = .014). Patients with complex abnormalities (n = 166) had a median survival of only 9 months. Median survival time with good-risk cytogenetics (n = 767) according to IPSS was 54 months, with intermediate-risk cytogenetics (n = 210) it was 31 months, and with high-risk cytogenetics (n = 216) it was 11 months.
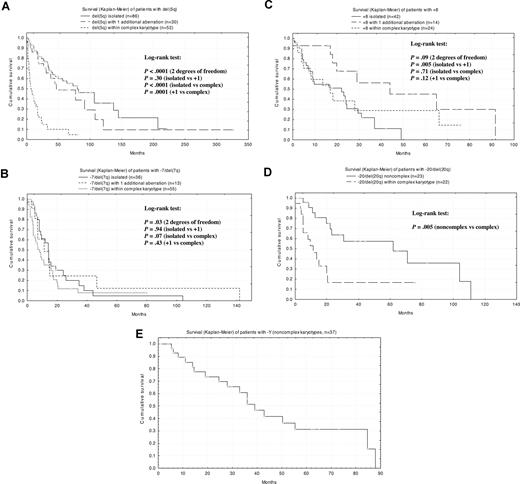
Among the more common karyotypic abnormalities (those affecting at least 3% of patients), prognosis was best for isolated del(5q), with a median survival of 80 months (Figure 2A). The presence of 1 additional karyotypic abnormality in conjunction with del(5q) decreased median survival to 47 months, and complex abnormalities including del(5q) further reduced median survival to 7 months. For monosomy 7 as an isolated deletion, median survival was 14 months; it was also 14 months with 1 additional abnormality and 8 months for the abnormality as part of a complex karyotype. Among patients with del(7q) (n = 18), there was a tendency toward better survival compared with patients with complete monosomy 7 as an isolated abnormality (19 months) and as a noncomplex aberration (14 months). Because the differences were not statistically significant, −7 and del(7q) was regarded as 1 category (Figure 2B). With trisomy 8 as an isolated deletion, median survival was 22 months, but it increased to 44 months with 1 additional abnormality, subsequently decreasing to 17 months again for trisomy 8 as a complex abnormality (Figure 2C). Isolated 20q deletion was associated with a very good prognosis (median survival, 71 months), but the prognosis was significantly worse when 20q deletion was part of a complex karyotype (median survival, 15 months; Figure 2D). Finally, loss of the Y chromosome in noncomplex karyotypes resulted in a median survival of 39 months (Figure 2E), but survival was reduced to 28 months with −Y in the context of a complex karyotype.
Kaplan-Meier survival curves for isolated anomalies with or without additional abnormalities. (A) del(5q). (B) −7/del(7q). (C) +8. (D) del(20q). (E) −Y
Kaplan-Meier survival curves for isolated anomalies with or without additional abnormalities. (A) del(5q). (B) −7/del(7q). (C) +8. (D) del(20q). (E) −Y
Rare cytogenetic findings.
Taking advantage of the large dataset, we also were able to study survival of patients with rare cytogenetic anomalies (Table 4). Translocations involving chromosome 5q within a noncomplex karyotype (n = 7) were associated with an extremely poor prognosis, with a median survival of only 4.4 months. Abnormalities with intermediate prognosis were 3q rearrangements (median survival 20 months; n = 13), translocations involving 11q23 as noncomplex changes (median survival 20 months; n = 6), del(11q) as noncomplex (n = 11) or isolated (n = 8) changes (median survival 26 and 16 months, respectively), and trisomy 19 as a noncomplex abnormality (median survival 20 months; n = 5). A favorable prognosis was seen for patients with translocations involving 1q with noncomplex karyotypes (median survival 35 months; n = 7), translocations involving 7q as noncomplex alterations (median survival 35 months; n = 7), all classes of del(12p) (median survival 66 months; n = 14), translocations involving 17q with noncomplex karyotypes (median survival 32 months; n = 6), monosomy 21 with noncomplex alterations (median survival 32 months; n = 6), trisomy 21 with 1 additional abnormality (median survival 80 months; n = 10) and with noncomplex alterations (median survival 101 months; n = 13), and noncomplex loss of an X chromosome (median survival 56 months; n = 6). Median survival times had not been reached for patients with the following anomalies: del(9q) overall (n = 12) and as a noncomplex abnormality (n = 5), del(12p) as a noncomplex abnormality (n = 7), del(15q) as a noncomplex abnormality (n = 5), and translocations involving chromosome 15 with t(15q) as a noncomplex abnormality (n = 5).
Frequency and median survival of cytogenetic prognostic subgroups
| Anomaly . | Frequency, % . | Median survival, mo . |
|---|---|---|
| Good-prognosis cytogenetics | ||
| del(9q), NC | 0.4 | NR |
| del(15q), NC | 0.4 | NR |
| t(15q), NC | 0.4 | NR |
| del(12p), NC | 0.8 | 108.0 |
| +21, NC | 1.1 | 100.8 |
| −Y, +1 | 0.4 | 84.6 |
| del(5q), isolated | 8.2 | 80.0 |
| +21, +1 | 0.8 | 80.0 |
| del(5q), NC | 10.7 | 77.2 |
| del(20q), isolated | 1.9 | 71.0 |
| del(20q), NC | 2.2 | 71.0 |
| −X, NC | 0.5 | 56.4 |
| No (normal karyotype) | 49.5 | 53.4 |
| del(5q), +1 | 2.5 | 47.0 |
| +8, +1 | 1.2 | 44.0 |
| −Y, NC | 2.7 | 39.0 |
| −Y, sole | 3.5 | 36.0 |
| +1/+1q, NC | 0.4 | 34.7 |
| t(1q), NC | 0.6 | 34.7 |
| t(7q), NC | 0.6 | 34.7 |
| t(11q), NC | 0.5 | 32.1 |
| −21, NC | 0.5 | 32.0 |
| Intermediate-prognosis cytogenetics | ||
| del(11q), NC | 0.9 | 26.1 |
| +8, NC | 5.0 | 23.0 |
| +8, isolated | 3.8 | 22.0 |
| t(11q23), NC | 0.5 | 20.0 |
| Rea 3q, NC | 0.5 | 19.9 |
| +19, NC | 0.4 | 19.8 |
| del(7q), isolated and NC | 0.6 | 19.0 |
| Any 3 abnormalities | 2.8 | 17.1 |
| del(11q), isolated | 0.6 | 15.9 |
| −7, +1 | 0.9 | 14.4 |
| −5, NC | 0.4 | 14.6 |
| −7, sole | 2.3 | 14.0 |
| −7, NC | 3.2 | 14.0 |
| Poor-prognosis cytogenetics | ||
| Complex, all | 13.4 | 8.7 |
| t(5q), NC | 0.4 | 4.4 |
| Four to 6 abnormalities | 5.3 | 9.0 |
| More than 6 abnormalities | 3.9 | 5.0 |
| Anomaly . | Frequency, % . | Median survival, mo . |
|---|---|---|
| Good-prognosis cytogenetics | ||
| del(9q), NC | 0.4 | NR |
| del(15q), NC | 0.4 | NR |
| t(15q), NC | 0.4 | NR |
| del(12p), NC | 0.8 | 108.0 |
| +21, NC | 1.1 | 100.8 |
| −Y, +1 | 0.4 | 84.6 |
| del(5q), isolated | 8.2 | 80.0 |
| +21, +1 | 0.8 | 80.0 |
| del(5q), NC | 10.7 | 77.2 |
| del(20q), isolated | 1.9 | 71.0 |
| del(20q), NC | 2.2 | 71.0 |
| −X, NC | 0.5 | 56.4 |
| No (normal karyotype) | 49.5 | 53.4 |
| del(5q), +1 | 2.5 | 47.0 |
| +8, +1 | 1.2 | 44.0 |
| −Y, NC | 2.7 | 39.0 |
| −Y, sole | 3.5 | 36.0 |
| +1/+1q, NC | 0.4 | 34.7 |
| t(1q), NC | 0.6 | 34.7 |
| t(7q), NC | 0.6 | 34.7 |
| t(11q), NC | 0.5 | 32.1 |
| −21, NC | 0.5 | 32.0 |
| Intermediate-prognosis cytogenetics | ||
| del(11q), NC | 0.9 | 26.1 |
| +8, NC | 5.0 | 23.0 |
| +8, isolated | 3.8 | 22.0 |
| t(11q23), NC | 0.5 | 20.0 |
| Rea 3q, NC | 0.5 | 19.9 |
| +19, NC | 0.4 | 19.8 |
| del(7q), isolated and NC | 0.6 | 19.0 |
| Any 3 abnormalities | 2.8 | 17.1 |
| del(11q), isolated | 0.6 | 15.9 |
| −7, +1 | 0.9 | 14.4 |
| −5, NC | 0.4 | 14.6 |
| −7, sole | 2.3 | 14.0 |
| −7, NC | 3.2 | 14.0 |
| Poor-prognosis cytogenetics | ||
| Complex, all | 13.4 | 8.7 |
| t(5q), NC | 0.4 | 4.4 |
| Four to 6 abnormalities | 5.3 | 9.0 |
| More than 6 abnormalities | 3.9 | 5.0 |
NC indicates noncomplex karyotype; and NR, not reached.
In addition, 21 further chromosomal aberrations were identified (Table S3) that were closely associated with complex abnormalities, and thus—even with this large database—it was not possible to delineate their own prognostic relevance.
Complex abnormalities.
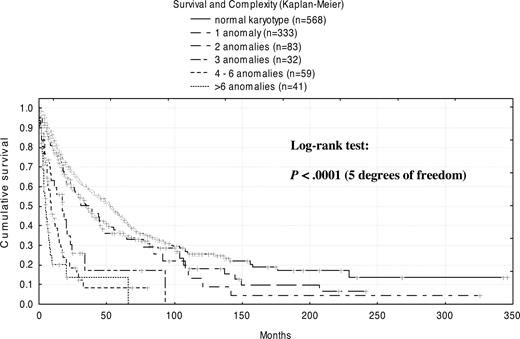
Regardless of the specific aberrations involved, an overall correlation was seen between prognosis and the extent of complexity (Figure 3). With no anomalies (normal karyotype), median survival was 53 months. The presence of 1 abnormality diminished survival to 35 months, whereas 2 abnormalities did not change outcomes significantly (38 months). In contrast, a profound decrease in median survival was observed for patients with 3 abnormalities (17 months) and with 4 to 6 abnormalities (9 months). Merged into 1 group, patients with more than 6 abnormalities showed the shortest survival, only 5 months. In detail, patients with 6 abnormalities had a median survival time of 8 months; patients with 7 abnormalities had a median survival time of 3 months; patients with 8 abnormalities had a median survival time of 6 months; and patients with more than 10 anomalies had a median survival time of only 4 months.
Survival by cytogenetics within FAB/WHO subgroups
To study the impact of the karyotype on median survival times (MSs) within WHO- and FAB-classified subgroups, patients were stratified according to the cytogenetic risk defined by the IPSS (Figure 4). When applied to the complete cohort of patients (supportive care only), survival curves of the respective IPSS risk groups could be separated clearly and showed significant differences of MS (P < .001). Subsequently, MSs for each IPSS cytogenetic risk groups were calculated for WHO- (Figure 5A-C) and FAB-classified (Figure 6A,B) cases. For WHO subgroups RA (n = 33; MS = 107 months) and RARS (n = 20; MS = 156 months), survival curves revealed no significant differences between cytogenetic subgroups (Figure 5A), which may be attributed to the relatively small number of patients. For RCMD (n = 116; MS = 37 months) and RSCMD (n = 48; MS = 44 months), survival was significantly different between good- and poor-risk groups as well as between patients with intermediate- and poor-risk cytogenetics (Figure 5B). Finally, in patients with RAEB-I (n = 63; MS = 17 months) and RAEB-II (n = 82; MS = 23 months), significant differences in survival could be observed between good and intermediate cytogenetic risk, and between good- and poor-risk cytogenetics (Figure 5C).
Kaplan-Meier survival curves for WHO morphologic subgroups according to IPSS cytogenetic risk group. (A) RA/RARS; (B) RCMD/RSCMD; (C) RAEB-I/RAEB-II.
Kaplan-Meier survival curves for WHO morphologic subgroups according to IPSS cytogenetic risk group. (A) RA/RARS; (B) RCMD/RSCMD; (C) RAEB-I/RAEB-II.
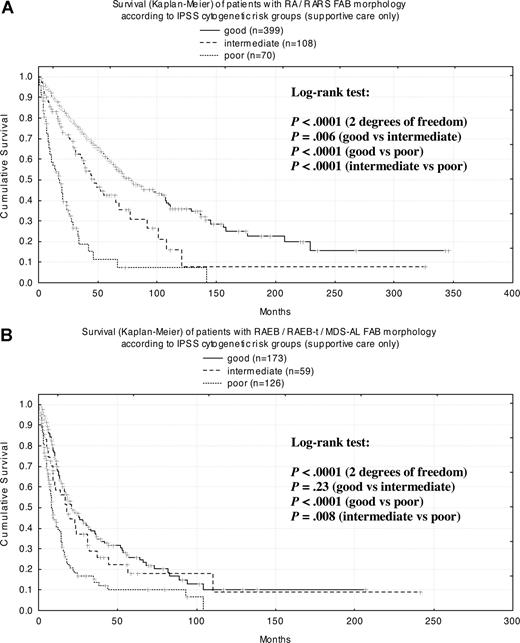
When cases were classified according to FAB criteria in patients with RA (n = 396; MS = 61 months) and RARS (n = 181; MS = 70 months) in every cytogenetic subgroup, survival curves were highly significantly different from each other (Figure 6A). In patients with RAEB (n = 241; MS = 18 months), RAEB-T (n = 100; MS = 9 months), and secondary AML (n = 17; MS = 4 months), there were highly significant differences between the good and poor cytogenetic risk groups as well as between intermediate and poor risk (Figure 6B).
Kaplan-Meier survival curves for FAB morphologic subgroups according to IPSS cytogenetic risk group. (A) RA/RARS; (B) RAEB/RAEB-t/MDS-AL.
Kaplan-Meier survival curves for FAB morphologic subgroups according to IPSS cytogenetic risk group. (A) RA/RARS; (B) RAEB/RAEB-t/MDS-AL.
Discussion
A large, well-characterized dataset is essential for analysis of patient outcomes as well as for an effective assessment and validation of current MDS risk stratification systems with respect to the significance of cytogenetics in general and of rarer cytogenetic abnormalities and specific combinations of karyotypic changes in particular. Our MDS registry, which includes data on 2124 patients with MDS and successful cytogenetic analyses on 2072 (97.6%) patients, is the largest cytogenetic dataset so far published in this field. In the entire cohort, the mean number of metaphases analyzed was 21.9, which is consistent with criteria for high-quality cytogenetics studies in MDS.38 By contrast, the frequencies of chromosomal abnormalities in MDS were previously reported by others using smaller datasets,14,29,–31 often based on analyses with fewer (≥ 10) metaphases, increasing the probability of missing an anomaly.38 Furthermore, our report for the first time includes survival curves associated with various chromosomal abnormalities in MDS.
The frequency of clonal cytogenetic abnormalities was 49.8% among patients with primary MDS (n = 1981) and 68.5% among those with secondary MDS (n = 143), which is in excellent agreement with published literature.27,28 Median survival for patients with normal karyotypes was 53 months, versus 9 months for patients with complex karyotypes (P < .001). The median survival was 54 months for good, 31 months for intermediate, and 11 months for poor IPSS cytogenetic risk subgroups. Our findings on survival in the intermediate and poor IPSS cytogenetic risk groups were comparable with previously reported data (29 months and 10 months, respectively).14
In our study, the frequencies of the common karyotypic anomalies are highly consistent with published data, but the study also adds a number of rarer karyotypic anomalies that were represented sufficiently to allow for determination of their prognostic relevance. Accordingly, the more abundant karyotypic abnormalities (del(5q), −7/del(7q), +8, del(20q), and −Y) were seen most often as isolated anomalies. Several anomalies, however, were seen predominantly as part of complex aberrations (Table S3) and/or with a relatively low incidence, so that the prognostic impact of the abnormalities themselves, independent of the complex abnormal status, could not be determined with statistical accuracy. These included der(1), all no. 2 abnormalities, all no. 4 abnormalities, −5, der(5), all no. 6 abnormalities, −8, all no. 10 abnormalities, +11, −12, t(12q), −13/del(13q), −14, all no. 14 abnormalities, −15, all no. 16 abnormalities, −17/del(17p), −18/del(18q), all no. 18 abnormalities, −20, and +Mar. For these abnormalities, no significant difference in median survival times was seen when comparing all patients with a given abnormality with those who only carried the respective abnormality as part of complex changes. Median survival ranged between 3.4 and 14.5 months (mean, 7.2 months) for all patients and between 4 and 10 months (mean, 6 months) for those with complex abnormalities.
The definition of complex abnormalities remains controversial, but in general, at least 3 independent clonal abnormalities are considered to constitute a complex karyotype. An exception is the Medical Research Council AML 10 trial, for which a complex karyotype was defined by the presence of at least 5 unrelated abnormalities.42 Our findings suggest 3 prognostically relevant thresholds of complexity. Since there were no significant differences between 1 and 2 anomalies (median survival, 35 and 38 months, respectively), but an additional abnormality significantly worsened prognosis (median survival, 17.1 months), we consider 3 anomalies as a distinct prognostic group. A second distinct group can be defined by 4 to 6 anomalies (median survival, 9 months), and a third by 6 or more karyotypic alterations (median survival, 5 months). Thus, we propose that 3 clonal abnormalities are associated with a more intermediate prognosis, whereas complex abnormalities in the narrower sense should be defined by 4 or more independent abnormalities, which could be further subdivided into 4 to 6 and more than 6 anomalies.
As expected, patients with isolated del(5q) fared best (median survival, 80 months) among the patients with clonal cytogenetic anomalies. Interestingly, in our cohort, patients with −7/del(7q) as an isolated deletion had a median survival of 14 months, but only 11 months with 1 additional abnormality and 8.3 months with −7/del(7q) as a complex abnormality. We were able to define median survival times for new cytogenetic groups, most of which correspond to the IPSS intermediate prognostic group. Most interestingly, translocations involving the long arm of chromosome 7, which are considered to be in the IPSS poor-risk group, actually had a favorable prognosis (median survival of 35 months) when these anomalies were part of noncomplex karyotypes. Chromosome 7 anomalies overall tended to have a better prognosis than described previously if not part of a complex karyotype. Some other rare abnormalities (as isolated changes) showed equally good prognoses, although case numbers are still comparably small (+1/+1q, t(1q), t(7q), del(9q), del(12p), chromosome 15 anomalies, t(17q), monosomy 21, trisomy 21, and −X). In particular, the del(12p) anomaly within a noncomplex karyotype was associated with a much better median survival (66 months for all patients, and the median not reached for patients with 12p deletion as a noncomplex abnormality) than expected. This observation is in good accordance with recent data from the Spanish group.31 Translocations involving the long arm of chromosome 5 (IPSS intermediate group) were associated with very poor prognosis in this study, even if they occurred in noncomplex karyotypes (median survival, 4 months). On the strength of these data, many aberrations can now be confirmed as intermediate-risk cytogenetics and constitute a group defined by comparable survival times and not solely by exclusion from the good- or poor-risk group. By increasing case numbers, international joint activities may corroborate these findings, especially for the very rare karyotypic findings.
We also studied the prognostic impact of additional chromosomal abnormalities on survival times. Whereas it appears that the addition of 2 or more aberrations overwhelms the impact of the initial anomaly by a quantitative effect, only 1 additional cytogenetic abnormality may modulate the prognostic value of the primary cytogenetic change in different ways. These need to be delineated specifically for each anomaly. We have observed 2 patterns: worsening of prognosis, as seen for del(5q) and −7/del(7q); and improvement of prognosis, as seen for trisomy 8 and loss of the Y chromosome.
Analysis of this dataset has yielded meaningful assessment and validation of current MDS risk stratification systems. Based on these findings, WHO-RARS was a better-defined entity with respect to cytogenetic homogeneity than was FAB-RARS, since WHO-RARS comprised only IPSS good- and intermediate-risk cytogenetics, whereas FAB-RARS contained good-, intermediate-, and poor-risk cytogenetics. Similarly, in FAB as well as WHO subgroups, the relevant blast count threshold separating subgroups with a high from those with a low incidence of poor-risk cytogenetics proved to be 5%. In contrast, this was not the case for the thresholds of 10% (WHO-RAEB-II), 20% (FAB-RAEB-t), and 30% (FAB-AML), for which no differences in the incidences could be seen. In the WHO subgroups, the introduction of multilineage dysplasia as a criterion defining distinct subgroups with fewer than 5% blasts proved its biological relevance by its association with poor-risk cytogenetics. This was underlined by the fact that the comparison between patients with multilineage dysplasia with blasts less than 5% and patients with 5% to 19% blasts failed to show statistical differences for the distribution of poor-risk cytogenetics.
Our examination of the impact of cytogenetic risk according to IPSS criteria on survival for patients grouped according to WHO and FAB criteria confirmed the very good prognosis of patients with less than 5% blasts and dysplasia limited to erythropoiesis (WHO-RA and -RARS), which was only moderately influenced by cytogenetics. For all other morphologic subgroups, regardless of the type of the classification system used, cytogenetics significantly influenced prognosis; the intermediate-risk group showed the least significant prognostic impact. This reflects the nature of the IPSS-defined intermediate-risk group as a merged exclusion group (neither good- nor poor-risk cytogenetics) and uncovers the fact that unknown poor- and good-risk cytogenetic subgroups are hidden within this group.
The only curative therapy for MDS is allogeneic stem cell transplantation, but this approach is suitable for only a small percentage of the MDS population and is associated with high treatment-related morbidity and mortality.4,43,,–46 An urgent need exists for therapies for patients with MDS who are ineligible for transplantation. Newer therapies such as 5-azacitidine,47,48 decitabine,49 and lenalidomide50,51 are changing the treatment landscape for patients with MDS. Certain therapies have also demonstrated particular efficacy in specific cytogenetic subgroups. For example, the immunomodulatory drug (IMiD) lenalidomide has shown high rates of hematologic remission and transfusion independence in transfusion-dependent Low/Int-1–risk MDS, with significantly greater response among patients harboring del(5q31).50,51 Lenalidomide was also associated with a high rate of cytogenetic remission among patients with del(5q31).48,49 Recently, it has been reported that patients with monosomy 7 might benefit predominantly from therapy with demethylating agents.26,52 Even complex abnormalities might be responsive future targets for demethylating agents and IMiDs such as lenalidomide.25,52 Thus, cytogenetic anomalies are not only an important component of the diagnosis and prognosis of MDS but are currently being used to identify appropriate therapies.
In summary, cytogenetic findings have an established role in the diagnosis and assessment of prognosis of MDS and are emerging as an important factor in treatment selection and monitoring response to therapy. Our analysis of a large cytogenetic dataset of patients with MDS provides a comprehensive overview of the cytogenetic profile of MDS, identifies additional previously undescribed, prognostically relevant cytogenetic subgroups, reveals correlations of blast count and multilineage dysplasia with cytogenetic risk groups, and highlights the need for a revised cytogenetic scoring system for prognostic models that considers our new findings and more adequately represents high-risk cytogenetics.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Helga Grüner for cytogenetic analyses and Rainer Steffens for technical assistance.
This work was supported by an unrestricted educational grant from Celgene Corporation and the German Kompetenznetz Akute und Chronische Leukämien. It was also supported in part by Tiroler Wissenschaftsfond and Tiroler Verein zur Förderung der Krebsforschung an der Universität Innsbruck (R.S.).
Authorship
Contribution: D.H. wrote the paper, designed and performed research, cytogenetic analyses and review, collected clinical data, and analyzed data; U.G. designed and performed research and morphologic review, collected clinical data, and gave final approval; J.S. designed and performed research, collected clinical data, performed data analyses and statistical analyses, and gave final approval; M.P., T.N, A.K., M.L., L.T., O.K., R.S., F.W., and P.V. collected clinical data and gave final approval; B.H., R.K., and C.F. performed cytogenetic analyses and gave final approval; A.G. and C.A. collected clinical data, performed morphologic review, and gave final approval; T.M. performed statistical analyses and gave final approval; C.S. designed and performed research and cytogenetic review, collected clinical data, performed statistical analyses, wrote the paper, and gave final approval.
Conflict-of-interest disclosure: D.H. is a member of the speakers' bureau at Celgene Corp, and the advisory boards at Novartis and Pharmion; A.G. is a member of the speakers' bureau and advisory board at Celgene Corp, and owns stock in Hoffmann La Roche AG. All other authors declare no competing financial interests.
Correspondence: Detlef Haase, Department of Hematology and Oncology, University of Göttingen, Robert-Koch-Str 40, Göttingen, 37075, Germany; e-mail:haase.onkologie@med.uni-goettingen.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal