HGAL is a newly identified germinal center (GC)–specific gene whose expression by the tumor cells correlates with a favorable prognosis in patients with diffuse large B-cell and classical Hodgkin lymphomas. The function of HGAL is unknown. Previous studies demonstrated that HGAL is dispensable for GC formation, immunoglobulin gene class-switch recombination, and somatic hypermutation. Herein, we identify a role for HGAL in the regulation of cell motility. We demonstrate that IL-6 induces the phosphorylation of the C-terminal tyrosine residue of the HGAL protein via the Lyn kinase, and promotes its relocalization from the cytoplasm to podosome-like structures. Further, IL-6–induced HGAL phosphorylation increases its interaction with myosin II and is associated with inhibition of cell migration. Knockdown of endogenous HGAL ameliorates IL-6–induced inhibition of cell migration, whereas overexpression of HGAL imparts inhibitory effects of IL-6 on cell migration. Taken together, our results suggest that HGAL is involved in negative regulation of lymphocyte migration, thus constraining lymphocytes to the GC. Inhibition of lymphocyte migration might contribute to the less aggressive clinical behavior of HGAL-expressing lymphomas.
Introduction
Gene-expression profiling studies identified an expressed sequence tag (UniGene cluster 49 614, Clone 814622, GI: 2210537) that was associated with better survival in patients with diffuse large B-cell lymphoma (DLBCL). We have cloned the full-length cDNA for this gene and named it human germinal center–associated lymphoma (HGAL).1 Later on, Pan et al also cloned the same gene and named it germinal center-expressed transcript 2 (GCET2).2 The HGAL gene is located on chromosome 3q13 and encodes a 178–amino acid (aa) protein with 51% identity and 62% similarity to the murine M17 protein. Like its murine counterpart, HGAL is specifically expressed in the cytoplasm of normal germinal center (GC) B cells. Studies in M17 knock-out mice3 revealed that this protein is dispensable for GC formation, immunoglobulin somatic hypermutation, and class-switch recombination, and for mounting of T cell–dependent antibody responses. However, in contrast to its wild-type littermates, M17-deficient mice exhibited reduced-sized Peyer patches. We have demonstrated that HGAL is also expressed in GC-derived lymphomas and distinguishes biologically distinct subgroups of classic Hodgkin lymphoma (cHL) associated with improved overall survival.1,4 These observations, coupled with the tightly regulated expression of the HGAL and M17 proteins during B-cell ontogeny, restricted to B lymphocytes in the GC compartment, and to their content of an immunoreceptor tyrosine-based activation motif (ITAM), usually implicated in signal transduction in B and T lymphocytes, suggests that these proteins have a specific signaling function
Herein, we report that HGAL mediates IL-6–induced inhibition of GC B-cell migration. We demonstrate that IL-6 induces Lyn-mediated phosphorylation of the HGAL C-terminal tyrosine and causes HGAL relocalization to podosome-like structures and spike-like filopodia. We show interactions between endogenous HGAL, actin, and myosin II, and delineate HGAL domains responsible for the interaction. We provide evidence that HGAL phosphorylation results in increased interaction with myosin II. Moreover, we demonstrate that knockdown of endogenous HGAL ameliorates the inhibitory effects of the IL-6 on cell migration. Taken together, these results identify HGAL as a physiologic mediator of IL-6 effects on lymphocyte migration and suggest that HGAL expression in GC lymphocytes, associated with previously reported down-regulation of IL-6 production by these cells,5 may contribute to the control of lymphocyte migration and localization of normal and malignant B cells in the GC microenvironment.
Materials and methods
Reagents and antibodies
Mouse monoclonal anti-HGAL antibody was generated in our laboratory, as reported previously.6 Mouse monoclonal antiphosphotyrosine (PY99) and rabbit polyclonal anti-WASP and Lyn antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antiphospho-Lyn (Tyr507) antibibody was from Cell Signaling Technology (Beverly, MA). Monoclonal anti-V5 antibody was from Invitrogen (Carlsbad, CA). Mouse monoclonal anti–β-actin and rabbit polyclonal anti–myosin IIa and IIb were from Sigma-Aldrich (St Louis, MO), anti–human IgM antibody was from Biosource (Biosource, Camarillo, CA), and anti–mouse immunoglobulin light-chain antibody from Jackson ImmunoResearch (West Grove, PA). Cy3/Cy2-conjugated goat anti–mouse immunoglobulin G and Cy3/Cy2-conjugated goat anti–rabbit immunoglobulin G were from Jackson ImmunoResearch. Rhodamine-labeled phalloidin and DAPI were from Molecular Probes (Invitrogen). Recombinant human IL-4, IL-6, and interferon-γ (INFγ) were from R&D Systems (Minneapolis, MN). Human plasma fibronectin was from Sigma-Aldrich, and G418 was from GIBCO (Invitrogen-GIBCO, Grand Island, NY). Cytochalasin D (Sigma-Aldrich) and Latrunculin B (BIOMOL Research Laboratories, Plymouth Meeting, PA) were used at a final concentration of 5 μM for 30 minutes to inhibit actin polymerization and disrupt microfilament organization.7,8 Sodium orthovanadate was from Sigma-Aldrich; sodium pervanadate was prepared immediately before use by mixing equimolar (100 mM) solutions of sodium orthovanadate and hydrogen peroxide for 10 minutes at room temperature, and was used at a final concentration of 1 mM.
Cells and cell culture
The human non-Hodgkin lymphoma (NHL) cell lines VAL, Ramos, Raji, and SUDHL6 were cultured at 37°C and 5% CO2 in RPMI 1640 medium (Fisher Scientific, Santa Clara, CA) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT), 2 mM glutamine, and 100 U/mL penicillin/100 μg/mL streptomycin (Invitrogen-GIBCO). The human cervical cancer cell line HeLa was grown in Dulbecco modified Eagle medium (DMEM; Invitrogen-GIBCO) and was similarly supplemented with FBS, glutamine, and penicillin/streptomycin.
Human CD77+ GC centroblasts and centrocytes (referred as GC B cells) were isolated from tonsils obtained during routine tonsillectomies. Informed consent was obtained in accordance with the Declaration of Helsinki from all patients, and tissue collection was approved by the institutional review committee of the Sylvester Comprehensive Cancer Center, University of Miami. The tonsils were mechanically minced, and mononuclear cells were isolated by Ficoll-Paque Plus density centrifugation (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's protocol. The GC B cells were enriched according to the protocol reported by Klein et al9 with minor modification: use of the AutoMACS system (Miltenyi Biotec, Auburn, CA) according to the manufacturer's protocol. The purities of the isolated cells were determined by LSRI flow cytometer (Becton Dickinson, Mountain View, CA) analysis. Briefly, 3 to 6 × 104 cells from the positive and negative selection fractions were stained with 2.5 μg mouse anti-rat IgM-FITC (Becton Dickinson), 1.5 μg rat anti-CD77 IgM (Beckman Coulter, Miami, FL), and 25 ng anti-CD38–PE (Becton Dickinson) antibodies, washed once with AutoMACS running buffer (Miltenyi Biotec), and analyzed by the flow cytometer. Purity ranged from 85% to 95% in the positive fractions. Less than 4% of CD77+ cells were detected in the negative fractions (CD77− fraction).
Plasmid constructs, cell transfection, and gene silencing
The full length wild-type human HGAL cDNA expression plasmid, based on the pcDNA3.1D/V5-His-TOPO vector (HGAL pcDNA3.1D; Invitrogen), was reported previously.6 The HGAL truncated mutant (HGALΔC), encoding the HGAL cDNA amino acids 1 to 118, was generated by polymerase chain reaction (PCR) from the original HGAL pcDNA3.1D plasmid (primers are available on request) and cloned in frame into the pcDNA3.1D/V5-His-TOPO vector (HGAL-ΔC pcDNA3.1D). The HGAL C-terminal tyrosine mutants (HGAL-Y128F and -Y148F) were generated by substitutions of tyrosines 128 and 148 to phenylalanine using the QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX) according to the manufacturer's instructions. The full-length wild-type human Lyn cDNA expression plasmid was amplified from cDNA of 293F cells with the following primers: forward, 5′-CACCATGGGATGTATAAAATCAAAAGGGAAAG-3′; reverse, 5′-AGGCTGCTGCTGGTATTGCCCTTCC-3′. A kinase-defective Lyn (K275D) was created by substituting Lys 275 in the ATP-binding pocket with Asp using the site-directed mutagenesis kit, and a constitutively active Lyn (Y508F) was created by substituting the conserved-tail Tyr residue with Phe, as previously reported by Dai et al and Shandary et al.10,11 These amplicons were cloned using standard protocols into the pcDNA3.1D/V5-His-TOPO vector. The structure of all plasmids was verified by sequencing.
HeLa cells were plated at 2.5 × 105 cells in 60-mm culture dishes (Nalge Nunc International, Rochester, NY) in 5 mL DMEM and grown overnight at 37°C and 5% CO2. Either HGAL expression or empty vectors were transfected into the cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The cells were subsequently expanded and grown in selection medium supplemented with 600 μg/mL G418 for 12 to 18 days. Single colonies were selected, expanded, and screened for the presence of the HGAL-V5 proteins by Western blot analysis with anti-HGAL and anti-V5 antibodies.
For knocking down the expression of the HGAL and Lyn proteins, we used specific Dharmacon siRNA smart pools (NM 152785 and NM 002350; Dharmacon, Chicago, IL). For control experiments, scrambled siRNA (Dharmacon) was used. Briefly, 4 × 105 HeLa-HGAL cells/well of the 6-well dish were transfected with 200 pmol of specific or control siRNA using Lipofectamine 2000. At 48 hours after transfection, the cells were trypsinized and replated in the 6-well dishes for indicated periods of time or used for preparation of whole-cell lysates for Western blotting. For knocking down the expression of these proteins in VAL cells, 1 to 5 × 106 cells per nucleofection sample were centrifuged for 10 minutes at 200g and then resuspended in 100 μL of the Nucleofector Solution V (Amaxa, Gaithersburg, MD) at room temperature. A total of 200 pmol of siRNA was added to the cell suspensions, and the mixtures were transferred to the Amaxa-certified cuvettes. Nucleofection was performed using the U-15 program. The cells were incubated for 48 hours in humidified 37°C/5% CO2 incubator and then used for either preparation of whole-cell lysates for Western blotting or chemotaxis assays.
Western blotting and immunoprecipitation
Whole human cell extracts were prepared as reported previously.12 Briefly, 5 × 106 cells were washed once with ice-cold phosphate-buffered saline (PBS) and homogenized in RIPA buffer (1 × PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 10 mM phenylmethylsulfonyl fluoride, 1 μg/mL aprotinin, and 100 mM sodium orthovanadate) on ice for 30 minutes. Cell lysates were centrifuged at 14 000g for 10 minutes at 4°C to remove insoluble material. Protein concentrations of lysates were determined using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL), and samples were adjusted to equal protein concentration. For Western blotting, 20 μg of whole-cell lysate was separated on 10% to 15% SDS–polyacrylamide gel electrophoresis (PAGE), transferred to polyvinylidene difluoride membranes (BioRad Laboratories, Hercules, CA), and immunoblotted with specific antibodies.
For immunoprecipitation (IP), 400 μg protein was precipitated for 1 to 2 hours with the indicated antibodies at 4°C with rotation. Protein G–agarose (Invitrogen) or TrueBlot anti-mouse Ig IP beads (eBioscience, San Diego, CA) were added, and the mixture was rotated for an additional 1 hour. Precipitated complexes were washed 4 times in RIPA buffer, boiled in Laemmli buffer (10 × concentrate: 0.25 M Tris, 1.92 M glycine, and 1% SDS-PAGE in aqueous solution), separated on 10% to 15% SDS-PAGE gel, and immunoblotted with the indicated antibodies. The membranes were then stripped for 30 minutes at room temperature with Restore Western Blot Stripping Buffer (Pierce), washed extensively in PBST (PBS with 0.1% Tween 20), blocked for 2 hours in 5% milk blocking buffer, and then reprobed with additional antibodies, as indicated.
For mass spectrometry, precipitated complexes were run on 10% SDS-PAGE gel and detected by Coomassie brilliant blue staining. Bands of selected size were cut out and submitted to Taplin Biological Mass Spectrometry Facility, Harvard Medical School (Boston, MA) for mass spectrometry analysis.
Immunofluorescence microscopy
The subcellular localization of HGAL protein and its colocalization with actin, myosin II, and WASP proteins were assessed by immunofluorescence microscopy of HeLa-HGAL, SUDHL6, and VAL cells. Briefly, HeLa-HGAL cells were seeded on glass coverslips in 6-well culture dishes while lymphoma cells were dropped on the slides and dried at room temperature. The cells were fixed with 4% paraformaldehyde in PBS for 20 minutes, permeabilized with PBS + 0.25% Triton X100 for 15 minutes and then blocked with 1% FBS in PBST for 30 minutes. After washing 3 times in PBST, the cells were incubated for 30 minutes at room temperature with the indicated antibodies at 1:200 to 1:500 dilutions. The cells were washed 4 × 5 minutes with PBST, and secondary Cy2-conjugated goat anti–mouse IgG and Cy3-conjugated goat anti-rabbit IgG antibodies at 1:200 dilution were added for 30 minutes. Actin filaments and nuclei were stained with rhodamine-labeled phalloidin and DAPI, respectively. The cells were visualized with a Zeiss LSM 510 laser scanning confocal unit on a Zeiss Axiovert 200 M microscope (Carl Zeiss, Thornwood, NY) with a plan neofluor 40×/1.3 NA DIC oil-immersion objective lens. The images were acquired using AxioVision 4.5 software (Carl Zeiss) and were processed using Adobe Photoshop CS2 (Adobe Systems, San Jose, CA).
Chemotaxis and wound assays
Chemotaxis assays were performed in 24-well plates (Nalge Nunc International) containing 3- to 5-μm porous polycarbonate membrane inserts (Nalge Nunc International), as was previously reported.13 Briefly, the inserts were coated with 10 μg/mL fibronectin for 30 minutes and equilibrated in RPMI 1640 medium. VAL and Ramos cells or human CD77+ GC centroblasts and centrocytes were washed twice with PBS and resuspended at 5 × 106 cells/mL in nonsupplemented RPMI 1640 medium. IL-6 at 10 ng/mL in a total volume of 300 μL was added to the bottom of the wells, and a similar total volume of cells with or without IL-6 was loaded on the inserts. The cells were allowed to migrate for 2 hours at 37°C and 5% CO2. The inserts were then fixed and stained with DiffQuik (Dade, Düdingen, Switzerland), and the number of migrated stained cells in 3 fields per well was counted under microscope (Olympus, Center Valley, PA) at 40× magnification. All the assays were performed in triplicate. Simultaneously, the number of cells that migrated to the bottom portion of the well was assayed by flow cytometry.
Wound assays were performed as previously described by Denker and Barber.14 Briefly, HeLa cells stably transfected with HGAL (HeLa-HGAL) or control vectors (HeLa-Neo) were grown to confluence in 35-mm dishes and wounded with a P200 pipette tip. Wounded monolayers were washed 3 times with complete growth medium and returned to the incubator for 1 hour to recover from wounding before experiments were begun. The cells were left untreated or incubated with 10 ng/mL IL-6 or a similar volume of PBS. At 0 and 24 hours, time-lapse images of the monolayers were photographed using 63×/1.4 NA objective lens of a Zeiss Axiovert 200M microscope equipped with the Zeiss AxioCam MRM camera (Carl Zeiss), and the wound width at the predetermined grid-marked points was measured using the AxioVision 4.5 measurement tool (Carl Zeiss). All the experiments were performed in duplicate and were repeated at least 3 times.
Cell proliferation assay
Cell proliferation response to IL-6 was assessed by [3H]thymidine incorporation assays. Briefly, cells at 10 × 103 per well in a 96-well plate were grown for 24 or 48 hours with and without IL-6 (10 ng/mL) before pulsing with 1 μCi (0.037 MBq) [3H]thymidine (Amersham) for 4 hours. Cells were then harvested onto glass filters using a microtiter plate cell harvester, and incorporation of radiolabeled thymidine into DNA was determined by scintillation counting using the Packard Microplate Scintillation & Luminescence Counter (Packard, Shelton, CT). All assays were done in quadruplicate, and the mean and standard deviations were calculated.
RNA isolation and reverse transcription reaction
Total cellular RNA was isolated from 5 × 106 to 1.0 × 107 cells using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. Total RNA isolated from the cells was quantified using spectrophotometric OD260 measurements. RNA (2 μg) was reverse transcribed using the High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol with a minor modification that consisted of the addition of RNase inhibitor (Applied Biosystems) at a final concentration of 1 U/μL. Samples were incubated at 25°C for 10 minutes and 37°C for 120 minutes.
Real-time PCR measurement of HGAL mRNA expression
Real-time PCR measurement of HGAL mRNA expression was performed as previously described,15 and was normalized to the 18S endogenous control.
Yeast 2-hybrid
Full-length HGAL cDNA was cloned into the 2-hybrid vector pGBKT7 (Clontech, Palo Alto, CA) containing a c-MYC epitope tag to produce a Gal4 DNA-binding domain (DBD) fusion to HGAL. Protein extracts prepared from AH109 transformed with the Gal4-HGAL construct were analyzed by Western blotting with an anti–c-MYC epitope tag antibody to confirm that the expected Gal4DBD/HGAL fusion protein was produced. Gal4DBD/HGAL was used to screen a human B-cell cDNA library prepared from a normal human tonsil (a generous gift of Dr Glen Barber, University of Miami) and expressed from the pACT vector. AH109 cells expressing the fusion protein were transformed with the pACT library. Ade2- and His3-expressing cells were initially selected on SD/Ade−/Leu−/Trp plates following by a second selection on SD/Ade−/His−/Leu−/Trp− plates. DNA sequence was obtained from the 5′ end of each HGAL interacting clone.
Statistical analysis
To test the differences in IL-6–induced chemotaxis, wound distance and cell proliferation between HGAL-expressing and -nonexpressing cells, we used the 2-tailed Student t test. A P value less than .05 was considered statistically significant.
Results
HGAL protein is phosphorylated in response to IL-6 stimulation
The HGAL protein contains 6 tyrosine residues, including the 2 C-terminal tyrosines within the modified ITAM. Since proteins containing ITAM are commonly tyrosine-phosphorylated in response to extracellular stimuli, we initially examined the HGAL protein after exposing cells to pervanadate, a potent phosphatase inhibitor that alters the balance between continuous endogenous tyrosine phosphorylation by kinases and dephosphorylation by phosphatases. To this end, HGAL-expressing lymphoma cell lines (Ramos, Raji, VAL, and SUDHL6) and HGAL-transfected Hela cells (HeLa-HGAL) were incubated in medium containing 1 mM pervanadate for up to 1 hour. The cells were lysed, and the HGAL protein was immunoprecipitated and immunoblotted with an antiphosphotyrosine antibody (Figure 1A). Under these conditions, HGAL was found to be tyrosine phosphorylated. To determine what physiologic stimuli might induce HGAL tyrosine phosphorylation, SUDHL6 lymphoma cells were treated for 5 to 60 minutes with a series of potential stimuli, including IL-4, IL-6, interferon- γ, and anti-IgM (Figure 1B). Among the tested stimuli, anti-IgM induced phosphorylation of significantly larger number of proteins as assessed by antiphosphotyrosine immunoblotting (data not shown); however, only IL-6 induced HGAL tyrosine phosphorylation, which was transient and peaked at 5 to 15 minutes. Identical results were observed in Raji, Ramos, and VAL lymphoma cell lines as well as in stably transfected HeLa-HGAL cells (data not shown). These results indicate that IL-6 can induce tyrosine phosphorylation of HGAL protein.
Pervanadate and IL-6 induce tyrosine phosphorylation of HGAL protein. SUDHL6 lymphoma cells were incubated for up to 60 minutes with pervanadate (1 mM) (A) or stimulated with IL-6 (10 ng/mL), IL-4 (10 ng/mL), anti-IgM (100 ng/mL), and IFNγ (1 μM) (B). Cellular lysates were extracted at the indicated time points, immunoprecipitated with anti-HGAL antibody, and blotted for phosphotyrosine (PTyr) and HGAL. Unconjugated beads served as a control (B). Representative blots of 3 independent experiments are shown.
Pervanadate and IL-6 induce tyrosine phosphorylation of HGAL protein. SUDHL6 lymphoma cells were incubated for up to 60 minutes with pervanadate (1 mM) (A) or stimulated with IL-6 (10 ng/mL), IL-4 (10 ng/mL), anti-IgM (100 ng/mL), and IFNγ (1 μM) (B). Cellular lysates were extracted at the indicated time points, immunoprecipitated with anti-HGAL antibody, and blotted for phosphotyrosine (PTyr) and HGAL. Unconjugated beads served as a control (B). Representative blots of 3 independent experiments are shown.
IL-6 stimulation induces HGAL localization to podosomes
We have previously demonstrated that in unstimulated cells, HGAL protein is localized mainly in the cytoplasm.6 Upon stimulation of the SUDHL6 and VAL lymphoma and HeLa-HGAL cell lines by IL-6, podosome-like structures and spike-like protrusions were detected on the cell surface. Noticeably, immunofluorescence (Figure 2) and confocal miscroscopic studies (data not shown) demonstrated relocalization of a fraction of the cytoplasmic HGAL protein to these structures. Since podosomes are formed by actin and WASP proteins, and the latter serves as a podosome marker,16 we examined whether HGAL colocalizes with these proteins. Unstimulated and IL-6–stimulated SUDHL6 and HeLa-HGAL cells were fixed on slides and stained with rhodamine-labeled phalloidin and with anti-HGAL and anti-WASP antibodies that were detected by Cy2- or Cy3-labeled secondary antibodies. Visualization with immunofluorescence (Figure 2) and confocal miscroscopy (data not shown) demonstrated that HGAL indeed colocalizes with actin and WASP proteins in the podosomes.
IL-6 stimulation induces colocalization of HGAL with actin and WASP proteins in podosomes. Unstimulated or IL-6–stimulated Hela-HGAL (A) and SUDHL6 (B) cells were fixed with 4% paraformaldehyde and stained with anti-HGAL antibody (green), anti-WASP antibody (green or red, as indicated), rhodamine-labeled phalloidin (red), and DAPI (blue), and were visualized on a Carl Zeiss LSM510 microscope. Arrows indicate podosome-like structures. Representative photographs of 3 independent experiments are shown (see “Immunofluorescence microscopy” for image acquisition information).
IL-6 stimulation induces colocalization of HGAL with actin and WASP proteins in podosomes. Unstimulated or IL-6–stimulated Hela-HGAL (A) and SUDHL6 (B) cells were fixed with 4% paraformaldehyde and stained with anti-HGAL antibody (green), anti-WASP antibody (green or red, as indicated), rhodamine-labeled phalloidin (red), and DAPI (blue), and were visualized on a Carl Zeiss LSM510 microscope. Arrows indicate podosome-like structures. Representative photographs of 3 independent experiments are shown (see “Immunofluorescence microscopy” for image acquisition information).
HGAL interacts with actin and myosin II
Colocalization of the HGAL protein with actin and WASP may be due to a spatial colocalization with or without direct physical interaction between these proteins. To ascertain whether HGAL and actin or WASP proteins might interact directly, coimmunoprecipitation experiments were performed. Direct physical interaction between HGAL and actin was demonstrated in the unstimulated HeLa-HGAL and in SUDHL6 cells (Figure 3A), confirming the immunofluorescence findings. The physical interaction between HGAL and actin was further confirmed by yeast 2-hybrid experiments in which HGAL was used as the bait (data not shown). In contrast, despite spatial colocalization, no direct physical interaction was detected between HGAL and WASP proteins when assessed by coimmunoprecipitation and by yeast 2-hybrid experiments (data not shown).
HGAL interacts with actin and myosin II proteins, and the interaction with the myosin is increased by IL-6–induced HGAL phosphorylation. (A) Cellular lysates of unstimulated HeLa-HGAL and SUDHL6 cells were immunoprecipitated with either anti-HGAL or anti-actin antibodies and blotted with the indicated antibodies. Unconjugated beads served as a control (“B”). The size of HGAL in the HeLa cells is 26 kDa, since the cloned protein is a fusion protein with V5 tag. (B) Cellular lysates were extracted from unstimulated and IL-6–stimulated SUDHL6 cells, immunoprecipitated with anti-HGAL antibody, resolved by SDS-PAGE, and stained with Coomassie blue. Protein bands of 250 kDa, 230 kDa, and 140 kDa (→) were excised and analyzed by mass spectrometry analysis and sequencing that revealed that all the 3 proteins represent myosin II, the expected size of which is 250 kDa. (C) Cellular lysates were extracted from unstimulated SUDHL6 and VAL lymphoma cell lines, immunoprecipitated with anti-HGAL antibody, and blotted with indicated antibodies. Unconjugated beads served as a control (“B”). (D) Cellular lysates were extracted from unstimulated or IL-6–stimulated SUDHL6 cells and immunoprecipitated with either anti-HGAL or antimyosin antibodies, resolved on the SDS-PAGE, and blotted with the indicated antibodies. (E) HeLa-HGAL cells were treated with cytochalasin D (5 μM) and latrunculin B (5 μM) for 30 minutes, respectively, to inhibit actin polymerization and disrupt actin filaments. The whole-cell lysates were extracted, immunoprecipitated with anti-HGAL antibody, and blotted with the indicated antibodies. (F) SUDHL6 and HeLa-HGAL cells were stimulated with IL-6 for up to 60 minutes. At the indicated time points, cellular lysates were extracted, immunoprecipitated with anti-HGAL antibody, and blotted with the indicated antibodies. Arrows indicate immunoglobulin light chains. (G) Unstimulated or IL-6–stimulated Hela-HGAL (left) and SUDHL6 (right) cells were fixed with 4% paraformaldehyde, stained with anti-HGAL antibody (green), antimyosin antibody (red), and DAPI (blue), and visualized with the Carl Zeiss LSM510 microscope. (H) HGAL and myosin II colocalization in podosome-like structures of IL-6–stimulated SUDHL6 cells, as visualized with the confocal Carl Zeiss LSM510 microscope (see “Immunofluorescence microscopy” for detailed image acquisition information). Colors as described in panel G. In panels A,C-E, representative blots of 3 independent experiments are shown.
HGAL interacts with actin and myosin II proteins, and the interaction with the myosin is increased by IL-6–induced HGAL phosphorylation. (A) Cellular lysates of unstimulated HeLa-HGAL and SUDHL6 cells were immunoprecipitated with either anti-HGAL or anti-actin antibodies and blotted with the indicated antibodies. Unconjugated beads served as a control (“B”). The size of HGAL in the HeLa cells is 26 kDa, since the cloned protein is a fusion protein with V5 tag. (B) Cellular lysates were extracted from unstimulated and IL-6–stimulated SUDHL6 cells, immunoprecipitated with anti-HGAL antibody, resolved by SDS-PAGE, and stained with Coomassie blue. Protein bands of 250 kDa, 230 kDa, and 140 kDa (→) were excised and analyzed by mass spectrometry analysis and sequencing that revealed that all the 3 proteins represent myosin II, the expected size of which is 250 kDa. (C) Cellular lysates were extracted from unstimulated SUDHL6 and VAL lymphoma cell lines, immunoprecipitated with anti-HGAL antibody, and blotted with indicated antibodies. Unconjugated beads served as a control (“B”). (D) Cellular lysates were extracted from unstimulated or IL-6–stimulated SUDHL6 cells and immunoprecipitated with either anti-HGAL or antimyosin antibodies, resolved on the SDS-PAGE, and blotted with the indicated antibodies. (E) HeLa-HGAL cells were treated with cytochalasin D (5 μM) and latrunculin B (5 μM) for 30 minutes, respectively, to inhibit actin polymerization and disrupt actin filaments. The whole-cell lysates were extracted, immunoprecipitated with anti-HGAL antibody, and blotted with the indicated antibodies. (F) SUDHL6 and HeLa-HGAL cells were stimulated with IL-6 for up to 60 minutes. At the indicated time points, cellular lysates were extracted, immunoprecipitated with anti-HGAL antibody, and blotted with the indicated antibodies. Arrows indicate immunoglobulin light chains. (G) Unstimulated or IL-6–stimulated Hela-HGAL (left) and SUDHL6 (right) cells were fixed with 4% paraformaldehyde, stained with anti-HGAL antibody (green), antimyosin antibody (red), and DAPI (blue), and visualized with the Carl Zeiss LSM510 microscope. (H) HGAL and myosin II colocalization in podosome-like structures of IL-6–stimulated SUDHL6 cells, as visualized with the confocal Carl Zeiss LSM510 microscope (see “Immunofluorescence microscopy” for detailed image acquisition information). Colors as described in panel G. In panels A,C-E, representative blots of 3 independent experiments are shown.
We next examined whether HGAL phosphorylation affects its interaction with the actin and WASP proteins. Cellular lysates prepared from SUDHL6 cells either stimulated with IL-6 or incubated with pervanadate were used for coimmunoprecipitation experiments. HGAL phosphorylation by either IL-6 or by pervanadate did not affect the extent of its interaction with the actin protein (data not shown). Further, even upon HGAL phosphorylation, no physical interaction between HGAL and WASP was observed (data not shown).
To further interrogate HGAL protein interaction partners in unstimulated and IL-6–stimulated cells, HGAL was immunoprecipitated from SUDHL6 cell lysates (Figure 3B). IL-6 stimulation for 5 and 10 minutes markedly increased HGAL interaction with proteins approximately 250 kDa, 230 kDa, and 140 kDa in size (Figure 3B). Mass spectrometry analysis and sequencing revealed that the largest protein represents the myosin II heavy chain. Noticeably, the 2 smaller proteins of 230 kDa and 140 kDa also represented myosin II heavy chain, presumably formed by proteolytic cleavage of the full-length protein.
To further confirm the physical interaction between HGAL and myosin II, coimmunoprecipitation experiments and immunofluorescence studies were performed in unstimulated and IL-6–stimulated SUDHL6 and VAL cells (Figure 3C,D). HGAL immunoprecipitates contained myosin II protein, confirming the physical interaction between HGAL and myosin II (Figure 3C). However, repeated immunoprecipitation experiments of myosin II protein in different cells failed to detect coimmunoprecipitated HGAL protein (Figure 3D). Since actin and myosin II interact with each other, it is possible that HGAL interaction with myosin II or actin are indirect and are mediated by actin or myosin, respectively. To rule out the possibility that interaction between HGAL and myosin II is mediated by actin, and to confirm a direct interaction between these proteins, coimmunoprecipitation experiments were repeated following treatment of cells with cytochalasin D or latrunculin B, which inhibit actin polymerization and disrupt microfilament organization. As is shown in Figure 3E, HGAL interacted with myosin II even in the absence of actin polymers. To find out whether HGAL also directly interacts with actin needs further confirmation studies.
IL-6 stimulation of SUDHL6 and HeLa-HGAL cells for 5 to 60 minutes led to transient increase in physical interaction between HGAL and myosin II, as demonstrated by increased quantities of myosin II protein coimmunoprecipitated with HGAL. Noticeably, the time course of IL-6 induced increased interaction between HGAL and myosin II proteins varied slightly between different cell lines (Figure 3F). Since a portion of cellular myosin II protein is insoluble, the observed results could stem from changes in myosin solubility and not only from IL-6–induced interaction between the HGAL and myosin II proteins. To rule out this possibility, we examined the levels of myosin II in the soluble whole-cell lysates and in the insoluble cell pellets before and after IL-6 stimulation. No significant changes in the myosin II levels in the soluble and insoluble cell fractions were observed upon cell stimulation with IL-6 (data not shown).
Myosin II protein exists in 2 isoforms: myosin IIA and IIB. Coimmunoprecipiatation studies demonstrated that HGAL interacted with both isoforms (data not shown). Immunofluorescence and confocal studies further demonstrated HGAL protein colocalization with both isoforms of myosin II in unstimulated HeLa-HGAL and SUDHL6 cells. IL-6 stimulation led to colocalization of HGAL and myosin II proteins in podosomes, similar to what we observed for actin protein (Figure 3G,H; data not shown).
IL-6–induced phosphorylation of the HGAL C-terminal tyrosine residue mediates increased HGAL interaction with myosin II protein
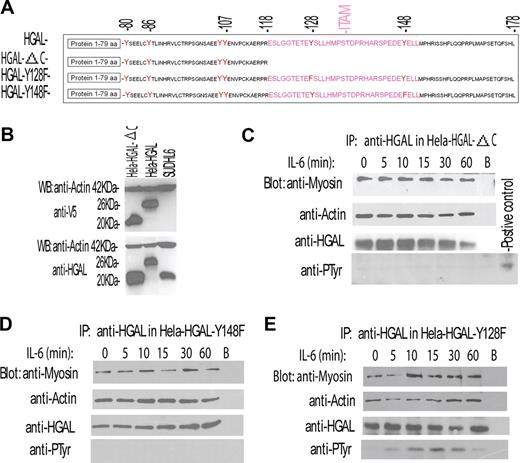
These results demonstrated that IL-6 cytokine induces both HGAL protein tyrosine phosphorylation and increases its interaction with myosin II. Taken together, these results suggested that the former might mediate the latter. HGAL protein contains 6 tyrosine residues, including the 2 C-terminal tyrosines of the ITAM. To determinate which of the HGAL tyrosine residues are phosphorylated upon IL-6 stimulation and to assess whether tyrosine phosphorylation mediates increased interaction with myosin II, we generated 3 HGAL mutant constructs (Figure 4A). The first construct, HGAL-ΔC, encodes a truncated HGAL protein (amino acids 1-118) in which the 2 C-terminal ITAM tyrosine residues are deleted. This mutant protein could be expressed, and was detected by either our monoclonal anti-HGAL or anti-V5 tag antibodies (Figure 4B). The remaining 2 HGAL mutant constructs, HGAL-Y128F and HGAL-Y148F, encode a full-length HGAL protein in which the C-terminal ITAM tyrosine residues 128 and 148 were substituted by phenylalanine.
IL-6 induces phosphorylation of the C-terminal tyrosine residue of the HGAL protein that mediates increased interaction with myosin protein. (A) Schematic representation of HGAL protein, its deletion mutant missing the C-terminal ITAM (HGAL-ΔC), and substitution mutants in which C-terminal tyrosine residues of the ITAM were substituted with phenylalanine (HGAL-Y128F and HGAL-Y148F). (B) Mutant missing the C-terminal ITAM (HGAL-ΔC) can be expressed in the cellular context. Whole-cell lysates were extracted from SUDHL6, HeLa-HGAL, and HeLa cells transiently transfected with the HGAL-ΔC vector and blotted with the indicated antibodies. (C-E) HeLa cells, transiently transfected with HGAL-ΔC (C), HGAL-Y148F (D), or HGAL-Y128F (E) vectors, were stimulated with IL-6 (10 ng/mL) for up to 60 minutes. Cellular lysates were extracted at the specified time points, immunoprecipitated with anti-HGAL antibody, resolved on SDS-PAGE, and blotted with the indicated antibodies. In panels C-E, representative blots of 3 independent experiments are shown.
IL-6 induces phosphorylation of the C-terminal tyrosine residue of the HGAL protein that mediates increased interaction with myosin protein. (A) Schematic representation of HGAL protein, its deletion mutant missing the C-terminal ITAM (HGAL-ΔC), and substitution mutants in which C-terminal tyrosine residues of the ITAM were substituted with phenylalanine (HGAL-Y128F and HGAL-Y148F). (B) Mutant missing the C-terminal ITAM (HGAL-ΔC) can be expressed in the cellular context. Whole-cell lysates were extracted from SUDHL6, HeLa-HGAL, and HeLa cells transiently transfected with the HGAL-ΔC vector and blotted with the indicated antibodies. (C-E) HeLa cells, transiently transfected with HGAL-ΔC (C), HGAL-Y148F (D), or HGAL-Y128F (E) vectors, were stimulated with IL-6 (10 ng/mL) for up to 60 minutes. Cellular lysates were extracted at the specified time points, immunoprecipitated with anti-HGAL antibody, resolved on SDS-PAGE, and blotted with the indicated antibodies. In panels C-E, representative blots of 3 independent experiments are shown.
HeLa cells transiently transfected with the 3 mutant constructs were stimulated with IL-6. HGAL protein was immunoprecipitated and immunoblotted with antibodies for myosin II, actin, and phosphotyrosine (Figure 4C-E). Truncation of the C-terminal portion of HGAL protein containing the ITAM abrogated IL-6–induced HGAL tyrosine phosphorylation (Figure 4C). Further, although the truncated protein could physically interact with both actin and myosin II in unstimulated cells, IL-6 stimulation failed to increase HGAL–myosin II interaction (Figure 4C), in contrast to what was observed with the wild-type HGAL protein (Figure 3F,G). Similarly, substitution of the C-terminal 148 tyrosine residue by phenylalanine did not abrogate HGAL interaction with actin and myosin II proteins in unstimulated cells, but prevented IL-6–induced HGAL tyrosine phsphorylation and precluded an increase in the HGAL–myosin II interaction (Figure 4D). In contrast, the HGAL-Y128F mutant that contains an intact C-terminal tyrosine residue at position 148 was tyrosine phosphorylated upon IL-6 stimulation and exhibited increased interaction with myosin II (Figure 4E), similar to what was observed with the wild-type HGAL protein. All the HGAL mutants relocalized to the podosomes upon IL-6 stimulation (data not shown). Overall, our results indicate that the N-terminal portion of HGAL (amino acids 1-118) is sufficient for the interaction with actin and myosin in unstimulated cells. Further, our data demonstrate that IL-6 induces HGAL phosphorylation at the C-terminal tyrosine residue at position 148, and that the latter is necessary for increased physical interaction between HGAL and myosin II proteins, but not for HGAL relocalization to podosomes. It is possible that IL-6 induces additional alterations in the N-terminal portion of HGAL that mediate its relocalization irrespective of its phosphorylation status. Structural analysis of the N-terminal portion of HGAL reveals presence of putative myristoylation (1MGNS) and palmytoylation (43CFC) sites. Indeed, in the cellular context HGAL protein can be myristoylated (our unpublished observations, February 2007) and palmitoylated, as recently shown by Pan et al17 while our manuscript was under review. Whether relocalization of the HGAL protein to the podosome-like structures upon IL-6 stimulation is mediated by the effects of the cytokine on HGAL myristoylation and palmitoylation is under investigation.
Lyn kinase psosphorylates HGAL protein
Our results demonstrate that IL-6 induces phosphorylation of HGAL C-terminal tyrosine residue that mediates increased physical interaction with the myosin II protein. Classically, IL-6 activates at least 2 major intracellular pathways, the Janus kinase (JAK)/signaling transducer and activation of transcription (STAT) pathway and the Ras/mitogen-activated protein kinase (MAPK) signaling cascade.18 However, it was also reported that IL-6 may activate the Src family protein tyrosine kinase (SFK) Lyn.19 Noticeably, Lyn is the predominant SFK in B cells that was previously reported to phosphorylate ITAM tyrosines in protein substrates, to colocalize with actin filaments, and to be involved in F-actin filament polymerization.20,–22 Based on this, we hypothesized that Lyn might mediate IL-6–induced HGAL tyrosine phosphorylation. To address this possibility, we initially confirmed Lyn phosphorylation upon IL-6 stimulation in HeLa-HGAL and SUDHL6 lymphoma cell lines (Figure 5A). Next, we examined the effects of Lyn knockdown on IL-6–induced HGAL phosphorylation. siRNA-induced decrease in Lyn expression in the VAL (Figure 5B) and HeLa-HGAL cells (data not shown) was associated with loss of the IL-6–induced HGAL phosphorylation. Concordantly, expression of the constitutively active Lyn Y508F mutant increased HGAL phosphorylation (Figure 5C). These results indicate that Lyn activation mediates IL-6–induced HGAL phosphorylation.
Lyn kinase mediates IL-6–induced tyrosine phosphorylation of the HGAL protein. (A) VAL and HeLa-HGAL cells were treated with IL-6 for up to 15 minutes. The whole-cell lysates were extracted, separated by SDS-PAGE, and detected with the indicated antibodies. (B) VAL cells were transfected with either siRNA for Lyn or scrambled control, respectively. At 48 hours after transfection, the cells were stimulated with IL-6 (10 ng/mL) for 10 minutes and cellular lysates were extracted, immunoprecipitated with anti-HGAL antibody, separated by SDS-PAGE, and immunoblotted with antiphosphotyrosine or anti-HGAL antibodies. (C) HeLa-HGAL cells were transiently transfected with plasmids encoding for Lyn-Y508F and Lyn-K275D. At 48 hours after ransfection, whole-cell lysates were prepared, immunoprecipitated with anti-HGAL antibody, separated by SDS-PAGE, and immunoblotted with antiphosphotyrosine or anti-HGAL antibodies. Unstimulated and IL-6–stimulated HeLa-HGAL cells served as controls. In panels A-C, representative blots of 2 independent experiments are shown.
Lyn kinase mediates IL-6–induced tyrosine phosphorylation of the HGAL protein. (A) VAL and HeLa-HGAL cells were treated with IL-6 for up to 15 minutes. The whole-cell lysates were extracted, separated by SDS-PAGE, and detected with the indicated antibodies. (B) VAL cells were transfected with either siRNA for Lyn or scrambled control, respectively. At 48 hours after transfection, the cells were stimulated with IL-6 (10 ng/mL) for 10 minutes and cellular lysates were extracted, immunoprecipitated with anti-HGAL antibody, separated by SDS-PAGE, and immunoblotted with antiphosphotyrosine or anti-HGAL antibodies. (C) HeLa-HGAL cells were transiently transfected with plasmids encoding for Lyn-Y508F and Lyn-K275D. At 48 hours after ransfection, whole-cell lysates were prepared, immunoprecipitated with anti-HGAL antibody, separated by SDS-PAGE, and immunoblotted with antiphosphotyrosine or anti-HGAL antibodies. Unstimulated and IL-6–stimulated HeLa-HGAL cells served as controls. In panels A-C, representative blots of 2 independent experiments are shown.
HGAL modulates IL-6 effects on cell migration
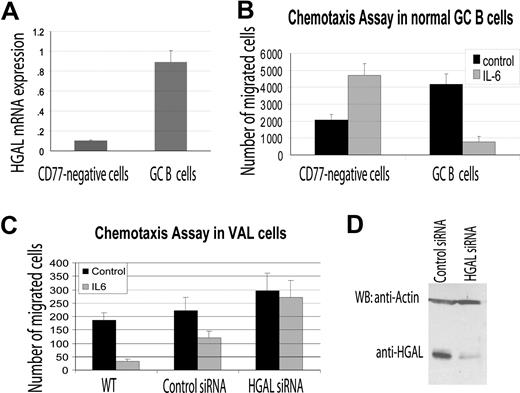
Podosomes are known to be involved in cell migration. Previous studies demonstrated that depending on the cellular context, IL-6 may either induce or inhibit cell chemotactic migration.18,23 Since our results demonstrated that IL-6 did induce HGAL phosphorylation and relocalization to podosome-like structures, and that HGAL physically interacts with the myosin II protein, we hypothesized that HGAL mediates IL-6 effects on cell migration. To examine this possibility, we isolated HGAL expressing CD77+ GC B cells from reactive human tonsils (Figure 6A) and used them in IL-6 chemotaxis assays on fibronectin-coated surfaces. Under these conditions, IL-6 significantly inhibited migration of the CD77+ GC B cells. In contrast, IL-6 increased the migration of the CD77− fraction of tonsilar cells that do not express high levels of HGAL (Figure 6A,B). Similarly, IL-6 inhibited the migration of HGAL-expressing VAL and Ramos lymphoma cells. When HGAL protein expression was knocked down with siRNA, the inhibitory effect of the IL-6 on the VAL and Ramos lymphoma cell migration was reversed (Figure 6C,D; data not shown).
HGAL mediates the inhibitory effects of IL-6 on cell migration. (A) CD77+ GC B cells and CD77− cells were enriched from normal tonsils as described in “RNA isolation and reverse transcription reaction.” RNA was extracted and HGAL RNA expression was measured by real-time reverse transcription–PCR in triplicates as described in “Real-time PCR measurement of HGAL mRNA expression.” (B) Enriched CD77+ GC B cells and CD77− tonsilar cells were evaluated in triplicate in IL-6 chemotaxis assay, as described in “Materials and methods.” Means and standard deviations of 2 independent experiments are demonstrated. (C) VAL lymphoma cells were transfected with control or HGAL siRNA. At 48 hours after siRNA transfection, the cells were used for IL-6 chemotaxis assay performed in triplicate, as described in “Chemotaxis and wound assays.” Means and standard deviations of 3 independent experiments are demonstrated. (D) Western blot analysis of HGAL protein in siRNA-transfected VAL lymphoma cells used for chemotaxis assays shown in panel C.
HGAL mediates the inhibitory effects of IL-6 on cell migration. (A) CD77+ GC B cells and CD77− cells were enriched from normal tonsils as described in “RNA isolation and reverse transcription reaction.” RNA was extracted and HGAL RNA expression was measured by real-time reverse transcription–PCR in triplicates as described in “Real-time PCR measurement of HGAL mRNA expression.” (B) Enriched CD77+ GC B cells and CD77− tonsilar cells were evaluated in triplicate in IL-6 chemotaxis assay, as described in “Materials and methods.” Means and standard deviations of 2 independent experiments are demonstrated. (C) VAL lymphoma cells were transfected with control or HGAL siRNA. At 48 hours after siRNA transfection, the cells were used for IL-6 chemotaxis assay performed in triplicate, as described in “Chemotaxis and wound assays.” Means and standard deviations of 3 independent experiments are demonstrated. (D) Western blot analysis of HGAL protein in siRNA-transfected VAL lymphoma cells used for chemotaxis assays shown in panel C.
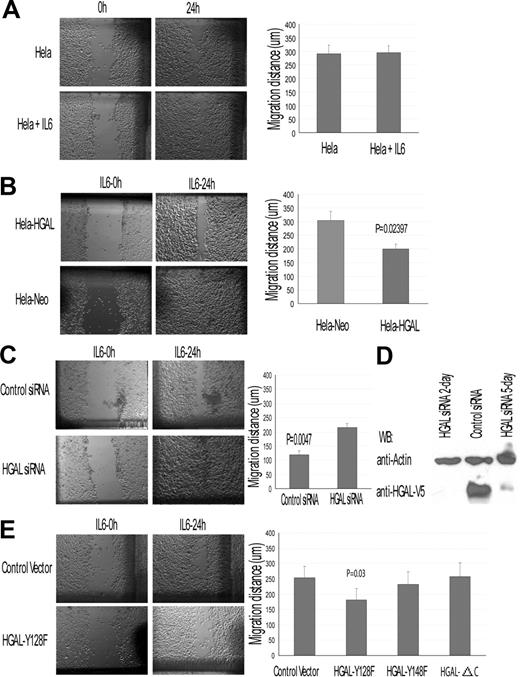
To further evaluate the effect of HGAL on IL-6–induced cell migration, we used HeLa-HGAL cells in wound assays. In HeLa (Figure 7A) and control HeLa-Neo cells (data not shown) IL-6 had no marked effect on cell migration. In contrast, HGAL expression in HeLa cells decreased their migration in response to IL-6 (Figure 7B). This result could not be attributed to IL-6 effects on cell proliferation (data not shown). siRNA knock-down of the HGAL protein expression in the HeLa-HGAL cells markedly reversed the inhibitory effects of IL-6 on cell migration (Figure 7C,D). Further, IL-6 did not affect the migration of HeLa cells expressing the HGAL-ΔC or HGAL-Y148F mutants, but did inhibit the migration of cells expressing the HGAL-Y128F mutant (Figure 7E). Overall, our results indicate that HGAL protein mediates the inhibitory effects of IL-6 on cell chemotaxis and migration. Further, our data suggest that HGAL phopshorylation at its tyrosine residue 148 is necessary to mediate the migration-inhibitory effects of IL-6.
HGAL modulates IL-6 effects on cell migration: wound assay in HeLa-HGAL cells. Wound assays were performed as described in “Chemotaxis and wound assays.” (A) IL-6 effect on HeLa cell migration in wound assay. (B) IL-6 effect on migration of HeLa-HGAL and HeLA-Neo cells. HGAL expression significantly decreased migration of the cells. (C) siRNA knock-down of HGAL in the HeLa-HGAL cells markedly reversed the HGAL-mediated, IL-6–induced inhibition of cell migration. (D) Western blot analysis of HGAL protein expression in HeLa-HGAL cells following siRNA knockdown. (E) IL-6 effect on migration of HeLa-HGAL mutants. Left panel exhibits results of a representative wound assay in HeLa-HGAL Y128F and control cells. Right panel summarizes the effect of IL-6 on migration of HeLa-HGAL-Y128F, HGAL-128F, and HGAL-ΔC mutants. P value represents difference between HeLa-HGAL Y128F and control cells. Means and standard deviations of 3 independent experiments are demonstrated. See “Immunofluorescence microscopy” for image acquisition information.
HGAL modulates IL-6 effects on cell migration: wound assay in HeLa-HGAL cells. Wound assays were performed as described in “Chemotaxis and wound assays.” (A) IL-6 effect on HeLa cell migration in wound assay. (B) IL-6 effect on migration of HeLa-HGAL and HeLA-Neo cells. HGAL expression significantly decreased migration of the cells. (C) siRNA knock-down of HGAL in the HeLa-HGAL cells markedly reversed the HGAL-mediated, IL-6–induced inhibition of cell migration. (D) Western blot analysis of HGAL protein expression in HeLa-HGAL cells following siRNA knockdown. (E) IL-6 effect on migration of HeLa-HGAL mutants. Left panel exhibits results of a representative wound assay in HeLa-HGAL Y128F and control cells. Right panel summarizes the effect of IL-6 on migration of HeLa-HGAL-Y128F, HGAL-128F, and HGAL-ΔC mutants. P value represents difference between HeLa-HGAL Y128F and control cells. Means and standard deviations of 3 independent experiments are demonstrated. See “Immunofluorescence microscopy” for image acquisition information.
Discussion
In response to antigen encounter, uncommitted naive B cells are activated and undergo a complex maturational process yielding phenotypically distinct subpopulations that form highly organized GCs. Within the GC, B cells undergo high rate proliferation and affinity maturation, are selected by antigen, switch toward advanced isotypes, and finally differentiate into either memory B cells or plasma cells. Recent studies demonstrated that antigen-specific GC B cells are restricted to the GCs, where they move with lower velocity and greater confinement compared with non-GC B cells.24 The mechanism underlying the altered motility of the GC B cells is unknown. However, it is well established that the maturation process leading to the generation of GC lymphocytes is characterized by tightly regulated suppression or increased expression of specific genes, resulting in a distinctive “GC gene expression signature.”25 The genes comprising the “GC expression signature” mediate the unique processes taking place in the GC microenvironment. Since several of the processes are exclusive to GC reaction, it is not surprising that expression of some of the genes are restricted to the GC lymphocytes.
HGAL is 1 of these GC-specific genes, the expression of which is correlated with better survival of patients with DLBCL and cHL, but the function of which is presently unknown.1,3,4 The mouse homolog of HGAL, M17 protein, was shown to be dispensable for GC formation, immunoglobulin somatic hypermutation, and efficient class-switch recombination.3 In the present study, we demonstrate that HGAL plays a role in modulating cell motility. We showed that HGAL functions as an adaptor binding actin and myosin II proteins, and demonstrated that its N-terminal portion is sufficient for these interactions. We report that IL-6 induces Lyn-mediated HGAL phosphorylation at its C-terminal tyrosine residue, leads to HGAL relocalization to podosomes, and increases its interaction with myosin II. Moreover, we demonstrate that HGAL mediates IL-6–inhibitory effects on cell migration. The latter may contribute to the recently in vivo demonstrated confinement of antigen-specific GC B cells to the GC microenvironment.24 Previously, IL-6 was shown to either stimulate or inhibit cell migration, depending on the cellular context.18,23 Cell lineage–specific HGAL expression may at least partially contribute to the reported variability in IL-6 effects on cell migration. Mice rendered deficient for IL-6 exhibited markedly smaller GCs with decreased numbers of GC B cells in response to T-cell–dependent antigens.26,27 Further, IL-6 production markedly changes during B lymphocyte maturation: while naive and memory B cells produce and secrete IL-6, GC lymphocytes do not.5 Interestingly, the expression of HGAL is complementary to the IL-6 production, since it is expressed at high levels only in GC lymphocytes, at intermediate to low levels in memory B cells, and not at all in the naive B cells. Similarly to the IL-6 knock-out animals, mice rendered deficient for M17 exhibited reduced-sized Peyer patches.3 Compilation of the previously reported data with the results presented herein suggests that HGAL up-regulation and IL-6 down-regulation in the GC lymphocytes are imperative for paracrine control of GC cell migration and localization by IL-6 that can be secreted by follicular dendritic cells, T cells, fibroblasts, and tangible body macrophages.5,27 Inhibition of GC lymphocyte migration by IL-6–induced HGAL phosphorylation may be necessary for confining B cells to GCs, thus allowing the completion of GC reaction. Indeed, in IL-6 knock-out mice, smaller GCs led to alterations in antigen-specific serum antibody titers.27 However, similar changes were not observed in M17 knock-out mice, suggesting that additional IL-6–induced intracellular mechanisms may be involved in the regulation of the antibody repertoire.
Although we demonstrate that IL-6–induced inhibition of GC B-cell migration is associated with increased binding of phosphorylated HGAL to myosin, but not to actin, the precise mechanism of the migration inhibition is presently unclear. According to the “swinging lever-arm” model for myosin force generation,28 small conformational changes in the myosin head are amplified by a lever-arm mechanism to a working stroke of several nanometers that leads to a directed actin-based movement. In the unstimulated cells, the N-terminal portion of HGAL binds to both actin and myosin II heavy chain. Upon IL-6 stimulation, HGAL is phosphorylated at its C-terminal tyrosine residue and increasingly binds to myosin II. It is possible that HGAL phosphorylation may change its conformation and transiently cross-link actin and myosin, thus preventing the directed sliding of the actin over myosin and inhibiting cell motility. Further studies are needed to confirm this model and to elucidate the mechanism underlying HGAL-mediated inhibition of cell migration.
Although our results reveal the first function of HGAL in GC lymphocytes that may contribute to confinement of the antigen-specific B cells to GCs,24 it is possible that this adaptor is involved in additional processes taking place in these cells. Further, the functions of HGAL protein in the malignant counterparts of GC lymphocytes are still unclear. We have demonstrated that high HGAL expression correlates with prolonged survival in patients with DLBCL and cHL. Whether the latter merely reflects the GC origin of these tumors, or that HGAL has a specific function that predisposes to improved outcome, is unknown. It is possible that HGAL-mediated inhibition of lymphocyte migration may contribute to the less-aggressive clinical behavior of HGAL-expressing tumors. In our initial observations, no correlation was detected between tumor stage and HGAL expression in DLBCL and cHL.1,4,6 Additional studies are needed to examine the function of HGAL in lymphomas. Furthermore, it is possible that persistent HGAL overexpression may predispose to lymphomagenesis by confining cells to the GC microenvironment and thus exposing them to high proliferation rates and active mutational machinery. Studies addressing these questions are presently under way in our laboratory.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by RO1 CA109335 from the United States Public Health Service–National Institutes of Health (I.S.L.) and the Dwoskin Family Foundation (I.S.L.). E.G.C. was supported by the fellowship of Fundación Alfonso Martin Escudero.
National Institutes of Health
Authorship
Contribution: X.L. performed research; J.C. performed research, analyzed data, and wrote the paper; R.M. performed research; E.C.G. performed research; D.M.H. designed the study and analyzed the data; and I.S.L. designed the study, performed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Izidore S. Lossos, Sylvester Comprehensive Cancer Center, Department of Hematology and Oncology, University of Miami, 1475 NW 12th Ave (D8-4), Miami, FL 33136; e-mail:ilossos@med.miami.edu.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal