Abstract
To date, there is no consensus regarding the influence of different CD45 isoforms during peripheral B-cell development. Examining correlations between surface CD45RO expression and various physiologic processes ongoing during the germinal center (GC) reaction, we hypothesized that GC B cells, like T cells, that up-regulate surface RO should progressively acquire phenotypes commonly associated with activated, differentiating lymphocytes. GC B cells (IgD−CD38+) were subdivided into 3 surface CD45RO fractions: RO−, RO+/−, and RO+. We show here that the average number of mutations per IgVH transcript increased in direct correlation with surface RO levels. Conjunctional use of RO and CD69 further delineated low/moderately and highly mutated fractions. Activation-induced cytidine deaminase (AID) mRNA was slightly reduced among RO+ GC B cells, suggesting that higher mutation averages are unlikely due to elevated somatic mutation activity. Instead, RO+ GC B cells were negative for Annexin V, comprised mostly (93%) of CD77− centrocytes, and were enriched for CD69+ cells. Collectively, RO+ GC B cells occupy what seems to be a specialized niche comprised mostly of centrocytes that may be in transition between activation states. These findings are among the first to sort GC B cells into populations enriched for live mutated cells solely using a single extracellular marker.
Introduction
CD45, also known as the common leukocyte antigen, is a protein tyrosine phosphatase that is present on the surface of all lymphoid lineage cells. Despite its nearly ubiquitous importance in proper cell activation, development, and differentiation,1-4 in the past 3 decades the characterization of CD45-dependent processes in lymphocytes has been confounded by at least 2 major observations. First, CD45 exists as several different isoforms, each with different, though not necessarily exclusive functions.4-8 The 2 major isoforms include the short, approximately 180-kDa CD45RO (RO) and the long, approximately 220-kDa CD45RA (RA) proteins, which are produced by alternative splicing of the single long RA pre-mRNA transcript.9-11 Second, the influence of a given isoform can vary between different lymphoid cell types. For example, transgenic CD45ROonly mice develop functional, mature peripheral T cells, but fail to restore their mature peripheral B cell pool.6,12,13 This differential effect in B and T cells prompted our investigation into the potential correlations between RO expression and key B-cell biosynthetic processes executed during development and differentiation, specifically during the germinal center (GC) reaction
The overwhelming majority of investigations regarding CD45 isoforms have focused on T cells. For example, the RA+RO− T-cell pool is enriched for resting naive T cells, whereas RA−RO+ T cells are primarily activated and are comprised of memory and other effector cells.14 Interestingly, the intermediate RA+/−RO+/− fraction is enriched for primed T cells that are in bidirectional transition between the resting RO− and activated RO+ stages.15-17 Collectively, these studies suggest that CD45 isoform expression patterns can be used to subdivide T cells into different subsets based both on their developmental stage (naive vs memory) and functional phenotypes such as activation, gene transcription, and protein synthesis. Unfortunately, correlations between B-cell development, differentiation, and RO expression remain largely undetermined, though some studies have previously evaluated CD45 isoform expression among B cells isolated from patients with leukemia or autoimmune diseases.18,19
Dawes et al20 and Tchilian et al21 have suggested that the quality and quantity of cellular activity can be significantly influenced by the level of CD45 isoform expression. RO has also been implicated in T-cell deactivation and may affect signaling thresholds.22,23 Factors that potentially modify B-cell receptor (BCR)–induced signaling and threshold levels are likely to affect B-cell activation, selection, development, and even the propensity for disease onset. CD45 has been implicated in each of these processes.18,23-26 It is therefore important to determine for B cells, as has been done for T cells, any reproducible correlations between CD45 isoform expression and functional phenotypes. This would facilitate the identification of potential CD45-related targets for preventing or reversing anomalous cellular responses leading to the onset of B-cell malignancies and autoimmunity.21,27
In the past 2 decades, our laboratory has extensively characterized events involved in human B-cell development, differentiation, diversification, and selection within secondary lymphoid tissues, especially tonsils.28-31 This work has been enhanced by our ability to subdivide B cells into major groups (ie, naive, GC, and memory) according to their differential expression of key surface markers. Taking advantage of the function/activation-based discriminatory power of CD45 isoforms (demonstrated in T cells), we set out to determine whether surface RO expression, in addition to our current strategies, would enable us to further subdivide core B-cell subsets into distinct fractions that are differentially engaged in 1 or more of the dynamic processes involved in B-cell differentiation. Our investigations focused on somatic hypermutation (SHM) for several reasons. First, CD45 regulates activation-induced cytidine deaminase (AID) expression,32 which is required for SHM (and class switch recombination [CSR]).33-36 In addition, SHM is unique to B cells and provides insight into the differential influence of RO between different lymphoid cell types. Finally, the number of mutations within an immunoglobulin (Ig) sequence is easily quantified and directly correlates with the incidence of, or length of time, a cell is exposed to the SHM machinery.
Potential correlations between RO expression and SHM were assessed by first sorting tonsil-derived GC B cells into 3 pools based on their surface RO expression (RO−, RO+/−, RO+). Each fraction was independently characterized by quantifying AID RNA transcript levels and the average number of mutations per VH4 family Ig transcript (as previously described in Jackson and Capra37 ). Results from each RO fraction were then compared. We observed that the number of mutations increased concordantly with RO expression. We therefore investigated whether the degree of other SHM-related processes, including cell activation and cell division, also varied with respect to RO expression. Potential correlations were quantified by staining RO-subdivided GC B cells with antibodies against CD69 (early activation marker, function uncertain),38,39 CD71 (activation marker; transferrin receptor),40 and Ki67 (marker of proliferation; stains all non-G0 stages of the cell cycle).29,41
After documenting that surface expression of the RO and RA isoforms correlated with cellular mRNA levels of the 2 major isoforms, we show here that RO expression is a powerful discriminator of the activation state of normal human B cells. Specifically, GC B cells with increased surface levels of the RO isoform were enriched for CD69 and contained the highest levels of somatic mutations in their heavy chain transcripts. These studies document that the addition of surface RO expression to the panel of reagents currently used to discriminate between human mature B-cell subsets provides new insights into B-cell differentiation pathways and should allow further purification of key developmental states of these critically important cells.
Materials and methods
Approval for this research was granted by the University of Oklahoma Health Science Center's Institutional Review Board.
Lymphocyte isolation and cytometric gating
Lymphocytes were isolated from normal children's tonsils (1-12 years) and prepared for cytometric analysis as previously described.37 Peripheral blood was from healthy adults. All protocols were Institutional Review Board approved. Antibodies against CD19 and CD4 were used to distinguish between B (CD19+CD4−) and T (CD19−CD4+) cells. Doublet discrimination (forward scatter [FSC] width (W) vs FSC area (A) and side scatter [SSC]–W vs SSC-A) was applied to all analyses. B-cell subsets were stained using antibodies listed in Table 1. Anti-IgD and anti-CD38 were used to identify GC (IgD−CD38+) B cells (Figure 1A). The tonsillar IgD+CD38− B-cell pool is enriched for naive B cells, which were ultimately identified by additional staining with anti-IgM and anti-CD27 (IgM+IgD+CD38−CD27−). Incubations with conjugated antibodies were for 20 minutes at 4°C in the dark. Intracellular staining with Ki67 was performed by fixing cells with Fix & Perm Cell Permeabilization Kit (Invitrogen, Carlsbad, CA). Stained cells were analyzed and sorted using the LSR II (BD Biosciences, San Diego, CA), FACSAria (BD Biosciences), or MoFlo (Dako Cytomation, Glostrup, Denmark) cytometers.
List of antibodies (cytometry)
| Antibody . | Fluorophore . | Manufacturer . |
|---|---|---|
| CD4 | FITC | Caltag (Burlingame, CA) |
| CD19 | PE-Cy7 | Caltag |
| CD27 | APC-Alexa Fluor 750 | Caltag |
| CD38 | APC Cy5.5 | Caltag |
| CD45RA | PE-Cy5.5 | Caltag |
| CD45RO | PE, TC | Caltag |
| CD69 | TC | Caltag |
| CD71 | FITC, TC | Caltag |
| Streptavidin | Red613 | Caltag |
| Ki67 | PE | Caltag |
| IgM | APC | Southern Biotech (Birmingham AL) |
| IgD | Biotin | BD Pharmingen (San Diego, CA) |
| Antibody . | Fluorophore . | Manufacturer . |
|---|---|---|
| CD4 | FITC | Caltag (Burlingame, CA) |
| CD19 | PE-Cy7 | Caltag |
| CD27 | APC-Alexa Fluor 750 | Caltag |
| CD38 | APC Cy5.5 | Caltag |
| CD45RA | PE-Cy5.5 | Caltag |
| CD45RO | PE, TC | Caltag |
| CD69 | TC | Caltag |
| CD71 | FITC, TC | Caltag |
| Streptavidin | Red613 | Caltag |
| Ki67 | PE | Caltag |
| IgM | APC | Southern Biotech (Birmingham AL) |
| IgD | Biotin | BD Pharmingen (San Diego, CA) |
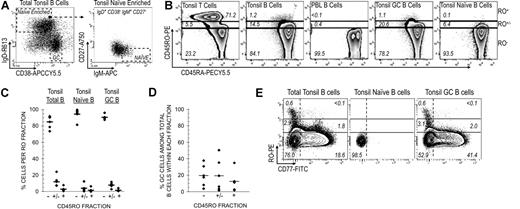
Cytometric identification of B and T cells. (A) GC B cells were identified from among CD19+ total tonsillar B cells using IgD and CD38 as previously described.39 Naive B cells were isolated by first gating on IgD+CD38− cells (naive-enriched) followed by IgM+ and CD27− gating (IgM+IgD+CD38−CD27−). (B) Tonsillar T and B cells, PBL B cells, and tonsillar GC and naive B cells were stained with RO and RA antibodies. T-cell RO expression levels were used to subdivide B cells into RO−, RO+/−, and RO+ fractions. Numbers represent the percentage of cells within each RO fraction. (C) RO expression levels were determined for tonsillar total B, GC, and naive B-cell pools. Results from 5 tonsils are presented, including the average value data point (horizontal crossbar). Percentages represent the distribution of each subset across each RO fraction. (D) The percentage of GC B cells among the total number of B cells within a given RO fraction. Equivalent data were collected from 5 tonsils. For example, GC B cells account for only 12.6% of all RO+ tonsillar B cells. (E) Tonsillar GC B cells were stained with the addition of anti-CD77. The correlation between RO and CD77 expression was quantified among gated tonsillar total, naive, and GC B cells. Note that most naive B cells are double-negative for both markers.
Cytometric identification of B and T cells. (A) GC B cells were identified from among CD19+ total tonsillar B cells using IgD and CD38 as previously described.39 Naive B cells were isolated by first gating on IgD+CD38− cells (naive-enriched) followed by IgM+ and CD27− gating (IgM+IgD+CD38−CD27−). (B) Tonsillar T and B cells, PBL B cells, and tonsillar GC and naive B cells were stained with RO and RA antibodies. T-cell RO expression levels were used to subdivide B cells into RO−, RO+/−, and RO+ fractions. Numbers represent the percentage of cells within each RO fraction. (C) RO expression levels were determined for tonsillar total B, GC, and naive B-cell pools. Results from 5 tonsils are presented, including the average value data point (horizontal crossbar). Percentages represent the distribution of each subset across each RO fraction. (D) The percentage of GC B cells among the total number of B cells within a given RO fraction. Equivalent data were collected from 5 tonsils. For example, GC B cells account for only 12.6% of all RO+ tonsillar B cells. (E) Tonsillar GC B cells were stained with the addition of anti-CD77. The correlation between RO and CD77 expression was quantified among gated tonsillar total, naive, and GC B cells. Note that most naive B cells are double-negative for both markers.
Lymphocyte populations were simultaneously stained with antibodies against CD45RA and CD45RO. Surface RO expression was subsequently correlated with 1 or more additional markers, including CD69, CD71, Ki67, and Annexin V (AnV). B and T cells can express RO at similar fluorescent intensities (Figure 1B). To more accurately compare results between patients and account for potential intersample variations when drawing cytometric gates, we developed a standardized gating strategy, “TB45,” that uses T-cell CD45 isoform expression levels to assign equivalent gates among B cells. Depicted in Figure 1B, T-cell RO−, RO+/−, and RO+ gating limits were applied to B cells from the same tonsil (peripheral blood lymphocyte [PBL] T cells were used for blood samples). Using this system, we reproducibly analyzed surface expression and other ex vivo phenotypes on equivalent subsets of heterogeneous lymphocyte pools isolated from multiple individuals.
RNA isolation and RT-PCR
Total RNA was isolated from sorted cells using the RNAqueous kit (Ambion, Austin, TX) according to the manufacturer's protocol. VH4 IgM and IgG, CD45, and AID RNA transcripts were amplified using One Step RT-PCR (Qiagen, Valencia, CA). Amplification parameters were performed as previously described.37,42 Primers are listed in Table 2. β-actin was used to normalize RNA levels between samples for semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) amplifications. PCR products were gel purified using Qiaquick (Qiagen), ligated into the pDrive vector (Qiagen PCR Cloning Kit), and used to transform Escherichia coli bacteria grown on agar plates containing kanamycin. Plasmid DNA was isolated and purified using the Qiagen QIAprep Spin Miniprep Kit and submitted for DNA sequencing (ABI 3730 DNA sequencer; Applied Biosystems, Foster City, CA).
Primers for RT-PCR amplification
| Gene . | Forward . | Reverse . |
|---|---|---|
| VH4 IgM | 5′-ATGAAACACCTGTGGTTCTT-3′ | 5′-CACGTTCTTTTCTTTGTTGC-3′ |
| VH4 IgG | 5′-ATGAAACACCTGTGGTTCTT-3′ | 5′-AGTAGTCCTTGACCAGGCAGCCCAG-3′ |
| β-actin | 5′-ATCCACACGGAGTACTTGCGCTCA-3′ | 5′-GTACCACTGGCATCGTGATGGACT-3′ |
| CD45* | 5¢-GGCAAAGCCCAACACCTT-3′ | 5′-TGTGGTTGAAATGACAGCG-3′ |
| AID† | 5′-ACTTGCAGGGAGGCAAGAAGACACTCT-3′ | 5′-CAAAAGGATGCGCCGAAGCTGTCTGGAG-3′ |
| Gene . | Forward . | Reverse . |
|---|---|---|
| VH4 IgM | 5′-ATGAAACACCTGTGGTTCTT-3′ | 5′-CACGTTCTTTTCTTTGTTGC-3′ |
| VH4 IgG | 5′-ATGAAACACCTGTGGTTCTT-3′ | 5′-AGTAGTCCTTGACCAGGCAGCCCAG-3′ |
| β-actin | 5′-ATCCACACGGAGTACTTGCGCTCA-3′ | 5′-GTACCACTGGCATCGTGATGGACT-3′ |
| CD45* | 5¢-GGCAAAGCCCAACACCTT-3′ | 5′-TGTGGTTGAAATGACAGCG-3′ |
| AID† | 5′-ACTTGCAGGGAGGCAAGAAGACACTCT-3′ | 5′-CAAAAGGATGCGCCGAAGCTGTCTGGAG-3′ |
DNA sequence and statistical analysis
A total of 547 IgVH4 family IgM and IgG transcripts were amplified and sequenced from RO-subdivided GC B cells. VH4 family gene segment use was analyzed using IgBlast44 and IMGT/V-QUEST (http://imgt.cines.fr:8104)45 databases. Sequence comparisons and alignments were performed using VectorNTI (Invitrogen, Carlsbad, CA). Statistical analyses including 2-tailed heteroscedastic T-tests were performed using Microsoft Excel for Windows (Redmond, WA).
Results
“TB45” gating strategy standardizes CD45 isoform expression levels among lymphocytes
In this report, the accuracy of quantifying RO-associated physiologic changes rests inextricably on the ability to reproducibly subdivide B cells according to their CD45 isoform expression level(s). T cells (both tonsillar and peripheral blood) have clear cytometrically defined RO− and RO+ fractions (RO+/− by noninclusion; Figure 1A). However, accurate assignment of RO gates for B cells is complicated by their spectral or bell-curve histogram pattern. We therefore standardized the assessment of B-cell–surface RO levels by assigning RO− and RO+ levels in accordance with those observed among T cells from the same tissue source. This provides a reference point enabling reliable and consistent RO quantification, even between samples collected from different patients. Described in detail here (“Materials and methods”), we call this strategy the “TB45” system, which can be used for all CD45 isoforms.
Only a small fraction of tonsillar B cells express surface RO
We examined total tonsillar cells for RO and RA isoforms and found that while more than 75% of T cells express modest (RO+/−) or substantial (RO+) amounts of RO, only about 15% of B cells were positive, and most of these were only modestly positive (approximately 10% RO+/−; Figure 1B,C). The RO+/− and RO+ tonsillar B-cell fractions appear to be constituted by a heterogeneous group of B-cell subsets, of which GC B cells account for only a small percentage (19% and 12%, respectively; Figure 1D). Therefore, high RO expression is not unique to GC B cells. Comparable percentages were observed in more than 30 tonsils analyzed. RO+ B cells were predominantly observed in tonsils in contrast to peripheral blood, where B cells were uniformly RO− (Figure 1B). There are both inter- and intrasubset differences in the maximal amount of surface RO expressed among mature B cells. For example, RO+ cells were readily identified among GC cells, but not among tonsillar naive B cells. In addition, most (93%) of all RO+ GC cells were CD77− centrocytes (Figure 1E).
A fraction of RO+/− and RO+ GC B cells concomitantly down-regulated surface RA in a pattern commonly associated with “transitioning” RA+/−RO+/− T cells (Figure 1B). However, it is uncertain whether increased RO expression among GC B cells correlates with differentiation (as in T cells) and receptor diversification processes, many of which initiate in response to activation-inducing stimuli and are therefore potentially influenced by CD45. We therefore investigated whether GC B cells that up-regulate surface RO exhibit phenotypes consistent with diversifying (SHM) and/or activated (CD69, CD71, and Ki67) B cells.
CD45RO mRNA levels directly correlate with surface RO in GC B cells
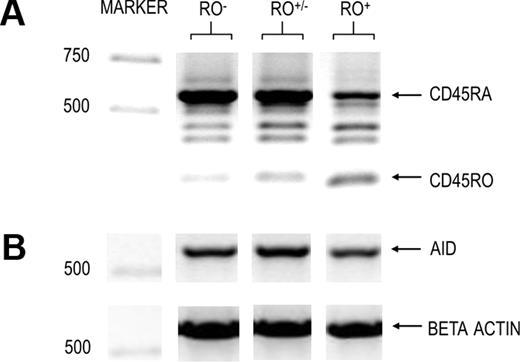
CD45RO is generated by alternative splicing of the long pre-mRNA RA transcript.9-11 Therefore, RO and RA transcript levels are inversely related. However, to determine whether increased surface RO protein levels correlated with changes at the mRNA level (increased RO and reduced RA transcripts) in the population under study, we performed semiquantitative RT-PCR analysis of CD45 isoform transcripts from sorted RO−, RO+/−, and RO+ GC B cells. Results are shown in Figure 2A. As expected, RO mRNA and surface protein levels increased in direct proportion to each other. For example, RO mRNA was highest among surface RO+ cells, RO+/− cells showed a slight increase in RO mRNA, and RO− cells showed only traces of RO mRNA. Again, as expected, RO+ cells showed a substantial decrease in RA mRNA, which parallels lower RA surface expression (Figure 1B). Finally, we found no decrease in RA mRNA in RO+/− GC B cells (relative to RO− cells) even though this fraction had a lower concentration of surface RA proteins (Figure 1B).
Semiquantitative RT-PCR of RO-subdivided GC B cells. IgD−CD38+ GC B cells were cytometrically sorted based on their surface RO levels as defined in Figure 1A,B. RNA was immediately isolated and used in semiquantitative RT-PCR using β-actin to normalize RNA levels. Amplified targets included (A) CD45 isoforms highlighting RA and RO and (B). AID molecular weight markers are indicated to the left.
Semiquantitative RT-PCR of RO-subdivided GC B cells. IgD−CD38+ GC B cells were cytometrically sorted based on their surface RO levels as defined in Figure 1A,B. RNA was immediately isolated and used in semiquantitative RT-PCR using β-actin to normalize RNA levels. Amplified targets included (A) CD45 isoforms highlighting RA and RO and (B). AID molecular weight markers are indicated to the left.
There is a direct correlation between RO expression and somatic hypermutation among GC B cells
The hallmark phenotype of GC B cells is their prodigious rate of cell division, during which cells diversify their antigen receptors (BCR) through AID-dependent SHM and CSR.33-36 To determine whether SHM was more likely to occur during a given RO stage, we first compared AID transcript levels between the various RO fractions. As shown in Figure 2B, AID transcripts were positively detected in each RO fraction.
The number of somatic mutations progressively accumulates over time as cells continue through cyclic stages of proliferation (incorporation of mutations) and nonproliferation (selection and eventual differentiation). Using the average number of mutations per sequence as a temporal gauge, we investigated whether surface RO expression could be used to enrich for GC B cells that are in early (germ-line or few mutations) or later (higher mutation averages) stages of the GC reaction. A total of 547 IgM and IgG transcripts were sequenced in order to get large enough sample sizes for a comprehensive analysis.
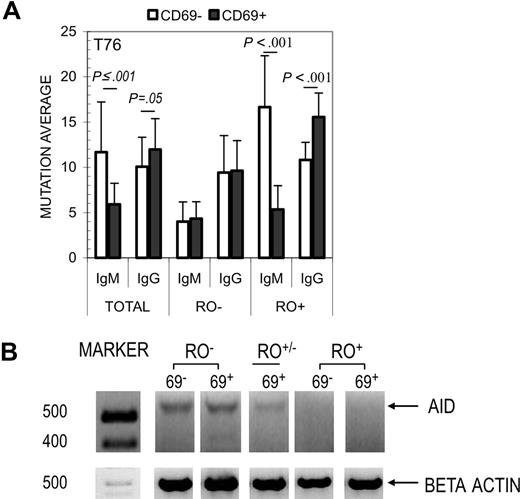
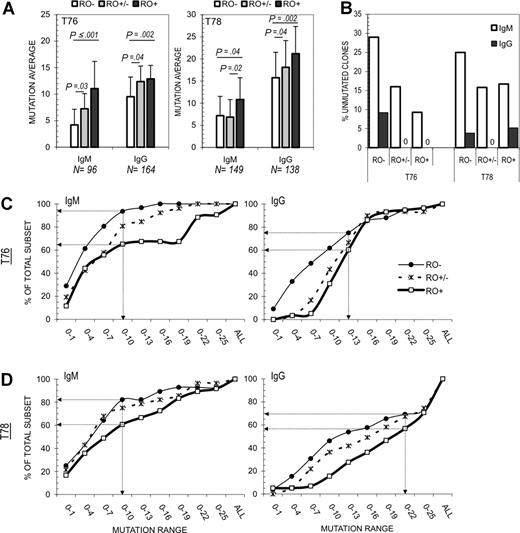
Results from 2 independent tonsils (T76 and T78) are shown in Figure 3A. For both tonsils, the average number of mutations per VH4 sequence increased in direct proportion to surface RO expression. The trend of increased mutations among RO+ GC B cells was repeatedly observed in both IgM and IgG sequence pools. For example, T76 averages increased from 4.2 to 11.4 (IgM) and 9.5 to 12.9 (IgG) for RO− versus RO+ cells, respectively. Statistical significance was high in each case, with P values less than .002 (except T78 IgM; P = .04). Mutation averages among RO+/− GC B cells were significantly different from RO− or RO+ values in 5 of 8 comparisons (ie, RO+/− vs RO− and RO+/− vs RO+ for IgM and IgG sequences from both tonsils). Interestingly, RO+/− mutation averages were closest to the averages for their respective total pools (ie, T76 total IgM, 7.9; RO+/− IgM, 7.3; total IgG, 11.2, and RO+/− IgG, 12.4).
Correlation between SHM and CD45RO expression levels. A total of 547 IgVH4 family IgM and IgG transcripts were amplified from RO-subdivided GC B cells. (A) The average number of mutations per VH4 sequence was calculated and compared for each RO fraction. Standard deviations are represented by error bars. P values are included where intersample differences were statistically significant (P ≤ .05). N values for RO− (□), RO+/− (▩), and RO+ (■) fractions: T76 IgM (31, 24, 41), T76 IgG (76, 30, 58), T78 IgM (28, 38, 83), and T78 IgG (26, 55, 57), respectively. Total IgM and IgG mutation averages were, respectively, 7.9 and 11.2 for T76 and 9.1 and 18.9 for T78. (B) The number of unmutated (germ-line) IgM (□) and IgG (■) sequences among each RO fraction was quantified for T76 and T78. (C-D) The cumulative distribution of mutations among IgM and IgG sequences was determined for RO− (thin line; ●), RO+/− (dashed line; ×), and RO+ (thick line; □) fractions. Percentages represent the number of samples that have mutations within the indicated range divided by the sample size n value (range increases in 3-bp increments). Arrows highlight the percentage difference between RO− and RO+ fractions at the RO+ average. For example, only 5% of T76 RO−IgM sequences have more than 10 mutations compared with 35% of RO−IgM sequences. Results are presented for (C) T76 and (D) T78. Note that RO+/− values are typically medial to RO− and RO+ values, suggesting that mutations progressively accumulate with respect to RO level.
Correlation between SHM and CD45RO expression levels. A total of 547 IgVH4 family IgM and IgG transcripts were amplified from RO-subdivided GC B cells. (A) The average number of mutations per VH4 sequence was calculated and compared for each RO fraction. Standard deviations are represented by error bars. P values are included where intersample differences were statistically significant (P ≤ .05). N values for RO− (□), RO+/− (▩), and RO+ (■) fractions: T76 IgM (31, 24, 41), T76 IgG (76, 30, 58), T78 IgM (28, 38, 83), and T78 IgG (26, 55, 57), respectively. Total IgM and IgG mutation averages were, respectively, 7.9 and 11.2 for T76 and 9.1 and 18.9 for T78. (B) The number of unmutated (germ-line) IgM (□) and IgG (■) sequences among each RO fraction was quantified for T76 and T78. (C-D) The cumulative distribution of mutations among IgM and IgG sequences was determined for RO− (thin line; ●), RO+/− (dashed line; ×), and RO+ (thick line; □) fractions. Percentages represent the number of samples that have mutations within the indicated range divided by the sample size n value (range increases in 3-bp increments). Arrows highlight the percentage difference between RO− and RO+ fractions at the RO+ average. For example, only 5% of T76 RO−IgM sequences have more than 10 mutations compared with 35% of RO−IgM sequences. Results are presented for (C) T76 and (D) T78. Note that RO+/− values are typically medial to RO− and RO+ values, suggesting that mutations progressively accumulate with respect to RO level.
We next compared the percentage of unmutated sequences in each RO fraction to determine whether higher RO+ mutation averages were due to a reduced number of unmutated cells in this fraction (ie, a greater ratio of mutated vs unmutated sequences). Graphed in Figure 3B, the percentage of unmutated IgM sequences in RO− and RO+ fractions respectively decreased from 29% to 9.3% (T76) and 25% to 16.7% (T78). Unmutated IgG sequences were predominantly identified among RO− fractions for both tonsils. For example, all T76 RO+ and RO+/− IgG+ sequences analyzed were mutated. Although 3 (n = 57) unmutated RO+ IgG sequences were identified in T78, collectively (T76 + T78) unmutated IgG sequences were 3 times less likely to be RO+ (3 of 115) than RO− (8 of 102).
The near exclusion of cells containing unmutated Ig heavy chain transcripts, and small size of the RO+ pool (< 10% of all GC cells) suggested that only a select group of cells up-regulate surface RO at any given time. Therefore, we generated cumulative distribution graphs to determine whether higher RO+ mutation averages were due to skewing by a few sequences with extremely high mutation frequencies (Figure 3C,D). Percentages represent the number of samples that have mutations within the indicated range divided by the sample size n value (range increases in 3-bp increments). Arrows highlight the percentage difference between RO− and RO+ fractions at the RO+ average. RO+ GC B cells (independent of isotype) were enriched for more sequences with higher mutation averages. For example, only 5% of T76 RO-IgM+ sequences have more than 10 mutations compared with 35% of RO+IgM+ sequences, representing a 7-fold increase (Figure 3C). Note that RO+/− values are typically medial to RO− and RO+ values, suggesting that mutations progressively accumulate with respect to RO level. Similar results were observed for T76 IgG sequences, with the largest differences occurring in the lower mutation range (ie, 95% of RO+ IgG sequences have 8 or more mutations, compared with only 50% of RO− sequences).
RO− GC B cells express the highest level of AnV
It has been reported that cells with increased autoreactive potential can have extremely high mutation averages. Lui and Arpin suggest that incorporating more mutations would increase the chances of ablating BCR autospecificity, which if uncorrected, could result in cell death.46,47 To determine whether the RO+ fraction was enriched for apoptotic cells, total tonsillar B cells were stained for GC and naive B-cell subsets as before with the addition of anti-AnV–APC (a marker of cell death).37,48 Illustrated in Figure 4A, 94.5% of all AnV+ GC B cells (0.6%) are RO−. Only 0.5% of RO+ GC cells were AnV+. The close proximity of most AnV+ cells to the RO−/RO+/− boundary suggests that the RO− fraction can be further subdivided to highlight an RO−/lo fraction. These data suggest that essentially all RO+ GC cells are alive and nonapoptotic. To determine whether AnV was more likely a marker of cell death or perhaps a marker expressed by differentiating cells (reported in Dillon et al49 ), CD69 mean fluorescence intensities were calculated for AnV− and AnV+ cells within each RO fraction (Figure 4B). Due to the dearth of RO+AnV+ cells, comparisons were made by gating on the highest and lowest 5% of cells under the AnV bell curve. Independent of RO fraction, CD69 mean fluorescence intensity (MFI) values were 3- to 4-fold lower in all AnV+ GC cells, which is more consistent with cells that are in the process of down-regulating many homeostatic processes.
Correlation between AnV and CD45RO expression levels. Total tonsillar lymphocytes were stained for GC and naive B-cell subsets as before with the addition of anti-AnV–APC. (A) RO versus AnV contour plots are shown for each population, including the APC-isotype control. The total percentage of AnV+ cells is indicated. *Distribution of AnV+ cells across the RO spectrum (ie, 94.5% of all AnV+ GC B cells [0.6%] are RO− compared with only 0.5% of RO+ GC cells). The proximity of most AnV+ cells to the RO−/RO+/− boundary suggests the RO− fraction can be further subdivided, highlighting an RO−/lo fraction. (B) CD69 MFIs were calculated for AnV− (□) and AnV+ (■) cells within each RO fraction. In the case of RO+ where there was a dearth of AnV+ cells, comparisons were made by gating on the highest and lowest 5% of cells under the AnV bell curve. × denotes no data point.
Correlation between AnV and CD45RO expression levels. Total tonsillar lymphocytes were stained for GC and naive B-cell subsets as before with the addition of anti-AnV–APC. (A) RO versus AnV contour plots are shown for each population, including the APC-isotype control. The total percentage of AnV+ cells is indicated. *Distribution of AnV+ cells across the RO spectrum (ie, 94.5% of all AnV+ GC B cells [0.6%] are RO− compared with only 0.5% of RO+ GC cells). The proximity of most AnV+ cells to the RO−/RO+/− boundary suggests the RO− fraction can be further subdivided, highlighting an RO−/lo fraction. (B) CD69 MFIs were calculated for AnV− (□) and AnV+ (■) cells within each RO fraction. In the case of RO+ where there was a dearth of AnV+ cells, comparisons were made by gating on the highest and lowest 5% of cells under the AnV bell curve. × denotes no data point.
An increased percentage of RO+ GC B cells are CD69+ with minor changes in CD71 expression
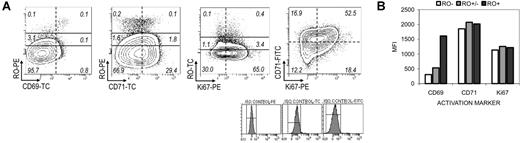
GC B cells become activated as they begin to somatically mutate.50,51 We investigated whether the RO-related increase in mutations correlated with signs of increased activation, including proliferation. GC B cells were costained with antibodies against RO and a panel of activation markers, including CD69, CD71, and Ki67. Isotype controls were used to demark positive and negative gates for each cytometric bell curve distribution. Results are depicted in Figure 5A. Each RO fraction contained both CD69+ and CD69− GC B cells. However, the highest ratio of CD69+ to CD69− cells was observed within the RO+ fraction (r = 1.0), compared with RO− (r < 0.1) and RO+/− (r < 0.1) fractions. Consistent with these findings, RO+ cells showed a 4-fold increase in CD69 MFI values compared with RO− cells (Fig 5B).
Correlation between GC B-cell CD45RO expression and activation markers CD69, CD71, and Ki67. (A) GC B cells were costained with antibodies against RO and a panel of activation markers, including CD69, CD71, and Ki67. Each activation marker generates a cytometric bell curve distribution. Therefore, isotype controls were used to demarcate positive and negative gates (dashed lines). Quadrant classification follows the standard Cartesian system such that RO is tri-sected into 3 expression levels (−, +/−, and +). All other markers are bisected into a “−” and “+” quadrant. The propensity of cell proliferation is indicated by a CD71-versus-Ki67 plot (greatest among CD71+Ki67+ double-positive population). (B) CD69, CD71, and Ki67 MFIs were calculated for each GC B-cell RO fraction (RO−, □; RO+/−, ▩; and RO+, ■).
Correlation between GC B-cell CD45RO expression and activation markers CD69, CD71, and Ki67. (A) GC B cells were costained with antibodies against RO and a panel of activation markers, including CD69, CD71, and Ki67. Each activation marker generates a cytometric bell curve distribution. Therefore, isotype controls were used to demarcate positive and negative gates (dashed lines). Quadrant classification follows the standard Cartesian system such that RO is tri-sected into 3 expression levels (−, +/−, and +). All other markers are bisected into a “−” and “+” quadrant. The propensity of cell proliferation is indicated by a CD71-versus-Ki67 plot (greatest among CD71+Ki67+ double-positive population). (B) CD69, CD71, and Ki67 MFIs were calculated for each GC B-cell RO fraction (RO−, □; RO+/−, ▩; and RO+, ■).
CD71 is up-regulated among cells that require additional iron uptake for metabolic (and other) processes, including cell differentiation and division.40,52 Similar to CD69, CD71+ and CD71− GC B cells were observed in each RO fraction. However the ratio of CD71+ cells did not significantly increase in the RO+ fraction (r = 0.5) compared with RO− (r = 0.4). RO+/− cells showed a 2-fold increase (r = 1.1), but comparison of CD71 MFI values revealed only a minor increase in this fraction (Figure 5B). Based on these data, RO+ cells showed increased signs of certain (CD69), but not all (ie, CD71) types of activation compared with their RO− counterpart. We therefore assessed whether activation markers that are more directly linked with the SHM process in GC B cells were differentially expressed in relation to surface RO.
Most proliferating GC B cells are RO−
Cell division is a prerequisite for SHM. Ki67 is routinely used to identify proliferating lymphocytes, positively stains cells in all stages except G0, and generally parallels CD71 expression.41,52 We compared Ki67 expression between RO-sorted GC B-cell fractions to determine whether proliferation (or nonproliferation) varied with respect to RO. Shown in Figure 5A, 71% of GC B cells were Ki67+ and most likely in a non-G0 stage of cell cycling (especially the 53% CD71+Ki67+ double positives). Like CD69 and CD71, Ki67+ GC B cells were observed in all 3 RO fractions. Both the ratio of Ki67+ to Ki67− cells (Figure 5A) and Ki67 MFI values (Figure 5B) varied little between RO fractions. These findings, combined with CD71 data, render it impossible to rule out the potential existence of proliferating cells within each RO fraction. Nonetheless, nearly 70% of RO− GC B cells are Ki67+, suggesting that the majority (but not necessarily the totality) of proliferation occurs during this stage.
Conjunct staining against CD69 and RO further subdivide GC B cells into isotype-influenced, differentially mutated subsets
Of the activation markers surveyed, CD69 provided the greatest MFI difference between RO+ and RO− GC B-cell fractions. We therefore investigated whether the combined use of anti-CD69 and anti-RO could be used to subdivide GC B cells within the same RO fraction into 2 pools with relatively low and relatively high mutation frequencies. GC B cells were first gated according to their surface RO expression and subsequently sorted into CD69− and CD69+ fractions (ie, RO+CD69− and RO+CD69+, RO−CD69−, etc). Results are shown in Figure 6. Like RO, CD69 alone separated GC B cells into subsets with significantly different mutation averages (Figure 6A). Interestingly, the correlation between CD69 expression and increased mutations were inversely related for IgM and IgG VH4 transcripts sequenced in this experiment. For example, sequences from CD69− IgM cells had higher mutation averages than CD69+ IgM cells (11.7 vs 5.9; P ≤ .001). In contrast, CD69− IgG cells were lower than their CD69+ counterpart (10.1 vs 12.0; P = .05). The same analysis (CD69− vs CD69+) was independently applied to RO− and RO+ fractions (ie, RO−CD69− vs RO−CD69+). Combined use of CD69 and RO resulted in the largest difference in mutation averages ([RO+CD69− − RO−CD69−] = [16.7 − 4] = 12.7) compared with either RO+ versus RO− (7.2) or CD69+ versus CD69− (5.8) alone. We next performed semiquantitative RT-PCR analyses on CD69− and CD69+ GC B cells from each RO fraction to determine whether surface CD69 could be used as a marker to enrich for GC B cells with higher or lower levels of AID expression (mRNA). Depicted in Figure 6B, AID was comparably expressed in CD69+ and CD69− GC B cells isolated from the same RO fraction. AID expression was inversely proportional to surface RO expression, with the highest level detected among RO− cells. These experiments show that while RO and CD69 are effective independently, when used in combination they provide the best discriminator for human GC cells that have the lowest or the highest mutation averages.
Correlation between SHM and dual expression of CD69 and CD45RO. (A) GC B cells from Tonsil 76 were costained with anti-RO and anti-CD69 and sorted into 6 resultant subsets. N values are listed for IgM: CD69−RO− (13), CD69−RO+ (20), CD69+RO− (18), and CD69+RO+ (21); and IgG: CD69−RO− (38), CD69−RO+ (38), CD69+RO− (33), and CD69+RO+ (25). (B) AID transcripts were semiquantitatively amplified (as in Figure 2B) from each of the above RO/CD69 fractions, including CD69+RO+/− (CD69−RO+/− cells were not collected). β-actin amplification was used to normalize RNA levels. Molecular weight markers are indicated to the left.
Correlation between SHM and dual expression of CD69 and CD45RO. (A) GC B cells from Tonsil 76 were costained with anti-RO and anti-CD69 and sorted into 6 resultant subsets. N values are listed for IgM: CD69−RO− (13), CD69−RO+ (20), CD69+RO− (18), and CD69+RO+ (21); and IgG: CD69−RO− (38), CD69−RO+ (38), CD69+RO− (33), and CD69+RO+ (25). (B) AID transcripts were semiquantitatively amplified (as in Figure 2B) from each of the above RO/CD69 fractions, including CD69+RO+/− (CD69−RO+/− cells were not collected). β-actin amplification was used to normalize RNA levels. Molecular weight markers are indicated to the left.
Discussion
For more than a decade, we have reproducibly separated human B cells into distinct subsets, including naive, GC, and memory, each of which displayed characteristic mutation averages.29 However, differences were more attributable to their developmental stage such that antigen-inexperienced naive B cells were unmutated and high affinity (SHM-dependent) memory B cells were among the most mutated. In contrast, GC B cells spanned the entire spectrum of mutations. Previous attempts using anti-CD77 (centrocyte vs centroblast) did not reliably separate GC B cells into enriched pools of cells with low and high IgH mutation averages.29 Typically, centrocytes and centroblasts had virtually identical mutation averages. Here, we show that the level of somatic mutation among GC B cells increased concordantly with RO up-regulation. These results document for the first time that GC B cells can be reproducibly separated into pools enriched for low or higher mutation averages, based solely on an extracellular surface protein.
A significant increase in mutations was consistently observed among RO+ fractions (relative to RO−) for both IgM and IgG sequences. Cumulative distribution analyses show that average increases within each RO fraction were gradual and were not due to the overexpansion of a highly mutated clonal family (mutation increases were observed for multiple VH4 family members; sequence data not shown). RO+ fractions had both a greater percentage of cells with more mutations (as high as a 7-fold increase in some cases; Figure 3C) and a decreased number of unmutated germ-line sequences. The latter was especially true for the IgG pool derived from RO+ GC cells, where in 1 tonsil, all 58 IgVH IgG sequences were mutated.
Like RO, CD69 also subdivided GC B cells into subpopulations with different levels of somatic mutation (though with a narrower range of difference; ROonly IgM = 7.2, CD69only IgM = 5.8). However, the combination of both RO and CD69 provided the greatest separation between cells bearing IgVH4 transcripts with low and high mutation averages (RO and CD69 IgM = 12.7; Figure 6A). Interestingly, CD69 had no discriminatory effect on RO− GC B cells. It is uncertain what factor(s) contributed to the inverse mutation pattern observed between CD69-subdivided RO+IgM+ and RO+IgG+ sequences (Figure 6A). Results may point toward different influences of CD45 on AID-dependent SHM and CSR. Comparable mutation averages with RO+IgG+ sequences render it plausible that CD69−RO+IgM+ GC B cells represent those that will imminently class switch and/or differentiate into post-GC B-cell subsets. Alternatively, this fraction may represent highly mutated cells whose BCRs confers self-specificity and are therefore targeted for deactivation during their potential transition toward anergy or deletion. Further investigation is needed to address these 2 points.
In preliminary efforts to uncover what force(s) are likely to contribute to differences in mutation averages between RO+ and RO− fractions, we investigated whether other SHM-related processes were differentially expressed with respect to RO. The 3 major factors critical for the initiation and persistence of SHM include AID expression (also responsible for CSR), cell activation (ie, CD69, CD71), and proliferation (access to exposed nucleic acid; Ki67).33-36 AID, Ki67, and CD71 were detected in each fraction, making it impossible to rule out proliferation or SHM during any RO stage. Nonetheless, AID expression decreased slightly among RO-expressing GC B cells, suggesting that at least a fraction of these cells may have terminated, or are in the process of down-regulating AID-dependent processes, including SHM. The relative difference in AID expression between RO− and RO+ cells varied between tonsils (Figures 2B,6B). Further experiments are needed to better determine whether certain cell cycle stages, particularly S-phase, predominate in a given RO fraction. Nonetheless, the majority of G0 (Ki67−) cells were RO− (Figure 5A). Preliminary analyses using forward and side scatter showed that RO+/− GC B cells were larger (FSChi) with greater internal complexity (SSChi) than RO− cells (data not shown), which may be indicative of different stages of the cell cycle occurring during a given cell's occupancy of RO− (resting, early stage) and RO+/− (mid-late and/or S phase) fractions.53 Further experiments will be required to fully understand this distinction. Previous reports concluded that mutations are incorporated during the CD77+ centroblast stage.29,37,54 Here, we show that nearly all centroblasts (99%) were RO− or RO+/−, suggesting that most mutations are incorporated during 1 or both of these stages.
It is possible that proliferating RO+AID+ cells may be involved in other AID-associated processes (ie, CSR). Investigations by Liu et al suggest that SHM precedes CSR.55 While this order of events may occur in most cases, it does not appear to be obligatory. First, unmutated IgG sequences, though rare, are routinely isolated. Secondly, we show that IgG transcript mutation averages continue to increase with respect to RO expression. In addition, we identified several examples of clonally related IgG sequences that had different numbers of mutations per transcript (data not shown; manuscript in preparation). Collectively, these data provide sequence-related support that GC B cells that have undergone CSR can in some cases reinitiate the somatic hypermutation process, and that CSR is unlikely to occur solely at the end of the GC reaction.
The ability to subdivide B cells into physiologically relevant levels of RO expression is central to our understanding of these data. The accuracy of B-cell RO gating was evidenced by changes in CD45 isoform transcript levels. Despite a seemingly small increase in detectable surface RO levels, RO-expressing GC B cells showed increased RO transcripts, reduced RA transcripts (possibly due to increased RA mRNA splicing and/or reduced overall synthesis of the long isoform), and adopted unique physiologically relevant phenotypes. The low percentage of RO-expressing tonsillar B cells (10%-20%) suggests that expression, or at least sustained expression, is not required for cell survival or retention within the tonsil environment. Though readily observed in tonsils (and presumably in other lymphoid tissues), RO+ B cells were rare in peripheral blood, suggesting that factors that promote RO up-regulation are either unique to or differentially administered in tonsils.
RA+RO− resting naive T cells up-regulate surface RO (RA+/−RO+/−) in response to activation and differentiation-inducing stimuli during “transition” toward memory and effector cells. We therefore hypothesize that GC B cells that up-regulate surface RO may be in the midst of transition, whether from 1 physiologic state to the next (ie, proliferating to nonproliferating), or differentiation to other B-cell subsets. Further investigation is needed to address this issue more directly. Nonetheless, data presented in this report show that RO-expressing GC B cells, especially RO+, exhibited characteristics that are consistent with cells in transition, including increased activation (CD69), enrichment for CD77− centrocytes (poised to undergo selection), and a decreased propensity for cell death (AnV−).
In summation, by adding CD45RO (and CD69) to our current strategy for identifying human GC B cells, we introduce a “temporal gauge” whereby cells can be separated into pools with low (recent descendents of the founder GC B cells) and high mutation averages (distally related to founder cells evidenced by multiple cell divisions; ie, cells mutate at a rate of 1 mutation per 3 cell divisions). It is important to note however, that surface RO was also expressed by human B-cell subsets other than GC B cells. Since mutations are predominantly introduced during the GC stage, it is unlikely that RO correlates with SHM in these cells. Applying similar RO-based strategies to subdivide non-GC B cells may elucidate whether surface RO correlates with other physiologically relevant phenotypes as it does with mutation load in GC B cells.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Patrick Wilson (Oklahoma Medical Research Foundation Molecular Immunogenetics), Dr Diana Hamilton and Jacob Bass (Paul Kincade Lab and Flowlab, Oklahoma Medical Research Foundation), Dr J. Jireh) and members of the Building Young Scientists (BYS) Science Program (Oklahoma City, OK) for their timeless dedication and advice administered during this project.
This work was supported by grants AI 12127 and NCRR P20RR15577 from the National Institutes of Health.
National Institutes of Health
Authorship
Contribution: S.M.J. wrote the manuscript, designed the research project, performed the research, and analyzed the data. Research was also performed by N.H., D.P., J.Z., S.W., Y.J.K. and C.C. As the principal investigator, J.D.C. reviewed the data and final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: J. Donald Capra, Molecular, Immunogenetics, Oklahoma Medical Research Foundation, 825 Northeast 13th, Street, Oklahoma City, OK 73104; e-mail: jdonald-capra@omrf.ouhsc.edu.

![Figure 4. Correlation between AnV and CD45RO expression levels. Total tonsillar lymphocytes were stained for GC and naive B-cell subsets as before with the addition of anti-AnV–APC. (A) RO versus AnV contour plots are shown for each population, including the APC-isotype control. The total percentage of AnV+ cells is indicated. *Distribution of AnV+ cells across the RO spectrum (ie, 94.5% of all AnV+ GC B cells [0.6%] are RO− compared with only 0.5% of RO+ GC cells). The proximity of most AnV+ cells to the RO−/RO+/− boundary suggests the RO− fraction can be further subdivided, highlighting an RO−/lo fraction. (B) CD69 MFIs were calculated for AnV− (□) and AnV+ (■) cells within each RO fraction. In the case of RO+ where there was a dearth of AnV+ cells, comparisons were made by gating on the highest and lowest 5% of cells under the AnV bell curve. × denotes no data point.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/12/10.1182_blood-2007-05-087767/4/m_zh80220709000004.jpeg?Expires=1769260834&Signature=SIF1ZcIEBAmS~2C7nkWLBYSCE-0AbV2Ns40xKVrKnDO-tuC6PUGvja-Q-AdWUVMM5pTLHBlRuJcl7cwgLMAGf996NTwCMmY2wQeOJsDSwMM1VFv1OhhtdMBp-A7rNxY4HhqjEkLG6ns7FPTA~4xBZxaAxWayxt4DpYVgUGixXD5rWgGuptGvRRaRK4EOfTJRPlKx-cMpI6f3RE30FX8A0q9m-Kyg-PdFBmb9cTt0p61-TRVCRatofMfyw2RakkwZatW6m4z9lGzZIA4ygF6mZoepnzDRdEFfQX7OWSetMd6jlAZicwIS~XyIcFsrKYID~12l2GuRM5AjMQl6B8ldlg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)