Abstract
The bone morphogenetic protein (BMP) signaling pathway regulates multiple steps of hematopoiesis, mediated through receptor-regulated Smads, including Smad1 and Smad5. Here, we use loss-of-function approaches in zebrafish to compare the roles of Smad1 and Smad5 during embryonic hematopoiesis. We show that knockdown of Smad1 or Smad5 generates distinct and even opposite hematopoietic phenotypes. Embryos depleted for Smad1 have an increased number of primitive erythrocytes, but fail to produce mature embryonic macrophages. In contrast, Smad5-depleted embryos are defective in primitive erythropoiesis, yet have normal numbers of macrophages. Loss of either Smad1 or Smad5 causes a failure in the generation of definitive hematopoietic progenitors. To investigate the mechanism behind these phenotypes, we used rescue experiments and found that Smad5 is unable to rescue the Smad1 loss-of-function phenotype, indicating that the 2 highly related proteins have inherently distinct activities. Microarray experiments revealed that the 2 proteins redundantly regulate the key initiators of the hemato-vascular program, including scl, lmo2, and gfi1. However, each also regulates a remarkably distinct genetic program, with Smad5 uniquely regulating the BMP signaling pathway itself. Our results suggest that specificity of BMP signaling output, with respect to hematopoiesis, can be explained by differential functions of Smad1 and Smad5.
Introduction
Hematopoiesis initiates within the zebrafish embryo in the intermediate cell mass (ICM) associated with the ventral side of the tail.1 The ICM forms from the fusion of 2 bilateral stripes of posterior mesoderm that are marked as early as the 3-somite stage by expression of hematopoietic transcription factors, including lmo2, scl, and gata1. The definitive (adult stage) blood lineage precursors originate from the ventral wall of the dorsal aorta,2,3 and migrate 2 days after fertilization (dpf) to a caudal region of the tail referred to as the posterior blood island (PBI). In addition to the ICM, dorsal aorta, and PBI, a distinct hematopoietic program arises from the anterior lateral mesoderm, generating primitive macrophages and granulocytes. These cells are initially marked by expression of lmo2 and scl during early somitogenesis stages as 2 short stripes, but by 24 hours after fertilization (hpf), the cells express markers specific to macrophage or granulocyte lineages, such as l-plastin and myeloperoxidase, respectively, and enter circulation around 48 hpf.4,5
Bone morphogenetic proteins (BMPs) are growth factors of the TGF-β superfamily, essential for patterning mesoderm character.6 High levels of BMP signaling specify the most ventral mesoderm, from which the ICM is derived. The dependency of embryonic hematopoiesis on BMP signaling is a conserved feature in vertebrate embryos independent of the initial steps of mesoderm induction.7,8 BMP signaling continues to regulate hemato-vascular development during somitogenesis, although the pathway may either promote or restrict the expansion of hematopoietic progenitors, depending on the developmental stage.9 BMPs at high concentration can extend proliferative capacity of adult human hematopoietic stem cells, and at low concentrations induce differentiation.10 Thus, the outcome of BMP signaling can differ depending on the developmental stage or signal strength.
BMP receptors signal by phosphorylation of regulatory (R) Smads 1, 5, and 8. The activated R-Smad, with the co-Smad4, translocates to the nucleus to regulate BMP target genes.6 Mice with mutations in components of the signaling pathway die early with hematopoietic defects. The few Bmp4 knockout mice that survive long enough to form a yolk sac are deficient in primitive blood islands.11 The Smad5 knockout is embryonically lethal at about embryonic day (e) 10 and displays abnormal vasculature with defects in the amnion and allantois.12,13 There is a reduction in the number of primitive erythroid precursors generated from Smad5−/− embryonic stem (ES) cells using the embryoid body system.14 The Smad1 knockout mouse dies from chorio-allantois defects around e9. Although some hematopoietic precursors are found in the allantois, further analysis is hindered by the early lethality and lack of proper placental connection.15,16 Double trans-heterozygote mutations for Smad1 and Smad5 are also embryonically lethal, causing phenotypes similar to the single homozygous knockouts, showing that there are genetic interactions between Smad1 and Smad5.17
Genetic screens in zebrafish identified mutants for components of the BMP signaling pathway. The mutants for bmp2 (swirl) and bmp7 (snailhouse) fail to develop mature blood cells.18-20 The mutant for a type I BMP receptor (Alk8) called lost-a-fin fails to develop blood circulation.21,22 The somitabundtc24 (sbn) mutation confers dominant-negative function to Smad5, and causes a strongly dorsalized phenotype lacking most ventral tissues.23 Two other mutant alleles are characterized in zebrafish that along with sbn provide a smad5 allelic series. The piggytaildty40 (pgy) allele is a point mutation that produces a weak dominant-negative (antimorphic) protein. The captain hook (m169) mutant contains an insertion that generates a nonfunctional protein with less severe dorsalization.24,25 Each of these smad5 mutants is anemic at the time of embryonic lethality, around 3 to 4 dpf.26
Zebrafish Smad1 and Smad5 share 83.5% amino acid identity and the common structure of the receptor-regulated Smads, containing 2 highly conserved Mad-homology (MH) domains joined by a more divergent linker region.27,28 Specificity of the BMP response can occur in principle at many levels, including ligand antagonism, receptor conformation, interaction with inhibitory receptors or Smads, and a host of complex cofactor interactions within the cytoplasm and nucleus.29,30 Here, we focus on the specificity of R-Smad activity during embryonic hematopoiesis, and show that Smad1 and Smad5 have strikingly different functions during embryonic hematopoiesis. While both proteins redundantly regulate key initiators of hematopoiesis, Smad5 but not Smad1 is essential for regulating the known downstream components of the BMP signaling pathway, including the so-called BMP4 synexpression group.31
Materials and methods
Zebrafish and MOs
Zebrafish were raised and maintained according to Westerfield32 and staged according to Kimmel et al.33 In situ hybridization was performed as described.34 The Smad5 morpholino (MO) was purchased from Open Biosystems (Huntsville, AL). The optimal concentration that phenocopies the piggytaildty40 mutant was determined to be 1 ng/embryo. Two Smad1 MOs were purchased from GeneTools (Philomath, OR). Smad1 MO (S1MO) targets the ATG translational initiation site with an optimal dose of 2 ng/embryo. A second MO was designed to block a splice junction (S1MO1) by targeting the exon2-intron2 boundary. At 25 ng/embryo, this MO gives a phenotype similar to S1MO: Smad5 MO, 5′ACATGGAGGTCATAGTGCTGGGCTG; Smad1 MO, 5′AGGAAAAGAGTGAGGTGACATTCAT; and Smad1 MO 1, 5′TAACAATTTAGCCAC-GCTCACCTGG.
A Nikon SMZ 1500 (Nikon; Melville, NY) was used for all microscopy. For embryos from the bud stage to 18 hpf the magnification was 6× (with a 10× eyepiece), while for embryos at 24, 36, or 48 hpf the magnification was 4× (with a 10× eyepiece). The common objective was an HR Plan Apo1X, WD54 lens. Embryos were mounted (without counter-staining) either in 3% methylcellulose or (following in situ hybridization) in a 2:1 solution of benzyl benzoate-benzyl alcohol (BB/BA). Images were taken with a Spot Insight Firewire 2 Model 11.2 color mosaic camera (Camera Diagnostic Instruments, Sterling Heights, MI), acquired with Spot advanced 4.5 software, and processed using Adobe (San Jose, CA) Photoshop CS2 v.9 software.
In vitro transcription and translation
Reactions were performed using the TNT Coupled Rabbit Reticulocyte System (Promega, Madison, WI). Each reaction contained 0.5 μg plasmid and 500 ng MO and was incubated at 30°C for 2 hours. Without further manipulation, an equal amount of each sample was loaded onto a 10% SDS Nu-Page gel (Invitrogen, Carlsbad, CA) and electrophoresed; hemoglobin levels confirmed equal loading.
RT-PCR
Wild-type embryos were injected with 25 ng/embryo of the splice-blocking MO (S1MO1). At 32 hpf, RNA was purified using Tri-Reagent (Molecular Research Center, Cincinnati, OH). A total of 1 or 2 μg total RNA was used to generate cDNA with First Strand Moloney murine leukemia virus (M-MLV) reverse transcriptase and random hexamers (Invitrogen). The cDNA was used in a polymerase chain reaction (PCR) accordingly: 95°C for 10 minutes (95°C for 30 seconds, 57.5°C for 30 seconds, and 72°C for 30 seconds) for 25 cycles, and 72°C for 10 minutes. Primers used were the following: βactin35 forward: 5′AAGCAGGAGTACGATGAGTCTG and reverse: 5′GGTAAACGCTTCTTCTGGAATGAC; and Smad1 forward: 5′GGCCTCACGTCATCTACTGCC; reverse: 5′GGGTTACGGAA-GCGTGGCAGCAT.
Smad cDNA constructs
The full-length Smad5 cDNA was available in the pCS2+ vector. The full-length Smad1 cDNA in pExpress1 was purchased from Open Biosystems. The Smad1 cDNA insert (NarI-SpeI) was shuttled into the pCS2+ vector digested with ClaI-XbaI. Smad1 and Smad5 constructs, designated Smad1 * and Smad5*, contain the opening reading frame with mutations in the wobble codons (bold) surrounding the ATG. These were generated by PCR amplification using the following primers: Smad1* forward: 5′ATGAATGTGACCTCGCTGTTCTCCTTC; reverse: 5′AGTTGCTTTATGGTCAGCTCCATCTAGG; and Smad5* forward: 5′CCGGAATTCCAACCAAGCACTATGACATCTATG; reverse: 5′GCTCTAGAGCCTCCCAGGTCAGCCCATCATTAC.
The Smad1* PCR product was cloned into the pCR4 vector using the Invitrogen TOPO TA cloning protocol; the insert was transferred by EcoRI digest into the pCS2+ vector. The Smad5* PCR product was generated containing EcoRI and XbaI sites and cloned directly into pCS2+. Plasmids were digested with NotI enzyme, and capped RNA was synthesized in vitro using the Sp6 mMessage kit (Ambion, Austin, TX).
Flow cytometry and microarray experiments
Detailed experimental methodology is provided in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Quantitative real-time PCR
First-strand cDNA synthesis was performed using Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen). The cDNA was analyzed with Qiagen QuantiTect SYBR Green Mix (Qiagen, Valencia, CA) by quantitative reverse transcription (RT)–PCR using the Opticon DNA Engine 3 real-time PCR machine (Bio-Rad Laboratories, Hercules, CA). All samples were done in triplicate, and each experiment was repeated 3 times. The PCR cycle conditions were 95°C for 15 minutes, (94°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds) for 40 cycles. PCR primers are listed in Table S2. The CT value data were analyzed using the 2−ΔΔT method.36
Results
Targeting Smad1 or Smad5 with specificity in zebrafish
We sought to compare the role of Smad1 and Smad5 during embryonic hematopoiesis using loss-of-function approaches in the zebrafish model. Smad1 and Smad5 are predicted to be the major mediators of BMP signaling, since Smad8 has not been identified in the zebrafish genome. We could use the hypomorphic point mutant piggytaildty40 to evaluate specifically Smad5 function.26 This piggytail mutation is located in the MH2 domain of Smad5 (Figure 1A). Since a Smad1 zebrafish mutant has not been identified, we designed a MO knockdown strategy to compare the Smad1 loss-of-function phenotype with piggytail. In order to ensure parity in the study design, we also generated a smad5 morphant model for confirmation of piggytail phenotypes.
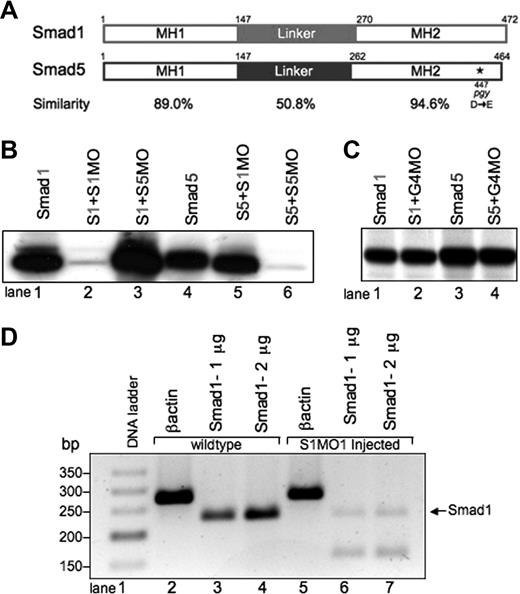
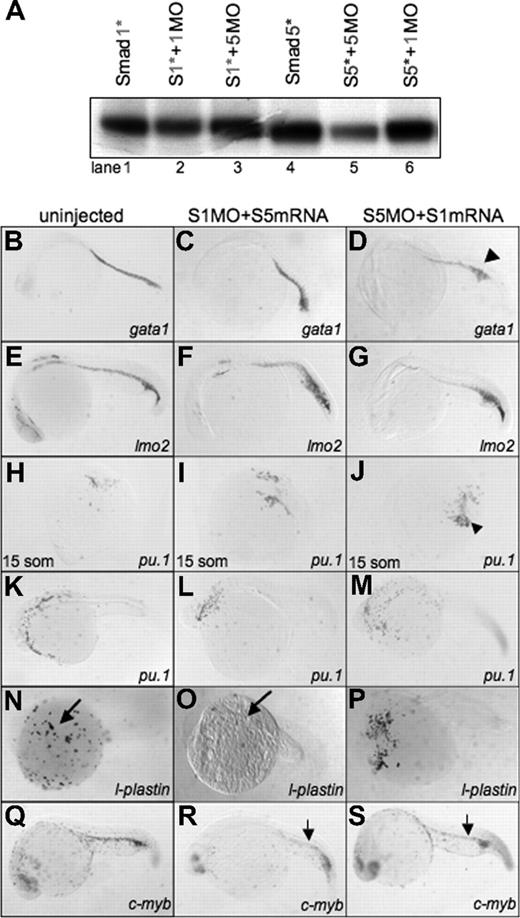
Smad1 and Smad5 are highly homologous, but can be individually targeted for loss of function studies using mutants and MO knockdown.(A) Comparison of zebrafish Smad1 and Smad5 protein structure. The MH1 and MH2 domains are highly similar, compared with the more divergent central linker region. The piggytail mutation (★) is located in the MH2 domain at amino acid 447 of Smad5. (B) In vitro transcription-translation of 35S-methionine–labeled Smad proteins. Smad1 (S1) can be efficiently and specifically targeted for knockdown by the addition of the Smad1 translation-blocking MO (S1MO; lanes 1 and 2). The reaction is not inhibited by the addition of the Smad5 MO (S5MO; lane 3). Smad5 (S5) can be targeted for inhibition by the addition of the Smad5 MO, but not the Smad1 MO (lanes 4-6). (C) In control reactions, a distinct MO (G4MO, specific for the gata4 gene) was used in equivalent reactions against Smad1 or Smad5. In each case, the control MO had no affect on translation, confirming that inhibition seen in panel B is specific. (D) RT-PCR analysis comparing RNA from uninjected embryos and embryos injected with the splice site–blocking MO (S1MO1) collected at 32 hpf. βactin serves as a positive control and generates a band of 277 bp. Primers that span exon2-intron2 generate for smad1 a 232-bp fragment. S1MO1 injection efficiently decreases the amount of properly spliced smad1 message. The smaller fragment at 170 bp was shown by sequencing to encode a misspliced smad1 isoform.
Smad1 and Smad5 are highly homologous, but can be individually targeted for loss of function studies using mutants and MO knockdown.(A) Comparison of zebrafish Smad1 and Smad5 protein structure. The MH1 and MH2 domains are highly similar, compared with the more divergent central linker region. The piggytail mutation (★) is located in the MH2 domain at amino acid 447 of Smad5. (B) In vitro transcription-translation of 35S-methionine–labeled Smad proteins. Smad1 (S1) can be efficiently and specifically targeted for knockdown by the addition of the Smad1 translation-blocking MO (S1MO; lanes 1 and 2). The reaction is not inhibited by the addition of the Smad5 MO (S5MO; lane 3). Smad5 (S5) can be targeted for inhibition by the addition of the Smad5 MO, but not the Smad1 MO (lanes 4-6). (C) In control reactions, a distinct MO (G4MO, specific for the gata4 gene) was used in equivalent reactions against Smad1 or Smad5. In each case, the control MO had no affect on translation, confirming that inhibition seen in panel B is specific. (D) RT-PCR analysis comparing RNA from uninjected embryos and embryos injected with the splice site–blocking MO (S1MO1) collected at 32 hpf. βactin serves as a positive control and generates a band of 277 bp. Primers that span exon2-intron2 generate for smad1 a 232-bp fragment. S1MO1 injection efficiently decreases the amount of properly spliced smad1 message. The smaller fragment at 170 bp was shown by sequencing to encode a misspliced smad1 isoform.
A S1MO was designed to block translation by binding to the sequences from +1 to +25 of the smad1 mRNA, relative to the ATG translational start site. We also designed an MO that should similarly block Smad5 translation by binding from −12 to +13 of the smad5 mRNA. We confirmed the activity and specificity of both MOs using an in vitro transcription-translation assay (Figure 1B). Addition of the Smad1 MO caused a significant block in the translation of Smad1, but not Smad5 protein (Figure 1B, lanes 2 and 5). Likewise, addition of Smad5 MO efficiently blocked Smad5 but not Smad1 translation (Figure 1B, lanes 3 and 6). Control reactions using a distinct MO that is targeted to the gata4 gene showed no inhibition of protein translation (Figure 1C).
Specificity of MO action is established when the same phenotype is replicated by 2 independent MOs. For this purpose, a second S1MO1 was designed to bind to the exon2-intron2 boundary and thereby block proper splicing of the smad1 pre-mRNA. RNA was prepared from embryos at 32 hpf derived from fertilized eggs injected with S1MO1 (or control normal embryos). The efficacy of S1MO1 was tested by RT-PCR assays using these samples (Figure 1D). In the samples derived from embryos injected with S1MO1 there is a near-complete loss of normal cDNA detected. These results indicate that S1MO1 efficiently targets the Smad1 pre-mRNA, thus validating the smad1 morphant phenotype. Therefore, using the mutant and morphant models, we compared the phenotypes caused by loss of Smad1 or Smad5 directly.
Loss of Smad1 or Smad5 generates distinct embryonic phenotypes
We determined that injection of 1 ng of Smad5-specific MO (S5MO) is sufficient to reproducibly phenocopy piggytaildty40 (Figure 2A). Smad5 morphants have a dorsalized phenotype evident around 18 hpf, with a curvature of the tail. This phenotype becomes more severe by 24 hpf with embryonic lethality at 3 to 4 dpf. Reduced numbers of embryonic red cells are seen in both piggytail and smad5 morphants at 24 hpf; by the time of death, there are no circulating blood cells. This block to primitive red cell production was noted previously in Smad5 mutants.26
Depletion of Smad1 or Smad5 causes distinct embryonic phenotypes. (A) Embryos were either control (wild-type [wt]; row 1) or injected with 2 ng translation-blocking Smad1 MO (S1MO), which causes a phenotype first visible at the bud stage by elongation of the embryo (row 2). This phenotype is more severe starting around 18 hpf, with a ragged dorsal edge, neural degeneration, and a widened notochord (row 2;  ). By 24 hpf, heart, brain, and endoderm defects are visible (
). By 24 hpf, heart, brain, and endoderm defects are visible ( indicate neural degeneration and yolk stalk regression). These embryos go on to die at 3 to 4 dpf. Alternatively, embryos were injected with 1 ng translation-blocking Smad5 MO (S5MO), which causes a known dorsalization phenotype (row 3) similar to that of characterized smad5 mutants. This phenotype is first visible around the 5-somite stage with a tail defect (
indicate neural degeneration and yolk stalk regression). These embryos go on to die at 3 to 4 dpf. Alternatively, embryos were injected with 1 ng translation-blocking Smad5 MO (S5MO), which causes a known dorsalization phenotype (row 3) similar to that of characterized smad5 mutants. This phenotype is first visible around the 5-somite stage with a tail defect ( ) and a more caudal arrangement of somites. By the 18 hpf stage, the curvature of the tail is visible, and by 24 hpf, the lack of ventral tissues is evident (
) and a more caudal arrangement of somites. By the 18 hpf stage, the curvature of the tail is visible, and by 24 hpf, the lack of ventral tissues is evident ( ). The Smad5 MO phenocopies the piggytail mutant (row 4). Each panel is a representative photograph of phenotype and stage shown. More than 200 embryos were observed for each panel. (B-E) Double knockdown of both Smad1 and Smad5 at half-dose causes lethality at 15 hpf. Shown are wt (B,D) and double-morphant (S1MO + S5MO) (C,E) embryos at 15 hpf and 24 hpf, as indicated. The phenotype just prior to death has combined features of each individual knockdown. Less than 1% of double morphant embryos survive to 24 hpf, as in panel E. Approximately 150 double-injected embryos were observed, and a representative is shown.
). The Smad5 MO phenocopies the piggytail mutant (row 4). Each panel is a representative photograph of phenotype and stage shown. More than 200 embryos were observed for each panel. (B-E) Double knockdown of both Smad1 and Smad5 at half-dose causes lethality at 15 hpf. Shown are wt (B,D) and double-morphant (S1MO + S5MO) (C,E) embryos at 15 hpf and 24 hpf, as indicated. The phenotype just prior to death has combined features of each individual knockdown. Less than 1% of double morphant embryos survive to 24 hpf, as in panel E. Approximately 150 double-injected embryos were observed, and a representative is shown.
Depletion of Smad1 or Smad5 causes distinct embryonic phenotypes. (A) Embryos were either control (wild-type [wt]; row 1) or injected with 2 ng translation-blocking Smad1 MO (S1MO), which causes a phenotype first visible at the bud stage by elongation of the embryo (row 2). This phenotype is more severe starting around 18 hpf, with a ragged dorsal edge, neural degeneration, and a widened notochord (row 2;  ). By 24 hpf, heart, brain, and endoderm defects are visible (
). By 24 hpf, heart, brain, and endoderm defects are visible ( indicate neural degeneration and yolk stalk regression). These embryos go on to die at 3 to 4 dpf. Alternatively, embryos were injected with 1 ng translation-blocking Smad5 MO (S5MO), which causes a known dorsalization phenotype (row 3) similar to that of characterized smad5 mutants. This phenotype is first visible around the 5-somite stage with a tail defect (
indicate neural degeneration and yolk stalk regression). These embryos go on to die at 3 to 4 dpf. Alternatively, embryos were injected with 1 ng translation-blocking Smad5 MO (S5MO), which causes a known dorsalization phenotype (row 3) similar to that of characterized smad5 mutants. This phenotype is first visible around the 5-somite stage with a tail defect ( ) and a more caudal arrangement of somites. By the 18 hpf stage, the curvature of the tail is visible, and by 24 hpf, the lack of ventral tissues is evident (
) and a more caudal arrangement of somites. By the 18 hpf stage, the curvature of the tail is visible, and by 24 hpf, the lack of ventral tissues is evident ( ). The Smad5 MO phenocopies the piggytail mutant (row 4). Each panel is a representative photograph of phenotype and stage shown. More than 200 embryos were observed for each panel. (B-E) Double knockdown of both Smad1 and Smad5 at half-dose causes lethality at 15 hpf. Shown are wt (B,D) and double-morphant (S1MO + S5MO) (C,E) embryos at 15 hpf and 24 hpf, as indicated. The phenotype just prior to death has combined features of each individual knockdown. Less than 1% of double morphant embryos survive to 24 hpf, as in panel E. Approximately 150 double-injected embryos were observed, and a representative is shown.
). The Smad5 MO phenocopies the piggytail mutant (row 4). Each panel is a representative photograph of phenotype and stage shown. More than 200 embryos were observed for each panel. (B-E) Double knockdown of both Smad1 and Smad5 at half-dose causes lethality at 15 hpf. Shown are wt (B,D) and double-morphant (S1MO + S5MO) (C,E) embryos at 15 hpf and 24 hpf, as indicated. The phenotype just prior to death has combined features of each individual knockdown. Less than 1% of double morphant embryos survive to 24 hpf, as in panel E. Approximately 150 double-injected embryos were observed, and a representative is shown.
The Smad1 knockdown in zebrafish shows a very different phenotype. Titration experiments using S1MO determined that 2 ng per injected embryo causes a consistent and penetrant phenotype, evident early in somitogenesis, and (like piggytail) the embryos survive until 3 to 4 dpf. The smad1 morphant phenotype is seen at the bud stage as a prominent embryonic elongation (Figure 2A; row 2). This, however, is resolved by the 5-somite stage, when the embryos round up and appear fairly normal. By 18 hpf, there is some neural degeneration and a delay in development of the otic placode. The dorsal edge of the embryo has a ragged appearance. In comparison, the smad5 morphant and piggytail mutant show apparently normal anterior development. Instead, there is a twisted and shortened tail, and somites are slightly irregular and caudally displaced (Figure 2A; rows 3 and 4, respectively).
By 24 hpf, the distinct phenotype caused by loss of Smad1 or Smad5 is obvious (Figure 2A). The smad1 morphants form normal tail tissues, but display 3 major features at this stage, with neural, yolk stalk, and cardiac defects. The neural degeneration seen at 18 hpf continues, and the otic placode is still not fully formed. The yolk stalk is not extended, indicating a potential problem with endoderm formation. The heart is also affected and fails to loop, although it is beating. The loss of Smad5 causes a similar cardiac “heart-string” phenotype, although this is the only commonality, since the other major defect is in tail formation without the brain or yolk stalk alterations. At 36 hpf and 48 hpf, the severity of the phenotype is increased for both morphants, and no new obvious features emerge. The ear is eventually formed by 48 hpf in smad1 morphants. Injection of the Smad1 splice-site MO (S1MO1) at 25 ng per embryo generates a very similar phenotype compared with S1MO at 48 hpf, although there is less yolk stalk regression and a greater curvature in the body at 24 hpf (data not shown).
Double knockdown of both Smad1 and Smad5 leads to early embryonic lethality
Injection of MOs that target both smad1 and smad5 in combination causes embryonic lethality before the onset of somitogenesis. Reducing the dose in half for each (1 ng for S1MO and 0.5 ng for S5MO) allows the embryos to survive to 15 hpf, when combined features of both individual morphants can be seen: neural degeneration with curvature of the tail (Figure 2E). Precisely between 15 and 16 hpf, the vast majority of double morphants consistently and predictably die with overall embryonic necrosis/apoptosis, indicating an essential and redundant cell-survival function for Smad1/5 at this stage of development. A very small number of double-injected embryos (less than 1%) survive to 24 hpf as shown (Figure 2D).
The loss of Smad1 or Smad5 causes opposite effects on primitive erythropoiesis
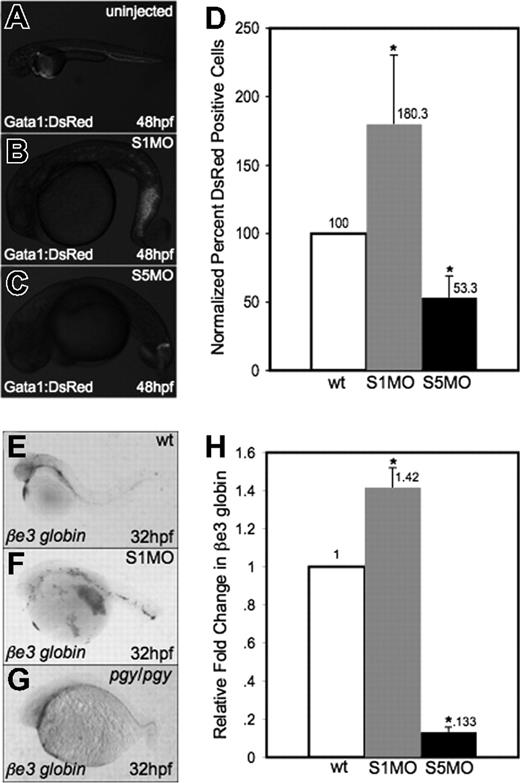
While the lack of blood in smad5 mutants/morphants is evident, the smad1 morphants appear by gross examination to have normal production of red blood cells. To confirm these observations, MOs specific to smad1 or smad5 were injected into fertilized eggs for a transgenic reporter fish expressing the DsRed fluorescent protein under the control of the gata1 promoter (Figure 3A-C). Injection of the Smad5 MO led to significantly decreased numbers of DsRed+ cells in the embryo at 48 hpf. In contrast, smad1 morphants appear to have normal or even increased numbers of DsRed+ cells present in the tail region at this stage. To quantify erythroid cells, we performed flow cytometry on cells isolated at 32 hpf from gata1:DsRed control and morphant embryos. The data (Figure 3D) confirm that smad1 morphants have a significant relative increase in Gata1+ erythroid cells compared with control siblings (approximately 2-fold). The opposite is seen in smad5 morphants, which as predicted show a significant decrease in Gata1+ cells (again, approximately 2-fold at this stage).
Smad1 knockdown enhances erythropoiesis. (A-C) Fertilized eggs derived from the gata1:DsRed reporter line were either uninjected controls (A) or injected with S1MO (B) or S5MO (C). There is an increase in erythrocyte production in the tail of smad1 morphants at 48 hpf (B), while smad5 morphants have a significant decrease in DsRed+ cells (C). Representative embryos are shown, and approximately 200 of each type of embryo were observed. (D) These phenotypes are quantified by flow cytometry of cells derived from control or morphant embryos at 32 hpf. Shown is the percentage of DsRed+ cells compared with control wt. *The difference compared with wt is significant by Student t test; P < .001. For each of 4 experiments, 30 embryos of each phenotype were collected for flow cytometry. (E-G) In situ hybridization was carried out for βe3 globin on wild-type (E), smad1 morphants (F), and pgy/pgy embryos (G). Shown are representative embryos at 32 hpf, showing that smad1 morphants are increased in erythrocyte production at 32 hpf (F); in contrast, piggytail (Smad5) mutants have a decrease in βe3 globin expression (G). In situ hybridization was done on a sample of 25 embryos for each; panels E-G are representative of the phenotypes seen. (H) Quantitative real-time PCR for βe3 globin was carried out on smad1 (S1MO) and smad5 (S5MO) morphant embryos at 32 hpf. Shown is the average fold change in expression calculated from 4 independent experiments, with samples analyzed each time in triplicate. Samples were normalized to β-actin, and wt set to 1. *The difference compared with wt is significant by Student t test; P < .001. Error bars in D and H indicate the standard deviation from the mean.
Smad1 knockdown enhances erythropoiesis. (A-C) Fertilized eggs derived from the gata1:DsRed reporter line were either uninjected controls (A) or injected with S1MO (B) or S5MO (C). There is an increase in erythrocyte production in the tail of smad1 morphants at 48 hpf (B), while smad5 morphants have a significant decrease in DsRed+ cells (C). Representative embryos are shown, and approximately 200 of each type of embryo were observed. (D) These phenotypes are quantified by flow cytometry of cells derived from control or morphant embryos at 32 hpf. Shown is the percentage of DsRed+ cells compared with control wt. *The difference compared with wt is significant by Student t test; P < .001. For each of 4 experiments, 30 embryos of each phenotype were collected for flow cytometry. (E-G) In situ hybridization was carried out for βe3 globin on wild-type (E), smad1 morphants (F), and pgy/pgy embryos (G). Shown are representative embryos at 32 hpf, showing that smad1 morphants are increased in erythrocyte production at 32 hpf (F); in contrast, piggytail (Smad5) mutants have a decrease in βe3 globin expression (G). In situ hybridization was done on a sample of 25 embryos for each; panels E-G are representative of the phenotypes seen. (H) Quantitative real-time PCR for βe3 globin was carried out on smad1 (S1MO) and smad5 (S5MO) morphant embryos at 32 hpf. Shown is the average fold change in expression calculated from 4 independent experiments, with samples analyzed each time in triplicate. Samples were normalized to β-actin, and wt set to 1. *The difference compared with wt is significant by Student t test; P < .001. Error bars in D and H indicate the standard deviation from the mean.
To confirm that this observation is not specific to the transgenic reporter gene, we performed in situ hybridization to evaluate expression of the erythroid-specific embryonic βe3 globin gene at 32 hpf. This showed an increase in red cell production in smad1 morphants and a decrease of the same cells in piggytail embryos compared with wild-type (Figure 3E-G). To measure quantitatively the observed changes in βe3 globin levels, real-time PCR was performed on cDNA from smad1 and smad5 morphant embryos at 32 hpf. Consistent with the qualitative analysis, smad1 morphants show an approximately 40% increase in βe3 globin expression, while smad5 morphants have an approximately 85% decrease (Figure 3H).
Initial hematopoietic programs are relatively unaffected by loss of Smad1 or Smad5
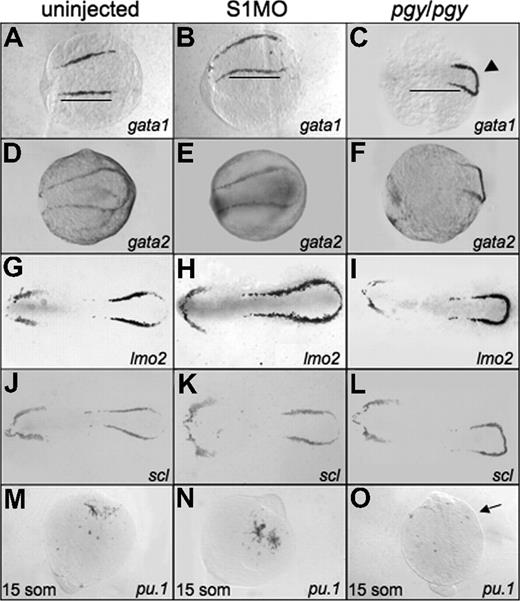
To determine if the increased erythroid cell production seen at 32 hpf in smad1 morphants is related to early expression of the hematopoietic regulatory program, we performed in situ hybridization on smad1 morphant embryos at the 5-somite stage. The expression patterns for the hematopoietic markers show a slight expansion, extending the boundary of expression to the caudal end of the embryo (scl and lmo2). The rostral border of the bilateral stripes (gata1, lmo2; Figure 4B,E) and the width of the pattern in lateral mesoderm (lmo2 and scl; Figure 4H,K) are also extended. None of the anterior lateral mesoderm markers show significant changes in the smad1 morphants, including the pattern of pu.1 expression at the 15-somite stage (Figure 4E,H,K,N).
Early hematopoietic markers are expressed in smad1 morphants and piggytail mutants at the 5-somite stage. Shown are representative embryos following in situ hybridization for various hematopoietic markers at 5 somites using control (uninjected) embryos (A,D,G,J), smad1 morphants (B,E,H,K), and piggytail (pgy/pgy) mutants (C,F,I,L). Transcripts were analyzed for gata1 (A-C), gata2 (D-F), lmo2 (G-I), and scl (J-L). Pgy/pgy embryos were identified by genotyping after hybridization. Smad1 morphants have a modest increase in gata1, lmo2, and scl expression, while pgy/pgy embryos have more caudal expression (◀) of these as well as gata2. In panels A and B, note the increase in gata1 expression in smad1 morphants; the length of the gata1 stripe is longer (B), indicated by the bars that denote the extent of expression in wt. In contrast, pgy/pgy embryos have a shorter region of gata1 expression (C). The anterior lateral plate mesoderm expression of these markers is not effected. In situ hybridization was also performed at the 15-somite stage to detect pu.1 transcripts. (M-O) Levels are equivalent in control embryos (M) and smad1 morphants (N), while pgy/pgy (O) do not express pu.1 (noted by  ). Each panel is a representative of 50 to 75 embryos analyzed per sample. Embryos are oriented: panels A-L are anterior to the left; panels A-F are whole mount; panels G-L are flat mount; and panels M-O are anterior to the top, whole mount.
). Each panel is a representative of 50 to 75 embryos analyzed per sample. Embryos are oriented: panels A-L are anterior to the left; panels A-F are whole mount; panels G-L are flat mount; and panels M-O are anterior to the top, whole mount.
Early hematopoietic markers are expressed in smad1 morphants and piggytail mutants at the 5-somite stage. Shown are representative embryos following in situ hybridization for various hematopoietic markers at 5 somites using control (uninjected) embryos (A,D,G,J), smad1 morphants (B,E,H,K), and piggytail (pgy/pgy) mutants (C,F,I,L). Transcripts were analyzed for gata1 (A-C), gata2 (D-F), lmo2 (G-I), and scl (J-L). Pgy/pgy embryos were identified by genotyping after hybridization. Smad1 morphants have a modest increase in gata1, lmo2, and scl expression, while pgy/pgy embryos have more caudal expression (◀) of these as well as gata2. In panels A and B, note the increase in gata1 expression in smad1 morphants; the length of the gata1 stripe is longer (B), indicated by the bars that denote the extent of expression in wt. In contrast, pgy/pgy embryos have a shorter region of gata1 expression (C). The anterior lateral plate mesoderm expression of these markers is not effected. In situ hybridization was also performed at the 15-somite stage to detect pu.1 transcripts. (M-O) Levels are equivalent in control embryos (M) and smad1 morphants (N), while pgy/pgy (O) do not express pu.1 (noted by  ). Each panel is a representative of 50 to 75 embryos analyzed per sample. Embryos are oriented: panels A-L are anterior to the left; panels A-F are whole mount; panels G-L are flat mount; and panels M-O are anterior to the top, whole mount.
). Each panel is a representative of 50 to 75 embryos analyzed per sample. Embryos are oriented: panels A-L are anterior to the left; panels A-F are whole mount; panels G-L are flat mount; and panels M-O are anterior to the top, whole mount.
Since piggytail embryos lack ventral tissues including blood by 3 dpf, we analyzed if these mutants initiate expression of the early hematopoietic-specific genes. For this purpose, in situ hybridization experiments were carried out using embryos at the 5-somite stage with probes specific for gata1, gata2, lmo2, and scl (Figure 4). Transcripts for each of these genes are present in piggytail embryos. While the overall levels are similar, the pattern appears shifted in the posterior lateral mesoderm of piggytail embryos, and the markers are expressed in a compacted region, with the bilateral stripes forming a connecting loop. Heterozygous embryos show an intermediate pattern (data not shown). In the anterior lateral mesoderm scl, lmo2, and gata2 are expressed normally at this stage in both heterozygous and homozygous embryos (Figure 4F,I,L). However, piggytail embryos, identifiable by morphology at the 15-somite stage, lack pu.1 transcripts compared with wild-type control siblings (Figure 4O). The in situ hybridization results suggest that the defect in embryonic erythropoiesis caused by loss of Smad5 is a relatively late event, since the normal early transcriptional program is activated, albeit displaced.
Initial mesoderm patterning markers are not affected by loss of Smad1 or Smad5, but hematopoietic progenitors are displaced
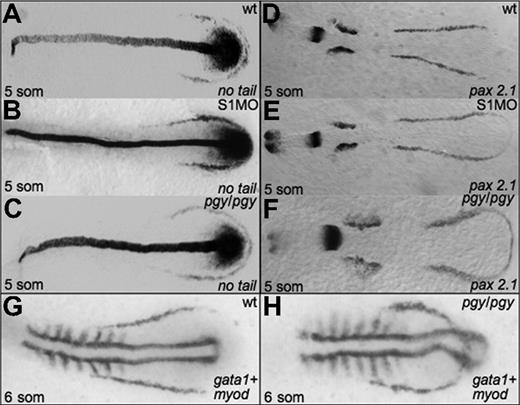
Given the alterations in hematopoiesis seen in both piggytail embryos and smad1 morphants, we analyzed if initial mesoderm markers are expressed normally in these embryos. In situ hybridization was performed at the 5-somite stage for smad1 morphants and piggytail mutants to evaluate transcripts encoded by the pax2.1 (lateral mesoderm), no tail (medial mesoderm), and myod (somitic mesoderm) genes. As shown in Figure 5, both Smad5 mutants and smad1 morphants display relatively normal patterns of pax2.1 and no tail transcripts, indicating that the alterations in erythropoiesis are not secondary to any major alteration in early mesoderm formation. However, the comparative patterns obtained by costaining piggytail embryos for gata1 and myod expression is revealing. Although the anterior boundary of the gata1 expression pattern is located normally in piggytail, the pattern is displaced to lie both more rostral and medial compared to normal, and thereby forms a loop that bisects through the caudal myod expressing region (compare Figure 5G and 5H).
Early mesoderm markers are not affected by Smad1 or Smad5 knockdown. Shown are representative embryos following in situ hybridization for no tail (A-C) or pax2.1 (D-F) at the 5-somite stage in control (A,D), smad1 morphants (B,E), and piggytail mutant embryos (C,F). Pgy/pgy embryos were identified by genotyping after hybridization. Neither Smad1 nor Smad5 loss causes a significant change in the expression pattern of these markers. However, in piggytail embryos, the expression of the hematopoietic marker gata1 is expressed in a compacted pattern that forms a loop bisecting the myod-expressing domain (G,H). A total of 50 embryos per sample were analyzed.
Early mesoderm markers are not affected by Smad1 or Smad5 knockdown. Shown are representative embryos following in situ hybridization for no tail (A-C) or pax2.1 (D-F) at the 5-somite stage in control (A,D), smad1 morphants (B,E), and piggytail mutant embryos (C,F). Pgy/pgy embryos were identified by genotyping after hybridization. Neither Smad1 nor Smad5 loss causes a significant change in the expression pattern of these markers. However, in piggytail embryos, the expression of the hematopoietic marker gata1 is expressed in a compacted pattern that forms a loop bisecting the myod-expressing domain (G,H). A total of 50 embryos per sample were analyzed.
Subsequent hematopoietic programs are altered substantially, but differently, due to loss of Smad1 or Smad5
By 24 hpf, piggytail embryos show a dramatic loss in expression of hematopoietic markers in the ICM. Compared with control embryos, gata1 and lmo2 transcripts are almost eliminated in the caudal region of the embryo (Figure 6C,F), explaining the subsequent lack of primitive erythropoiesis by 32 hpf. The opposite result is observed with the smad1 morphants. The posterior ICM at 24 hpf shows an increase in the levels of gata1 expression (Figure 6B), which correlates with enhanced erythropoiesis documented at 32 hpf. The lmo2 transcript levels are relatively unaffected in smad1 morphants at 24 hpf, indicating that the increase in erythrocytes in smad1 morphants is a specific change in erythropoiesis, and not a change in early progenitor numbers (Figure 6E).
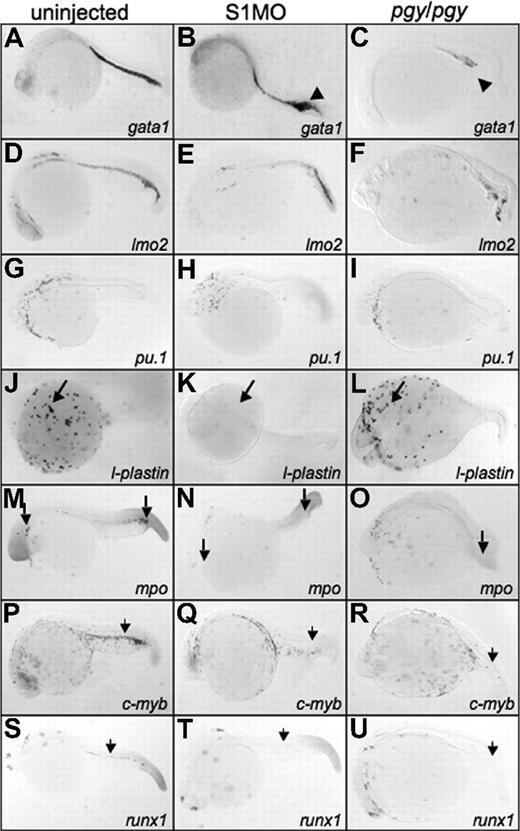
Smad1 morphants have an increase in gata1 expression, but lack l-plastin, while piggytail embryos lose most posterior hematopoietic marker expression at 24 hpf. Shown are representative embryos, 50 to 75 embryos per sample, following in situ hybridization. Uninjected control embryos (left column) (A,D,G,J,M,P,S) were compared with smad1 morphants (S1MO; middle column) (B,E,H,K,N,Q,T), and pgy/pgy mutant embryos (right column) (C,F,I,L,O,R,U) for primitive and definitive hematopoietic transcripts: gata1 (A-C), lmo2 (D-F), pu.1 (G-I), l-plastin (J-L), myeloperoxidase (mpo) (M-O), c-myb (P-R), and runx1 (S-U). At 24 hpf, smad1 morphants have an increase in gata1 expression in the tail (◀ in panel B), while pgy/pgy embryos have a significant decrease (◀ in panel C). In contrast, pgy/pgy have normal l-plastin (long arrow, panel L) and mpo (long arrow, panel O) expression, while smad1 morphants lack all l-plastin expression (long arrow, panel K) and most mpo expression (long arrow, panel N). Loss of either Smad1 or Smad5 leads to a lack of the definitive markers c-myb and runx1 (noted by  ). In all images, the embryos are shown whole mount with anterior to the left.
). In all images, the embryos are shown whole mount with anterior to the left.
Smad1 morphants have an increase in gata1 expression, but lack l-plastin, while piggytail embryos lose most posterior hematopoietic marker expression at 24 hpf. Shown are representative embryos, 50 to 75 embryos per sample, following in situ hybridization. Uninjected control embryos (left column) (A,D,G,J,M,P,S) were compared with smad1 morphants (S1MO; middle column) (B,E,H,K,N,Q,T), and pgy/pgy mutant embryos (right column) (C,F,I,L,O,R,U) for primitive and definitive hematopoietic transcripts: gata1 (A-C), lmo2 (D-F), pu.1 (G-I), l-plastin (J-L), myeloperoxidase (mpo) (M-O), c-myb (P-R), and runx1 (S-U). At 24 hpf, smad1 morphants have an increase in gata1 expression in the tail (◀ in panel B), while pgy/pgy embryos have a significant decrease (◀ in panel C). In contrast, pgy/pgy have normal l-plastin (long arrow, panel L) and mpo (long arrow, panel O) expression, while smad1 morphants lack all l-plastin expression (long arrow, panel K) and most mpo expression (long arrow, panel N). Loss of either Smad1 or Smad5 leads to a lack of the definitive markers c-myb and runx1 (noted by  ). In all images, the embryos are shown whole mount with anterior to the left.
). In all images, the embryos are shown whole mount with anterior to the left.
In piggytail embryos there is normal l-plastin transcript pattern at 24 hpf (Figure 6L), indicating that Smad5 is not required for the formation of blood derived from anterior lateral mesoderm (even though pu.1 expression is absent in piggytail at 15 somites; Figure 6O). In addition, mpo expression in the anterior lateral mesoderm of piggytail embryos is normal, indicating proper granulocyte production. In contrast, smad1 morphants are almost completely defective for anterior blood cells, shown by the lack of l-plastin and mpo expression at 24 hpf (Figure 6K,N). Therefore, Smad1 (and not Smad5) is specifically required in the anterior lateral mesoderm for the differentiation of macrophages and granulocytes.
Compared with control embryos, both piggytail embryos and smad1 morphants show a significant loss of transcripts encoding c-myb and runx1 in the region of the developing dorsal aorta that marks the emergence of the definitive hematopoietic progenitors (Figure 6Q,R,T,U). Therefore, while Smad1 and Smad5 regulate specific functions necessary for different aspects of primitive hematopoiesis, both factors are required for the proper formation of definitive hematopoietic derivatives.
Smad1 can replace Smad5 during embryogenesis and hematopoiesis, but Smad5 cannot function for Smad1
To test whether Smad1 and Smad5 function is exchangeable, we developed a strategy to rescue morphants by coexpression of specific RNAs encoding either smad1 or smad5. For this purpose, cDNA clones were generated that contain mutations in the MO target site for either Smad1 (Smad1*) or Smad5 (Smad5*). These were used in another round of in vitro transcription-translation assays to confirm that the MOs fail to bind functionally to the altered sequences (Figure 7A). We next tested if the morphants could be rescued by ectopic expression of the cognate target RNA (Table 1). When smad1* RNA is coinjected with S1MO, 69% of the embryos observed at 24 or 48 hpf are normal by gross morphology. The other 31% show a smad1 overexpression phenotype similar to that described by Dick et al27 also seen when smad1 or smad1* is injected in the absence of MO (not shown).27 Coinjection of the smad5* RNA was likewise fully capable of rescuing the phenotype caused by S5MO, with 93% of the embryos normal at 24 or 48 hpf. As expected, smad5 RNA is also capable of rescuing the dorsalization phenotype seen in 25% of the offspring from a cross between 2 heterozygote piggytail adults. However, coinjection of smad5 RNA is never able to rescue the smad1 morphant phenotype at any dose tested (0%). In contrast, smad1 RNA very efficiently (79%) rescues the smad5 morphant phenotype. Smad1 RNA is equally capable of rescuing piggytail mutants. Therefore, Smad1 is able to replace Smad5 during embryogenesis, while Smad5 cannot function in place of Smad1.
Smad1 can rescue the hematopoietic defect in smad5 morphants. (A) In vitro transcription-translation of 35S-methionine–labeled Smad proteins. Translation of wobbled-changed Smad1 and Smad5 (Smad1* or S1* and Smad5* or S5*) are not inhibited by their own MO or the opposite Smad MO (Smad1*, lanes 1-3; Smad5*, lanes 4-6). (B-S) Embryos were either uninjected control (left column) (B,E,H,K,N,Q), coinjected with Smad1 MO plus Smad5 mRNA (middle column) (C,F,I,L,O,R), or Smad5 MO plus Smad1 mRNA (right column) (D,G,J,M,P,S) and processed at 24 hpf for in situ hybridization to identify hematopoietic transcripts: gata1 (B-D), lmo2 (E-G), pu.1 (15 somites, panels H- J; 24 hpf, panels K-M), l-plastin (N-P), and c-myb (Q-S). Smad1 is capable of rescuing gata1 (D) (◀), lmo2 (G), pu.1 (J) (◀), and c-myb (S) (small arrow) in smad5 morphants. However, Smad5 is unable to rescue the loss of l-plastin expression in smad1 morphants (O) (long arrow). Each panel is a representative embryo of a 25-embryo sample. All images are shown whole mount. In panels B-G and L-S, anterior is to the left; in panels H-J, anterior is to the top.
Smad1 can rescue the hematopoietic defect in smad5 morphants. (A) In vitro transcription-translation of 35S-methionine–labeled Smad proteins. Translation of wobbled-changed Smad1 and Smad5 (Smad1* or S1* and Smad5* or S5*) are not inhibited by their own MO or the opposite Smad MO (Smad1*, lanes 1-3; Smad5*, lanes 4-6). (B-S) Embryos were either uninjected control (left column) (B,E,H,K,N,Q), coinjected with Smad1 MO plus Smad5 mRNA (middle column) (C,F,I,L,O,R), or Smad5 MO plus Smad1 mRNA (right column) (D,G,J,M,P,S) and processed at 24 hpf for in situ hybridization to identify hematopoietic transcripts: gata1 (B-D), lmo2 (E-G), pu.1 (15 somites, panels H- J; 24 hpf, panels K-M), l-plastin (N-P), and c-myb (Q-S). Smad1 is capable of rescuing gata1 (D) (◀), lmo2 (G), pu.1 (J) (◀), and c-myb (S) (small arrow) in smad5 morphants. However, Smad5 is unable to rescue the loss of l-plastin expression in smad1 morphants (O) (long arrow). Each panel is a representative embryo of a 25-embryo sample. All images are shown whole mount. In panels B-G and L-S, anterior is to the left; in panels H-J, anterior is to the top.
Smad1 rescues the Smad5 mutant or morphant, but Smad5 does not rescue the Smad1 morphant
| Mutant or morphant . | (Co-) Injected with . | n . | Phenotype at 24 and 48 hpf, % . |
|---|---|---|---|
| Smad1 MO | Smad1* RNA | 109 | 69 normal, 31 overexpression phenotype† |
| Smad5 MO | Smad5* RNA | 120 | 93 normal, 6.6 morphant phenotype |
| Smad1 MO | Smad5 RNA | 300 | 100 morphant phenotype |
| Smad5 MO | Smad1 RNA | 160 | 79 normal, 21 morphant phenotype |
| pgy/+ cross | Smad5 RNA | 200 | 100 normal |
| pgy/+ cross | Smad1 RNA | 205 | 78 normal, 1 pgy/pgy phenotype, 21 overexpression phenotype† |
| Mutant or morphant . | (Co-) Injected with . | n . | Phenotype at 24 and 48 hpf, % . |
|---|---|---|---|
| Smad1 MO | Smad1* RNA | 109 | 69 normal, 31 overexpression phenotype† |
| Smad5 MO | Smad5* RNA | 120 | 93 normal, 6.6 morphant phenotype |
| Smad1 MO | Smad5 RNA | 300 | 100 morphant phenotype |
| Smad5 MO | Smad1 RNA | 160 | 79 normal, 21 morphant phenotype |
| pgy/+ cross | Smad5 RNA | 200 | 100 normal |
| pgy/+ cross | Smad1 RNA | 205 | 78 normal, 1 pgy/pgy phenotype, 21 overexpression phenotype† |
Wild-type embryos were injected with the indicated combinations of MO, and capped RNAs or mutants were injected with capped mRNA at the 1-cell stage. The established doses of MOs were used (2 ng Smad1 MO, 1 ng Smad5 MO). The following doses of RNA were used for injection: Smad1*, 450 pg; Smad5*, 300 pg; Smad5, 75 to 300 pg; and Smad1, 75 to 150 pg. Each dose was titrated to avoid an overexpression phenotype and provide optimal rescue. The phenotypes were then scored at 24 and 48 hpf for gross morphology by microscopic examination. The following phenotypes were seen: normal (appearing wild-type), Smad1 overexpression (as described27 ), smad5 morphant (phenocopies pgy/pgy; Figure 2A; text), smad1 morphant phenotype (Figure 2A; text), and pgy/pgy mutant (Figure 2A). The number of embryos scored (n) and percentage displaying each phenotype was tabulated as shown.
Smad1 overexpression phenotype as described by Dick et al.27
All obvious aspects of embryogenesis are restored by the injection of smad1 RNA into smad5 morphant embryos. To evaluate hematopoiesis, in situ hybridization was performed at 24 hpf on embryos derived from coinjected fertilized eggs (Figure 7B-S). Injection of smad1 RNA into smad5 morphants rescued normal expression patterns for gata1, pu.1, and lmo2. In addition, the definitive marker c-myb was restored in the rescued smad5 morphants. These results indicate that Smad1 can function in place of Smad5 during primitive and at least initiation of definitive hematopoiesis in the embryo. In contrast, smad5 RNA was unable to rescue the loss of l-plastin, or fully rescue c-myb expression in the smad1 morphant, further indicating that Smad5 cannot function for Smad1 during embryonic hematopoiesis. We did observe some partial rescue of c-myb expression in the smad1 morphants injected with smad5 RNA (Figure 7R). Since runx1 is a more specific marker of hematopoietic progenitors in the dorsal aorta, we repeated the experiment and found that runx1 was not rescued under these conditions (data not shown), indicating that the partial c-myb signal is not hematopoietic. The lmo2 transcript levels might also be increased modestly in the smad1 morphants injected with smad5 RNA, and this might reflect a minor ventralization caused by Smad5 overexpression.27
The above results show that Smad1 and Smad5 are not exchangeable, and therefore suggest that their different activities regulate distinct sets of target genes. To resolve this question, we compared changes in the global transcript profiles caused by loss of Smad1 or Smad5. RNA was isolated from control or morphant embryos at precisely the 1-somite stage, a time when there are minimal morphologic alterations in morphants compared with wild-type embryos. In addition, we prepared similar cDNA samples from embryos injected with both Smad1 and Smad5 MOs (double-knockdown). The cDNA was used to interrogate Nimblegen (Madison, WI) gene expression arrays consisting of 60mer probes (9 probes per target) covering 43 422 targets. In each experiment, the morphant sample was compared with cDNA generated from stage-matched sibling control embryos. The data obtained from 3 independent experiments for each class was normalized together (Table S3; all data submitted to National Center for Biotechnology Information Gene Expression Omnibus, series accession no. GSE8903) and underwent statistical analysis. Thus, we compared 3 independent biological samples for Smad1, Smad5, and double-knockdown morphants in addition to the matched 9 control samples. Unbiased clustering grouped samples from each experiment according to wild-type, Smad1 morphant, Smad5 morphant, or double-knockdown, as expected (not shown). Taking as a cut-off transcript level changes of 2-fold or more relative to the control samples, we established gene sets that are overexpressed (normally repressed) or underexpressed (normally activated) for Smad1, Smad5, or double-knockdown morphants. The results were tested in an independent set of samples using quantitative RT-PCR to measure the relative expression levels for a set of 13 genes, which confirmed and thus validated the microarray data (Table 2). We next compared the gene sets to define specific gene subsets that are common (overlapped) or distinct (unique). The results are shown in Table 3, and demonstrate clearly that Smad1 and Smad5 single morphants are deregulated for strikingly distinct genetic networks. Of the 702 and 542 genes that are overexpressed in Smad1 or Smad5 morphants, respectively, only 160 (around 25%) are shared, and of the 718 and 461 genes that are underexpressed in the 2 morphants, respectively, only 154 (again around 25%) are in common.
qPCR validation of microarray results
| Gene . | Fold change . | |
|---|---|---|
| qPCR . | Microarray . | |
| Smad1 morphants | ||
| Cyclin G1 | 2.97 | 2.21 |
| Dazl | 3.59 | 9.29 |
| Rap2 inter. protein | 39.3 | 28.6 |
| Runx3 | 0.159 | −2.23 |
| Kruppel-like factor 12 | 0.489 | −2.00 |
| Smaf2 | 0.349 | −2.55 |
| Wnt16 | 0.415 | −2.67 |
| Smad5 morphants | ||
| GDNF receptor α-1a | 1.51 | 2.65 |
| HoxC1a | 1.56 | 2.08 |
| Angiopoietin-like 3 | 3.38 | 3.58 |
| Dlx5 | 0.216 | −10.0 |
| Bmp4 | 0.300 | −5.68 |
| Gata2 | 0.482 | −3.13 |
| Gene . | Fold change . | |
|---|---|---|
| qPCR . | Microarray . | |
| Smad1 morphants | ||
| Cyclin G1 | 2.97 | 2.21 |
| Dazl | 3.59 | 9.29 |
| Rap2 inter. protein | 39.3 | 28.6 |
| Runx3 | 0.159 | −2.23 |
| Kruppel-like factor 12 | 0.489 | −2.00 |
| Smaf2 | 0.349 | −2.55 |
| Wnt16 | 0.415 | −2.67 |
| Smad5 morphants | ||
| GDNF receptor α-1a | 1.51 | 2.65 |
| HoxC1a | 1.56 | 2.08 |
| Angiopoietin-like 3 | 3.38 | 3.58 |
| Dlx5 | 0.216 | −10.0 |
| Bmp4 | 0.300 | −5.68 |
| Gata2 | 0.482 | −3.13 |
A total of 13 genes shown by the microarray experiments to have altered expression in morphant embryos were chosen for validation by quantitative real-time PCR. The qPCR reactions were carried out on cDNA from smad1 or smad5 morphant embryos at the 1-somite stage. Shown is the average fold change in expression calculated from 3 independent experiments, with samples analyzed each time in triplicate. Samples were normalized to β-actin, and wild-type was set to 1. In every case, the differences compared with wild-type are significant by Student t test; P < .05. Also shown is the fold change as determined by microarray with negative fold change indicating underexpression in the morphant samples.
Number of genes altered by Smad1 and/or Smad5 knockdown
| Genes . | No. (%) . |
|---|---|
| Overexpressed | |
| Smad1 knockdown | |
| Total | 702 (100) |
| Unique | 542 (77.2) |
| Overlap with Smad5 knockdown | 160 (22.8) |
| Smad5 knockdown | |
| Total | 557 (100) |
| Unique | 397 (71.3) |
| Overlap with Smad1 knockdown | 160 (28.7) |
| Smad1 + 5 knockdown | |
| Total | 742 (100) |
| Unique | 360 (48.5) |
| Underexpressed | |
| Smad1 knockdown | |
| Total | 718 (100) |
| Unique | 564 (78.6) |
| Overlap with Smad5 knockdown | 154 (21.4) |
| Smad5 knockdown | |
| Total | 461 (100) |
| Unique | 313 (67.9) |
| Overlap with Smad1 knockdown | 154 (33.4) |
| Smad 1 + 5 knockdown | |
| Total | 1006 (100) |
| Unique | 579 (57.6) |
| Genes . | No. (%) . |
|---|---|
| Overexpressed | |
| Smad1 knockdown | |
| Total | 702 (100) |
| Unique | 542 (77.2) |
| Overlap with Smad5 knockdown | 160 (22.8) |
| Smad5 knockdown | |
| Total | 557 (100) |
| Unique | 397 (71.3) |
| Overlap with Smad1 knockdown | 160 (28.7) |
| Smad1 + 5 knockdown | |
| Total | 742 (100) |
| Unique | 360 (48.5) |
| Underexpressed | |
| Smad1 knockdown | |
| Total | 718 (100) |
| Unique | 564 (78.6) |
| Overlap with Smad5 knockdown | 154 (21.4) |
| Smad5 knockdown | |
| Total | 461 (100) |
| Unique | 313 (67.9) |
| Overlap with Smad1 knockdown | 154 (33.4) |
| Smad 1 + 5 knockdown | |
| Total | 1006 (100) |
| Unique | 579 (57.6) |
For each morphant class, the total number of genes is shown that are altered by a 2-fold or higher change in transcript level, as determined by microarray. The number of genes that are unique to each set (ie, those that are not expressed in the other single knockdown), and the number that are present in both sets (overlap) are also shown. For each of the single Smad knockdowns, approximately 75% of the genes altered by MO injection at the 1-somite stage are unique to that subset. For the double-knockdown experiment, the unique genes are those that are not expressed in either of the single knockdowns, and represent genes that are regulated redundantly by Smad1 and Smad5. In this case, around 50% of the genes are unique.
The unique gene sets comprising the most highly changed annotated genes for each morphant class are shown in Table S4. Analysis of these gene lists reveals 2 striking features. First, the loss of Smad5 alone causes a significant decrease in expression for many of the known components of the BMP signaling pathway itself and the known direct targets of the BMP synexpression group, including bmp4, vent2, ved, smad7, cdx2, and eve1. Other early genes known or suspected to be important for establishing a BMP-dependent developmental program are also unique to the Smad5 list, including draculin and the homeobox proteins msh-c, msh-d, and dlx-5. Loss of Smad1 has no effect on this set of genes, but instead targets an alternative set of genes, including histones H1 (decreased) and H3 (increased) as well as a number of genes associated with signaling, adhesion, and cell cycle.
Second, there is also a gene set identified that is unique to the Smad1 plus Smad5 double-knockdown morphants, revealing genes that are regulated redundantly by Smad1 and Smad5. Strikingly, at the top of this list are several known key early hemato-vascular regulatory genes, including scl, lmo2, runx3, and gfi1. This explains why hematopoiesis can be initiated in either set of single morphants, since expression of either Smad1 or Smad5 is sufficient for relatively normal levels of these early regulatory genes. Subsequent development of the erythroid or myeloid programs is dependent on Smad5 or Smad1, respectively, because each also regulates a unique gene set that is not shared or compensated by the other Smad protein.
Discussion
Smad1 and Smad5 are often referred to generically as mediators of the BMP signaling pathway. Using a loss-of-function approach, we found that there are shared functions but also clear differences in the normal roles of these 2 signaling molecules during early embryogenesis, and specifically with regard to hematopoiesis. Neither gene is required to initiate the hematopoietic regulatory program, since in either morphant gata1, gata2, scl, and lmo2 are activated at the appropriate stage, suggesting functional redundancy for this step. Indeed, our microarray experiments confirmed this for scl and lmo2, since only in the double-knockdown morphants were these genes deregulated significantly. However, in the absence of Smad5, the erythroid program is not maintained, and erythropoiesis fails. This defect correlates with the misplacement of the progenitors in the tail, including the “loop” region that connects the 2 bilateral stripes of lateral mesoderm across the region of myod expression that is not normally hematopoietic. This displacement may expose progenitors to an inappropriate stromal environment, leading to hematopoietic failure. Transplantation studies by Hild et al23 indicate that sbn/sbn (smad5 mutant) cells survive in a wild-type environment to form tail tissue and blood. This suggests that the embryonic hematopoietic defect we observe in the smad5 mutants/morphants is not cell autonomous. In adult steady-state hematopoiesis, the picture may be different. Singbrant et al37 reported that the conditional knockout of Smad5 in murine bone marrow causes no defect in adult hematopoiesis. It is possible that Smad1 may compensate for Smad5 in this situation. Our more global analysis of gene expression patterns explains why the program is not maintained in the smad5 morphants, since smad5 controls integral genes of the BMP signaling pathway itself.
Smad1 loss leads instead to enhanced erythropoiesis. While this could be an indirect effect, for example due to increased access of Smad5 with Smad4, an alternative possibility is that Smad1 functions normally to restrict erythropoiesis. With respect to myelopoiesis, macrophage development is dependent on Smad1, while Smad5 is dispensable. Both Smad1 and Smad5 are essential for the initiation of definitive hematopoiesis, as judged by the failure of either morphant to express runx1 or c-myb in progenitors associated with the dorsal aorta. Recently, runx1 was shown to be a direct target of Smad1 signaling.38 Gata2 might also be a direct target gene, based on our kinetic analysis using murine ES cells designed to express Smad1.39
Myeloid defects were described in lost-a-fin (laf) mutants (in which embryos lack expression of the BMP type I receptor Alk8) due to defects in pu.1 expression that is independent of scl.40 Therefore, embryos lacking Smad1, Smad5, or Alk8 express scl normally in the anterior lateral mesoderm, and both smad1 morphants and laf mutants lack anterior myeloid cells, but only laf mutants fail to express pu.1. It is possible that Alk8 activates both smad1 and smad5 redundantly to control expression of pu.1. We note that at 15 somites, piggytail embryos are defective for pu.1 expression, but the pattern recovers by 24 hpf (even 20 hpf; data not shown), which suggests compensation at the earlier stages by Smad1. Activation of either smad1 or smad5 appears sufficient to activate pu.1 in the anterior lateral mesoderm, but only smad1 controls myeloid differentiation.
Some aspects of the differential phenotypes could be explained by when and where (and at what levels) they are expressed. For example, smad5 has a maternal component, while smad1 is not expressed until the shield stage of development. Also, at the 5-somite stage, the smad1 expression is ubiquitous but enhanced in the eyes, somites, and rhombomeres. At 24 hpf, smad1 continues to be expressed in the brain, optic, and otic placodes as well as in the hemangiogenic region and tail tissues27,28 (L.J.M., unpublished data, December 2006).
However, in addition, the 2 proteins clearly have distinct transcriptional regulatory activities, since they are not exchangeable, and the transcript profiling experiments revealed markedly distinct differences in downstream genetic programs. We showed by rescue experiments that Smad1 can functionally replace Smad5, but Smad5 cannot replace Smad1. It was shown previously that injection of smad1 RNA partially rescues the Bmp2b mutant swirl, which again implies the ability of Smad1 to compensate for the loss of Smad5.27 Those authors proposed that smad1 acts later and is actually a target of smad5. This is supported by data indicating that smad5 can activate expression of smad1 in early ectoderm.41 Our transcript-profiling experiments suggest, however, that the epistatic relationship of Smad5 and Smad1 is complex. If Smad1 were fully dependent on the BMP pathway downstream of Smad5, one would have predicted the Smad1 targets to represent a subset of those controlled by Smad5, but this is not the case. Our data suggest that Smad1 regulates a large set of genes independent of Smad5 or the BMP pathway. This is consistent with the fact that Smad1 morphants are not dorsalized, and that hematopoiesis is defective for the anterior blood island but not the ventral blood island. It is also possible that Smad1 or Smad5 relay additional signals. Liu et al14 found that Smad5 is required to block induction of globin mRNA and effectively inhibit hematopoietic progenitor growth in day-4 embryoid body cultures treated with TGFβ1. These data suggest that Smad5 can mediate signaling from either TGFβ or BMP ligands. However, the situation might be more complicated, since loss of Smad5 could lead to additional availability of Smad4 and thus enhanced signaling through Smad2/3.
In this study, we show that Smad1 has a distinct biochemical activity for which Smad5 cannot substitute. Smad5 clearly regulates the BMP signaling pathway itself, while a large component of the Smad1 pathway lies outside of this network. The phenotypes and associated genetic networks regulated by these highly related proteins are different. Currently, there is considerable interest in exploiting the potential for the TGF-β and BMP signaling pathways to alter hemato-vascular development and function (and also tumor biology). Our results show that specificity is imparted at the level of the highly related Smads, which may need to be taken into account when manipulating the upstream pathway.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Nadia Severina for assistance with genotyping and Spartak Kalinin for zebrafish care. We are grateful for the help and advice of Jennifer Tucker with genotyping of the smad5 mutants. We also thank members of the Evans Laboratory for critical analysis of the data and helpful suggestions, and appreciate the help of Drs. Leonard Zon (Howard Hughes Medical Institute, Children's Hospital, Boston, MA), Yu Chen (Albert Einstein, Bronx, NY), Hanh Nguyen (Erlangen, Germany), Thomas Look (Dana Farber Cancer Institute, Boston, MA), and Andrew Perkins (Queensland, Australia), for reagents. Leonard Zon also provided the gata1:DsRed fish strain.
This work was supported by a grant to T.E. from the National Institutes of Health (HL056182) and to M.C.M. from the National Institutes of Health (GM56326). L.J.M. is supported by an MSTP training grant (T32 GM007288). M.E.F. is supported by an ASH Fellow Scholar Award in Basic Research.
National Institutes of Health
Authorship
Contribution: All authors contributed significantly to this work. L.J.M. designed the study, performed the experiments, and wrote the manuscript. S.G. carried out microarray experiments. M.E.F. analyzed the microarray data. M.C.M. designed the study, provided essential unpublished reagents, and edited the manuscript. T.E. designed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Todd Evans, Department of Developmental and Molecular Biology, Albert Einstein College of Medicine, 1300 Morris Park Ave, Chanin Rm 501, Bronx, NY 10461; e-mail: tevans@aecom.yu.edu.

![Figure 2. Depletion of Smad1 or Smad5 causes distinct embryonic phenotypes. (A) Embryos were either control (wild-type [wt]; row 1) or injected with 2 ng translation-blocking Smad1 MO (S1MO), which causes a phenotype first visible at the bud stage by elongation of the embryo (row 2). This phenotype is more severe starting around 18 hpf, with a ragged dorsal edge, neural degeneration, and a widened notochord (row 2; ). By 24 hpf, heart, brain, and endoderm defects are visible ( indicate neural degeneration and yolk stalk regression). These embryos go on to die at 3 to 4 dpf. Alternatively, embryos were injected with 1 ng translation-blocking Smad5 MO (S5MO), which causes a known dorsalization phenotype (row 3) similar to that of characterized smad5 mutants. This phenotype is first visible around the 5-somite stage with a tail defect () and a more caudal arrangement of somites. By the 18 hpf stage, the curvature of the tail is visible, and by 24 hpf, the lack of ventral tissues is evident (). The Smad5 MO phenocopies the piggytail mutant (row 4). Each panel is a representative photograph of phenotype and stage shown. More than 200 embryos were observed for each panel. (B-E) Double knockdown of both Smad1 and Smad5 at half-dose causes lethality at 15 hpf. Shown are wt (B,D) and double-morphant (S1MO + S5MO) (C,E) embryos at 15 hpf and 24 hpf, as indicated. The phenotype just prior to death has combined features of each individual knockdown. Less than 1% of double morphant embryos survive to 24 hpf, as in panel E. Approximately 150 double-injected embryos were observed, and a representative is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/12/10.1182_blood-2007-04-085753/4/m_zh80240710530002.jpeg?Expires=1769099017&Signature=xLUvhw-50J3c5wvACwIH5GwfZ1KP5wRbCyjGjBdkljYKjlskI-PZ8y24xKA9RL2h8S9TK6hVxMjFl2EtFWUAk1NNJOywkJoydJxNi2Ds5eibYauodRPnq0QR36RkbBbeaR61TP5erxCkkf4ewNLM6st1VeMGa-2u90JQbHqlfKIcudQvUgBER7FbG442~a10mYjICRAceDQU0u9rxDBvTqiJZc~-Z3vU1pbQIj1fzKTos4HeAwsiI7CHTvuZATvwEF9LeZcTb0h-HDHrRyXtFnVhDcSa3jNH1sZ~8GQppoqOMnAdOCXo~XO~fClDZR40m2bO3csnWKr5gJ7ObLjTIA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal