Abstract
Aim To develop a DNA-based method for measurement of minimal residual disease (MRD) in CML
Background Currrent RNA-based methods for quantification of the BCR-ABL translocation have a number of disadvantages, including the potential for RNA degradation, requirement for a reverse transcription step, uncertain sensitivity and specificity, and only an indirect relationship between assay result and cell number.
Methods Tagged PCR primers were designed to span the common 3kb breakpoint region of BCR and corresponding 140kb breakpoint region of ABL. Primers were spaced to bind at approximately 500bp intervals in order to limit the size of the target sequence and ensure efficient amplification. PCR was performed with pools of 1-6 BCR forward primers and pools of 24–282 ABL reverse primers. The first round products were diluted and amplified in a second round using similarly sized pools of nested primers. The resulting products were sequenced, the sites of breakpoint identified and 3 pairs of PCR primers were synthesised for each patient. A 3 round nested quantitative PCR was established, in which amplification remained exponential until the third round, which was a quantitative real-time PCR using a Taqman probe or EvaGreen to measure fluorescence. Dilutions of the diagnosis DNA were used to establish a standard curve. Test samples, corresponding to different levels of MRD, were artificial mixtures made by diluting diagnosis DNA in up to 25 mcg of normal peripheral blood DNA. Specificity was determined by quantifying BCR-ABL sequences in 25 mcg of normal peripheral blood DNA.
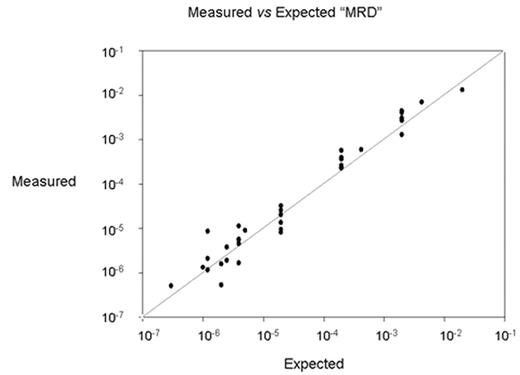
Results Samples from 23 patients are being studied and the translocation breakpoint has thus far been isolated and sequenced in 15. Study of the remaining patients is in progress. No favoured breakpoint site or sequence has as yet become evident, although the numbers are small. The results of assays of mixtures of DNA from 9 patients are shown in Fig 1.
There results follow a 1:1 linear relationship (line shown) between expected and measured MRD levels down to approximately 10−6. Use of a Taqman probe or EvaGreen for quantification gave similar results DNA samples from 15 normal individuals were tested against each of the set of BCR-ABL primers from 3 patients. No BCR-ABL translocations were detected.
Conclusions
Isolation and sequencing of the BCR-ABL translocation breakpoint is feasible in the majority of patients with CML. Short-range multiplex PCR is a promising strategy for doing this.
Sensitive and specific DNA-based measurement of BCR-ABL sequences is possible for CML patients in whom the BCR-ABL breakpoint sequence can be determined.
The clinical utility of this approach to measurement of MRD in CML remains to be determined.
Author notes
Disclosure:Ownership Interests: Morley and Flinders University hold equity in Monoquant P/L. Research Funding: Bartley, Martin-Harris and Latham are employed by Flinders University under a grant from Monoquant P/L. Membership Information: Morley is a Director of Monoquant P/L. Financial Information: Monoquant P/L has lodged a provisional patent covering the intellectual property described.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal