Abstract
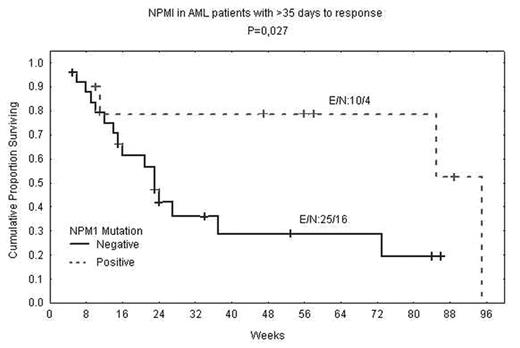
Nucleotides insertion in the nucleophosphamin (NPM1) gene has been reported in about one third of patients with acute myeloid leukemia (AML). Multiple studies showed that the presence of NPM1 mutations associated with better outcome in patients with AML. Studies reported to date have analyzed leukemic cells obtained from bone marrow or peripheral blood. We tested for mutations in the NPM1 gene using peripheral blood plasma and compared results with clinical outcome from a single institution. Analyzing plasma from 98 newly diagnosed patient with AML showed NPM1 mutation in 24 (23%) of patient while only one (4%) of 28 previously untreated patients with myelodysplastic syndrome (MDS) showed NPM1 mutation. Compared with peripheral blood cells, 2 (8%) of the 24 positive patients were negative by cells; none were positive by cells and negative by plasma. Most of the mutations detected (45%) were in patients with FAB classification M2, M4 and M5. In addition to the reported 4 bp insertion, we also detected 4 bp deletion in one patient in cells and plasma. Patients with NPM1 mutation had a significantly higher white blood cell count (P = 0.0009) and a higher blast count in peripheral blood (P = 0.002) and in bone marrow (P = 0.002). Blasts in patients with NPM1 mutant expressed lower levels of HLA-DR (P = 0.005), CD13 (P = 0.02) and CD34 (P < 0.0001), but higher CD33 levels (P = 0.0004). Patients with NPM1 mutation appear to have better chance of responding to standard therapy (P = 0.06). Event free survival of patients with NPM1 mutation was longer (P = 0.056) than in patients with intermediate cytogenetic abnormalities. The most striking difference in survival was in patients who required >35 days to respond to therapy (Figure). Survival was significantly longer in patients with NPM1 mutation requiring >35 days to respond (P = 0.027). This data not only support that NPM1 plays a significant role in the biology and clinical behavior of AML, but also show that plasma DNA is enriched with leukemia-specific DNA and is a reliable source for testing.
Author notes
Disclosure: No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal