HIV-1 recognition by, interaction with, and/or infection of CD4+CCR5+ tissue macrophages and dendritic cells (DCs) play important roles in HIV-1 transmission and pathogenesis. By comparison, circulating CD4+CCR5+ monocytes appear relatively resistant to HIV-1, and a fundamental unresolved question involves deciphering restriction factors unique to this precursor population. Not only do monocytes, relative to macrophages, possess higher levels of the innate resistance factor APOBEC3G, but we uncovered APOBEC3A, not previously associated with anti-HIV activity, as being critical in monocyte resistance. Inversely correlated with susceptibility, silencing of APOBEC3A renders monocytes vulnerable to HIV-1. Differences in promiscuity of monocytes, macrophages, and DCs can be defined, at least partly, by disparities in APOBEC expression, with implications for enhancing cellular defenses against HIV-1.
Introduction
The complex life cycle of HIV-1 involves critical functional interactions with CD4+ host-cell factors. In addition to CD4+ T cells, CD4+CCR5+ macrophages represent a primary target and host of HIV-1. Infected macrophages replicate copious amounts of virus at their surface and at intracellular membranes where the virions accumulate in vesicles.1 Macrophages are not typically subject to viral-induced death and may persist as reservoirs of virus in tissues for long periods of time.2,3 In addition, infected macrophages may be resistant to antiviral agents,3,,–6 and the unique attributes and factors that are enabling for this persistent viral host remain elusive. Critically, all monocytic cells are neither equally permissive to HIV-1 nor supportive of the viral life cycle. In vitro or in vivo, dendritic cells (DCs) may or may not become infected, depending on maturational status7 ; peripheral-blood monocytes are nearly impervious (< 1% HIV-1 DNA+)8,9 ; and of longstanding interest is the enhanced susceptibility of macrophages to HIV-1 compared with immature monocytes, the basis of which remains a mystery. The fact that macrophages, in culture and in tissues, are more susceptible to infection than monocytes cannot be attributed to levels of membrane CD4 or HIV-1 coreceptor expression10 or multiple other criteria that have been considered.11
This fundamental question has obvious significance in that if the monocyte-resistance factor(s) can be identified, opportunities may emerge for manipulating susceptible myeloid populations to impose a restrictive barrier to HIV-1. Limited evidence supports an early postentry block that occurs in association with or shortly after reverse transcription (RT).8 Beyond the well-established CD4 and CCR5/CXCR4 entry requirements, recent evidence implicates additional membrane and intracellular factors that influence early host-cell responsiveness to productive infection.11,,,,,–17 Among the potential innate intracellular viral antagonists, initially characterized in T cells18 and more recently in monocytes,19,20 are cytidine deaminases that edit viral RNA and mutate DNA. These cytoplasmic apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (APOBEC) subunits, particularly APOBEC3G (hA3G), become incorporated into virions, leading to mutation of nascent HIV-1 DNA formed during reverse transcription and its subsequent degradation.18,21,22 HIV-1, in turn, inhibits hA3G via HIV-1–encoded viral infectivity factor (Vif)–dependent and–independent pathways to thwart this antiviral defense within the target cell. Vif not only facilitates 26S proteasome–mediated degradation of hA3G but also diminishes its synthesis to collectively exclude hA3G incorporation into the budding virions.23,–25 Nonetheless, whether differential HIV-1 susceptibility in myeloid populations could be attributed to constitutively expressed hA3G or any other components of this defensive superfamily had not been addressed.
In this study, by oligonucleotide microarray analysis, we identify unique patterns of expression of these innate intracellular antiviral molecules dependent on myeloid maturation status. In fact, an apparent inverse correlation between certain APOBEC3 family proteins and susceptibility to productive HIV-1 infection emerged. Monocytes with high levels of hA3G were refractory to HIV-1, as were immature DCs, whereas macrophages with less hA3G were receptive hosts, consistent with the ability to reverse monocyte resistance by silencing hA3G. However, hA3G was not solely responsible for viral resistance in that we uncovered a previously unrecognized antiretroviral function for another member of the APOBEC3 superfamily, APOBEC3A (hA3A), which is highly expressed in monocytes and decreases during maturation. Inhibiting or enhancing hA3A dictates a differential response to HIV-1, overruling constitutive resistance and susceptibility patterns. Collectively, these cytidine deaminases represent a potent innate barrier to HIV-1 infection in monocytes, which unfortuitously diminishes during monocyte-to-macrophage transition, resulting in a defenseless population of macrophages that can become viral havens. Identification of agents capable of inducing or restoring expression of these pivotal molecules may provide opportunities for protecting macrophages from HIV-1 and its pathogenic sequelae.
Materials and methods
Monocyte isolation and differentiation to macrophages and dendritic cells
Human peripheral-blood mononuclear cells (PBMCs), obtained by leukapheresis of healthy volunteers (Department of Transfusion Medicine, National Institutes of Health [NIH], Bethesda, MD), were diluted in endotoxin-free PBS without Ca2+ or Mg2+ (BioWhittaker, Walkersville, MD) and separated by density centrifugation on lymphocyte sedimentation medium (Organon Teknika, Durham, NC) before elutriation.16 For monocyte cultures, the elutriated monocytes (30 × 106/mL) were suspended in 15-mL round-bottom tubes (Becton Dickinson, Franklin Lakes, NJ) in DMEM (BioWhittaker) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Invitrogen, Gaithersburg, MD), 2 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Sigma, St Louis, MO) at 37°C and 5% CO2. For differentiated macrophages, monocytes were adhered in serum-free DMEM in 6-well (6 × 106 cells/well) plates (Corning, Cambridge, MA) for 2 to 4 hours, 10% FBS was added, and the cells were cultured for 6 to 7 days.16,26 To generate DCs, monocytes were plated in 6-well (6 × 106 cells/well) plates in RPMI 1640 medium (BioWhittaker) supplemented with 10% FBS, l-glutamine, antibiotics, 1000 U/mL GM-CSF, and 500 U/mL IL-4 (PeproTech, Rocky Hill, NJ; National Cancer Institute–Frederick Cancer Research and Development Center [NCI-FCRDC], Frederick, MD) for 6 to 7 days (S.N., et al, manuscript in preparation). On alternate days, half of the media was removed and replaced with fresh media/cytokines, and cells collected by centrifugation at 300g of spent media were added back. DCs generated by this method were CD1a+, CD86+, HLA-DR+, DC-SIGN+, CD3−, CD19−, CD14low/−, and macrophages were CD14+, CD68+ prior to stimulation as monitored by flow cytometry (FACSCalibur; Becton Dickinson, Sunnyvale, CA). Monocytes, DCs, and macrophages were left untreated (medium control), stimulated with 100 ng/mL Escherichia coli LPS (strain 055:B5; Sigma) for 1 hour or with IFNα (NCI-FCRDC; R&D Systems, Minneapolis, MN) or IFNγ (10 ng/mL; NCI-FCRDC) for 4 hours to evaluate the primary transcriptional response.
Oligonucleotide microarrays
Cells were harvested and RNA was isolated using Trizol, and total RNA was further purified using the RNeasy Mini Columns (Qiagen, Valencia, CA). Preparation of biotin-labeled cRNA, hybridization, and scanning were performed according to protocol (Affymetrix, Santa Clara, CA). Briefly, 10 μg of total RNA was used to generate double-stranded cDNA using the One-Cycle cDNA Synthesis Kit and the oligo(dT)24 primer (Affymetrix) containing a 3′ T7 RNA polymerase promoter site. Biotin-labeled cRNA probes were produced from cDNA using the IVT labeling kit (Affymetrix). The probes were purified, fragmented, and hybridized to Affymetrix Plus 2.0 Microarrays that display more than 47 000 unique probe sets and expressed sequence tags (ESTs) for 16 hours. Microarrays were washed, stained using Affymetrix Fluidics Station 450, and fluorescence measured using the Affymetrix GeneChip scanner.
Oligonucleotide microarray data processing and analysis
Affymetrix MAS5 Signal and Present Call values were stored in the NIHLIMS, an internal NIH database for storage and retrieval of chip data. Data were statistically analyzed using the MSCL Analyst's Toolbox27 and the JMP statistical software package (SAS, Cary, NC; http://www.jmp.com). The results for the chips were retrieved, and the Affymetrix Signal values were subjected to an adaptive variance-stabilizing, quantile-normalizing transformation termed “S10.”28 A principal components analysis (PCA) of the transformed, normalized data was performed to visualize potential outlier chips and to assess the overall organization of results. Results for 11 probe sets on the U133Plus2 array with annotations related to APOBEC genes were collected (1553831_at - APOBEC3D, 206160_at - APOBEC2, 215579_at - APOBEC3G, 207158_at - APOBEC1, 214994_at - APOBEC3F, 243912_x_at - APOBEC3F, 204205_at - APOBEC3G, 214995_s_at - APOBEC3G, 209584_x_at - APOBEC3C, 206632_s_at - APOBEC3B, 210873_x_at - APOBEC3A), as were results for 45 probe sets indicated in Figure 2A (203547_at - CD4, 206991_s_at - CCR5, 205483_s_at - G1P2, 224731_at - HMGB1, 224734_at - HMGB1, 204645_at - CCNT2, 214638_s_at - CCNT2, 213743_at - CCNT2, 229184_at - CCNT2, 200680_x_at - HMGB1, 201872_s_at - ABCE1, 201873_s_at - ABCE1, 212888_at - DICER1, 202284_s_at - CDKN1A, 210705_s_at - TRIM5, 201758_at - TSG101, 206967_at - CCNT1, 214938_x_at - HMGB1, 213229_at - DICER1, 206061_s_at - DICER1, 216260_at - DICER1, 200679_x_at - HMGB1, 33304_at - ISG20, 204698_at - ISG20, 212808_at - FLJ14639, 212809_at - FLJ14639, 217526_at - FLJ14639, 217527_s_at - FLJ14639, 207416_s_at - NFATC3, 210555_s_at - NFATC3, 210556_at - NFATC3, 225137_at - NFATC3, 225139_at - NFATC3, 225141_at - NFATC3, 208003_s_at - NFAT5, 215092_s_at - NFAT5, 224984_at - NFAT5, 1405_i_at - CCL5, 1555759_a_at - CCL5, 205114_s_at - CCL3, 204103_at - CCL4, 209664_x_at - NFATC1, 210162_s_at - NFATC1, 211105_s_at - NFATC1, 209477_at - EMD). Hierarchical clustering of S10 values relative to probe-set mean for monocytes, DCs, and macrophages, either unstimulated or stimulated with LPS, was performed using the Ward method and a heat map calculated using JMP. Parallel line plots of the gene-expression values in S10 scale were prepared for the 3 cell types from each of 3 donors for 6 of the 11 APOBEC probe sets (210873_x_at, 206632_s_at, 209584_x_at, 214994_at, 204205_at, 214995_s_at) registering as “Present” by Affymetrix software in more than 50% of the samples. For each cluster in Figure 2A, corresponding parallel line plots were created using the relative gene-expression values. Bar graphs report fold changes that were calculated by taking the antilog of the difference between treatment group mean and control mean in S10 units.
Infection with HIV-1
Cells were incubated with HIV-1BaL(HIV-1) at 103/50% tissue culture infective dose (TCID50)/mL (ABI, Columbia, MD) in DMEM containing 10% FCS for 2 hours at 37°C. After infection, cells were washed with PBS and cultured in DMEM containing 10% FCS for up to 14 days.15,16 IFNα and IFNγ were added at indicated concentrations.19 To measure viral infectivity, supernatant p24 levels were determined with HIV-1 p24 enzyme-linked immunosorbent assay (ELISA) kits (Perkin Elmer Life Sciences, Boston, MA). For the polymerase chain reaction (PCR)–based assay for newly synthesized viral DNA,19,26 DNase-treated HIV-1BaL (100 μL; 104/mL) was added to macrophages for 2 hours and the cells were washed 3 times with PBS, treated with trypsin-EDTA (0.05% trypsin, 0.53 mM EDTA; 5 minutes, 24°C) to remove noninternalized virus particles, washed twice with DMEM and 10% FCS, and incubated 1 to 3 days. Extracted DNA (1 μg) was subjected to nested-primer PCR amplification (25 μL total volume). The first 35-cycle round of amplification used primers corresponding to the env gene (nucleotides [nt's] 8838 to 8358, HIV-1HXB2 sequence) and the U3 region of the 3′ long terminal repeat (nt's 9533 to 9558). Each cycle consisted of denaturation for 1 minute at 94°C, annealing for 1 minute at 55°C, and elongation for 1 minute at 72°C. PCR reactions (2.5 μL each) from the first amplification were then subjected to a second 30-cycle amplification using primers (5′ primer, nt's 8754 to 8782; 3′ primer, nt's 9436 to 9457) located within the nef gene. PCR products (∼730 bp) were visualized by ethidium bromide staining after agarose gel electrophoresis.
RNA isolation and RT-PCR
Total RNA was extracted and for RT-PCR, the GeneAmp RNA PCR kit (Perkin Elmer, Branchburg, NJ) was used with a total of 0.15 to 0.3 μg RNA from each sample for first-strand cDNA synthesis. The cDNA was then divided and used for PCR amplification of APOBEC3G and other APOBEC3 family genes. PKR and GAPDH were used as controls. All PCR was performed by 30-cycle amplification, except that 25 cycles were used for GAPDH (94°C for 4 minutes followed by 30 cycles of 94°C for 30 seconds, 55°C for 3 seconds, 72°C for 30 seconds, and finally, extension at 72°C for 10 minutes).19 Primers were as follows: APOBEC3A: 5′-TTC TTT GCA GTT GGA CCC GG-3′ (forward), 5′-CTC ATC TAG TCC ATC CCA GG-3′ (reverse); APOBEC3F: 5′-TAC GCA AAG CCT ATG GTC GG-3′ (forward), 5′-GCT CCA AGA TGT GTA CCA GG-3′ (reverse); APOBEC3G: 5′-TTA CCT GCT TCA CCT CCT GG-3′ (forward), 5′-TCA TCT AGT CCA TCC CAG GG-3′ (reverse); PKR: 5′-GCC TTT TCA TCC AAA TGG AAT TC-3′ (forward), 5′-GAA ATC TGT TCT GGG CTC ATG-3′ (reverse); GAPDH: 5′-CCT TGG AGA AGG CTG GGG-3′ (forward), 5′-CAA AGT TGT CAT GGA TGA CC-3′ (reverse).
Western blot for APOBEC proteins
Cells were lysed with ice-cold buffer containing 10 mM HEPES (pH7.9), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.5 mM PMSF, 1 mM DTT, and 0.5% NP40. Fifteen micrograms protein was used for electrophoresis (10% polyacrylamide gel) and blotted onto nitrocellulose membranes. After blocking with 5% milk in Tris-buffered saline with 0.05% Triton X-100 (TBS-T), membranes were probed with monoclonal anti-APOBEC3G at 1:1000 (Immunodiagnostics, Woburn, MA), polyclonal anti-PKR (1:1000; Cell Signaling, Beverly, MA), or anti-APOBEC3A (1:1000; SynPep, Dubin, CA) at 4°C overnight. Membranes were washed with TBS-T 3 times for 10 minutes, followed by secondary antibodies conjugated with HRP (Santa Cruz Biotechnology, Santa Cruz, CA) and detected by chemiluminescence (Pierce, Rockford, IL).
siRNA inhibition of APOBEC3A and APOBEC3G
Cells (10 × 106 cells) were treated with 12 μL specific or control small interfering RNA (siRNA; 20 nM; Qiagen) in 100 μL Dendritic Nucleofector Solution (Amaxa Biosystems, Rockville, MD) or siRNA buffer only at room temperature. hA3G siRNA was synthesized as described19 and 4 hA3A siRNAs were synthesized from the following sequences: hA3A-1 siRNA, r(GCAGUAUGCUCCCGAUCAA)dTdT/r(UUGAUCGGGAGCAUACUGC)dTdT; hA3A-2 siRNA r(AGUACACAAUAGUAAGAUU)dTdT/ r(AAUCUUACUAUUGUGUACU)dTdG; hA3A-3 siRNA, r(CGGUCAAGAUGGACCAGCA)dTdT/r(UGCUGGUCCAUCUUGACCG)dAdG; and hA3A-4 siRNA, r(GGCUUCAUAUCUAGACUAA)dTdT/ r(UUAGUCAGAUAUGAAGCC)dAdA. The targeted APOBEC3A sequences (NM_145699) are as follows: 798 to 818 bp, 1178 to 1198 bp, 206 to 226 bp, and 1254 to 1273 bp from 5′ end, respectively. One hundred–microliter aliquots were added to an electroporation cuvette for electroporation using the U-02 program.15,19 Following electroporation, cells were added to 24-well plates containing prewarmed RPMI 1640. After incubation in a humidified 37°C and 5% CO2 incubator for 3 hours, the medium was changed to complete DMEM (1.5 mL/well) for 6 days of culture prior to infection with HIV-1.
Results
Differential susceptibility of monocytes, macrophages, and DCs to HIV-1
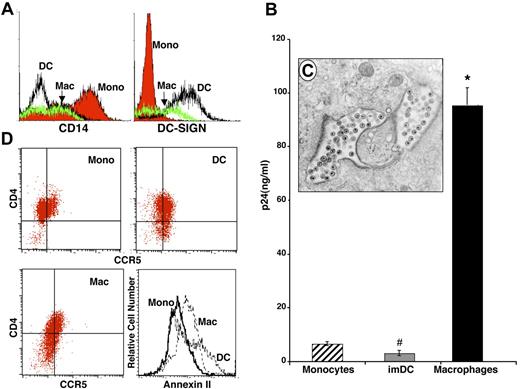
Immature, nonstimulated monocytes purified from PBMCs were phenotyped by flow cytometry (Figure 1A, CD14+) and, from the same donor monocyte pool, aliquots were cultured under conditions known to induce differentiation to macrophages (6 to 7 days adherence in 10% FBS) or DCs (6 to 7 days culture in media containing IL-4 and GM-CSF). At the time of harvest, these populations were morphologically and phenotypically DC-SIGN+ immature DCs and DC-SIGNlowCD14lowCD68+ macrophages (Figure 1A). In our system, after exposure to equivalent amounts of R5 HIV-1BaL, freshly isolated blood monocytes exhibited minimal evidence of infection, and immature DCs in suspension also typically resisted HIV-1 (Figure 1B; monocytes compared with DCs, P = not significant [NS]). However, as anticipated, the majority of donor-matched macrophages contained intracellular virions (Figure 1C) and expressed high levels of HIV-1 p24 (Figure 1B, day 14 shown, monocytes vs macrophages, P < .05). These differences in HIV-1 susceptibility cannot be attributed to expression levels of the primary HIV-1 receptor CD4 or the chemokine coreceptor CCR5, which appear not to be substantively different among the 3 populations (Figure 1D). In fact, CD4 and/or CCR5 typically decrease during macrophage maturation, despite increasing susceptibility to HIV-1, although the macrophage cofactor annexin II15 is expressed at higher levels on macrophages and DCs (Figure 1D).
Differential HIV infection of monocytes, DCs, and macrophages. Peripheral-blood monocytes were processed immediately or cultured as described (“Materials and methods, Monocute isolation and differentiation to macrophages and dendritic cells”) to generate immature DCs and differentiated macrophages. (A) By flow-cytometry analysis using antibodies specific to CD14 and DC-SIGN, the 3 populations were phenotypically distinct. (B) Cultures of monocytes, DCs, and macrophages were incubated with HIV-1BaL for 90 minutes, washed, and incubated for 12 to 14 days. Media aliquots were removed every third day for p24 ELISA and replaced with fresh DMEM containing antibiotics and serum. Representative of 4 experiments (day 14 after infection; *P < .001, #P < .05) (SEM). (C) Transmission electron microscopy (EM) of macrophage infected with HIV-1 for 10 days, demonstrating intracellular budding and accumulation of virions (original magnification, × 20,000) using a Zeiss EM10 Microscope (LEO Electron Microscopy, Oberkochen, Germany). (D) Dual-color flow-cytometry analysis of CD4 and CCR5 expression and single-color annexin II staining on monocytes, macrophages, and DCs.
Differential HIV infection of monocytes, DCs, and macrophages. Peripheral-blood monocytes were processed immediately or cultured as described (“Materials and methods, Monocute isolation and differentiation to macrophages and dendritic cells”) to generate immature DCs and differentiated macrophages. (A) By flow-cytometry analysis using antibodies specific to CD14 and DC-SIGN, the 3 populations were phenotypically distinct. (B) Cultures of monocytes, DCs, and macrophages were incubated with HIV-1BaL for 90 minutes, washed, and incubated for 12 to 14 days. Media aliquots were removed every third day for p24 ELISA and replaced with fresh DMEM containing antibiotics and serum. Representative of 4 experiments (day 14 after infection; *P < .001, #P < .05) (SEM). (C) Transmission electron microscopy (EM) of macrophage infected with HIV-1 for 10 days, demonstrating intracellular budding and accumulation of virions (original magnification, × 20,000) using a Zeiss EM10 Microscope (LEO Electron Microscopy, Oberkochen, Germany). (D) Dual-color flow-cytometry analysis of CD4 and CCR5 expression and single-color annexin II staining on monocytes, macrophages, and DCs.
APOBEC3 levels and susceptibility to HIV-1
In order to distinguish potentially unique maturation-dependent genotypic and functional profiles underlying cells' response to pathogens, we compared global gene expression among monocytes, macrophages, and DCs. Among the cellular genes previously associated with macrophage HIV resistance (high-mobility group box 1 [HMGB1], Dicer 1, TRIM5α)29,,,–33 and susceptibility (CD4, CCR5, nuclear factor–activated T cells [NFATs], tumor susceptibility gene [TSG101], emerin [EMD]),11,34,–36 many were not differentially expressed among the 3 cell types, although monocytes expressed higher levels of both resistance (HMGB1, CCL5, ISG20) and susceptibility (FLJ14639 [NFAT], cyclin T2 [CCNT2]) genes, and 1 probe set for Dicer, CCL4, and several probe sets for NFATC3 were significantly overexpressed in DCs (Figure 2A-B; hierarchical clustering of 45 probes with heat map and parallel plots). A second distinct, nonoverlapping probe set for Dicer appears to have higher expression in monocytes, suggestive of alternative splicing control of expression. Each of these genes individually or in concert may influence the differential susceptibility of myeloid populations to HIV, although the expression patterns did not clearly define maturation-dependent susceptibility.
HIV susceptibility and resistance gene expression. (A) Monocytes, DCs, and macrophages were prepared and RNA was extracted and processed for microarray analysis using the Affymetrix system. Relative transformed expression intensity values were calculated within probe set (row) and hierarchically clustered (“Materials and methods, Oligonucleotide microarray data processing and analysis”), and the corresponding heat map and dendrogram are shown here (red indicates above-average expression; black, average expression; and green, below-average expression for genes associated with HIV resistance and susceptibility). Some genes were represented by multiple probe sets that often fall within the same cluster. Those that show distinct expression behavior may suggest the presence of splice variants (eg, Dicer 1 in clusters 2 and 4). (B) Using the clusters from panel A, parallel line plots by cluster-compare gene-expression profiles in monocytes, DCs, and macrophages from 3 donors. (C) Monocytes, DCs, and macrophages were unstimulated or treated with LPS for 1 hour and RNA was extracted and processed for microarray analysis using the Affymetrix system. Data were treated as in panel A. Data shown are for all 11 probe sets corresponding to the APOBEC cytidine deaminase gene superfamily, represented on the U133Plus2 chip. (D) Parallel line plot comparing APOBEC3 family gene-expression profiles in monocytes, DCs, and macrophages from 3 donors for the 6 probe sets having more than 50% present calls (206632_s_at - 3B, 204205_at - 3G, 209584_x_at - 3C, 210873_x_at - 3A, 214995_s_at - 3G/ 3F, 214994_at - 3F).
HIV susceptibility and resistance gene expression. (A) Monocytes, DCs, and macrophages were prepared and RNA was extracted and processed for microarray analysis using the Affymetrix system. Relative transformed expression intensity values were calculated within probe set (row) and hierarchically clustered (“Materials and methods, Oligonucleotide microarray data processing and analysis”), and the corresponding heat map and dendrogram are shown here (red indicates above-average expression; black, average expression; and green, below-average expression for genes associated with HIV resistance and susceptibility). Some genes were represented by multiple probe sets that often fall within the same cluster. Those that show distinct expression behavior may suggest the presence of splice variants (eg, Dicer 1 in clusters 2 and 4). (B) Using the clusters from panel A, parallel line plots by cluster-compare gene-expression profiles in monocytes, DCs, and macrophages from 3 donors. (C) Monocytes, DCs, and macrophages were unstimulated or treated with LPS for 1 hour and RNA was extracted and processed for microarray analysis using the Affymetrix system. Data were treated as in panel A. Data shown are for all 11 probe sets corresponding to the APOBEC cytidine deaminase gene superfamily, represented on the U133Plus2 chip. (D) Parallel line plot comparing APOBEC3 family gene-expression profiles in monocytes, DCs, and macrophages from 3 donors for the 6 probe sets having more than 50% present calls (206632_s_at - 3B, 204205_at - 3G, 209584_x_at - 3C, 210873_x_at - 3A, 214995_s_at - 3G/ 3F, 214994_at - 3F).
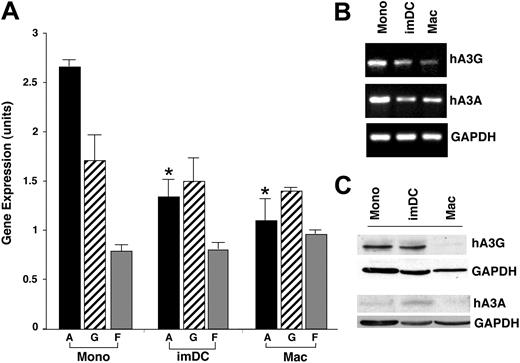
In further comparing differential gene expression among these populations, we noted changes in another molecule associated with cellular defense against HIV. The transcriptome of freshly isolated unstimulated monocytes revealed higher constitutive expression of 2 members of the APOBEC3 superfamily than differentiated macrophages (Figure 2C heat map, 2D parallel plots). Strikingly, hA3A appeared to be the highest expressed member of the APOBEC family in immature monocytes (Figure 2C-D; ∼20 fold, P < .002) and quantification of its expression in multiple donors revealed significantly higher levels in monocytes than macrophages (Figure 3; P < .001). Moreover, hA3G was also higher in monocytes but not significantly across all donors, and intermediate levels of hA3G and hA3A were seen in DCs. In comparison, hA3F was nondiscriminating (Figures 2C-D, 3A) and hA3B was elevated in 2 of 3 donor macrophages (Figure 2C).
Expression of APOBEC3 family genes during monocyte differentiation to macrophages. (A) Mean (± SEM) gene-expression level in S10 units (similar to log base 10 units) over 3 donors for 3 APOBEC3 family genes (A,G,F) in monocytes (Mono), dendritic cells (imDC), and macrophages (Mac). *P < .001 between monocyte hA3A and DCs or macrophage hA3A. (B) By RT-PCR, monocyte, DC, and macrophage expression of APOBEC3G and 3A in a representative donor. GAPDH was used as a control gene. (C) Protein levels of APOBEC3G and APOBEC3A were monitored in monocytes, DCs, and macrophages by Western blot using specific antibodies with GAPDH as the loading control.
Expression of APOBEC3 family genes during monocyte differentiation to macrophages. (A) Mean (± SEM) gene-expression level in S10 units (similar to log base 10 units) over 3 donors for 3 APOBEC3 family genes (A,G,F) in monocytes (Mono), dendritic cells (imDC), and macrophages (Mac). *P < .001 between monocyte hA3A and DCs or macrophage hA3A. (B) By RT-PCR, monocyte, DC, and macrophage expression of APOBEC3G and 3A in a representative donor. GAPDH was used as a control gene. (C) Protein levels of APOBEC3G and APOBEC3A were monitored in monocytes, DCs, and macrophages by Western blot using specific antibodies with GAPDH as the loading control.
Confirmation of this divergent pattern of basal expression of APOBEC3 genes was seen by RT-PCR for hA3G and hA3A, which are highest in monocytes (Figure 3B, representative donor). The protein levels for hA3G, detected in the lower–molecular mass form (Figure 3C; ∼46 kDa),20 reflected the transcriptional profile in these resting myeloid populations. In these unstimulated cells, hA3A protein was often difficult to detect with the available antibody, especially in macrophages, but typically seen in low levels in monocytes and DCs. This apparent inverse correlation between innate antiretroviral inhibitors and monocyte maturation, coupled with the differential susceptibility of immature and differentiated myeloid cells to infection with HIV-1, implicated a potential causative link.
Kinetics of APOBEC3 family expression during monocyte differentiation
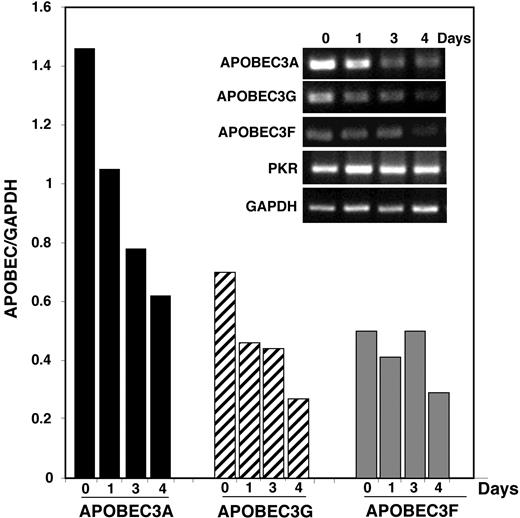
Examination of the individual members of the APOBEC gene family revealed that they were not necessarily coordinately expressed (Figures 2–3). Because hA3G and hA3F have been associated with neutralization of HIV-1, we focused on these cytidine deaminases along with hA3A, which has not previously been shown to have antiretroviral activity but which is clearly higher in monocytes than in macrophages. To establish the kinetics of decreasing APOBEC3, freshly isolated blood monocytes in suspension were harvested immediately (day 0) or adhered and harvested daily. Within 24 hours, a decrease in hA3A was apparent, which continued to decline (Figure 4). A similar profile was noted for hA3G expression, whereas hA3F appeared more stable, and, as a control, double-stranded RNA–activated protein kinase (PKR) did not diminish during this same interval. The kinetics of the decline of hA3G and/or hA3A was consistent with the time required for monocyte-to-macrophage differentiation associated with enhanced susceptibility to HIV-1 infection (Figure 1). During the in vitro evolution of peripheral-blood monocytes to immature DCs, there is a less substantive change in expression of APOBEC3 cytidine deaminases (data not shown), and the retention of these defensive molecules in unstimulated DCs may be associated with their resistance to HIV-1 (Figure 1B).
Kinetics of APOBEC3 family gene expression during monocyte differentiation to macrophages. Peripheral-blood monocytes were harvested immediately after isolation or cultured for indicated days as adherent cells and their RNA were analyzed by RT-PCR for the expression of hA3A, hA3F, hA3G, PKR, and GAPDH (inset). Graph represents the APOBEC-to-GAPDH ratio (representative kinetics, n = 2).
Kinetics of APOBEC3 family gene expression during monocyte differentiation to macrophages. Peripheral-blood monocytes were harvested immediately after isolation or cultured for indicated days as adherent cells and their RNA were analyzed by RT-PCR for the expression of hA3A, hA3F, hA3G, PKR, and GAPDH (inset). Graph represents the APOBEC-to-GAPDH ratio (representative kinetics, n = 2).
Effect of cell activation on APOBEC gene expression
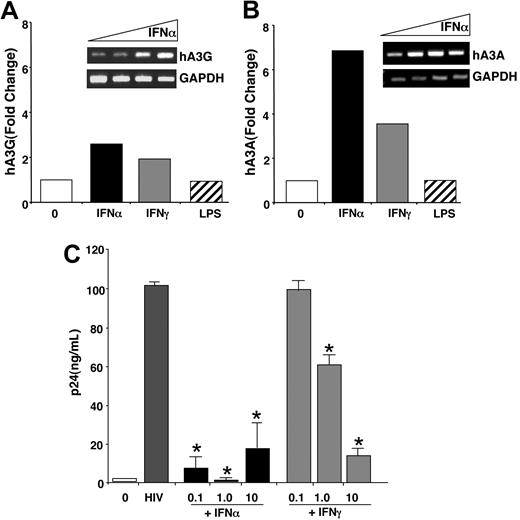
To establish whether activation of myeloid cells altered APOBEC expression, we stimulated the cells with the TLR4 agonist LPS or with IFN. Activation of monocytes, macrophages, and DCs with LPS initiated a dramatic transcriptional profile, with hundreds of genes induced 2-fold or more within 1 hour (S.N., P.M., Z.R., S.W., manuscript in preparation). Surprisingly, however, members of the APOBEC3 family with known or suspected activity against HIV-1 were not up-regulated by LPS as detected by microarray (Figure 2C) or by RT-PCR as shown for LPS-treated macrophage hA3G and hA3A (Figure 5A-B). To the contrary, IFNα and, to a lesser extent, IFNγ both amplified hA3G gene expression in differentiated macrophages 2-fold or more (n = 3), as recently described (Figure 5A-B),19 and increased hA3A, and this was associated with resistance to HIV-1 (Figure 5C). These composite findings support a maturation-dependent change in hA3G levels as a contributor to HIV-1 vulnerability, which could be reversed by IFN. However, the parallel expression and enhancibility of hA3A (Figures 2, 4, 5B), not previously detected nor associated with antiviral defense, might also protect against HIV-1.
IFN coordinately enhances macrophage APOBEC3 expression and decreases susceptibility to HIV-1. (A-B) Macrophages (n = 3 donors) were cultured in the presence or absence of 10 ng/mL IFNα or IFNγ for 4 hours or 100 ng/mL LPS for 1 hour and the RNA was processed for microarray analysis. The bar graph shows the fold change for hA3G (A) and hA3A (B) across treatments. (Insets) IFNα dose-response (0, 0.1, 1, 10 ng/mL; 4 hours) induction of hA3G (A) and hA3A (B) by RT-PCR. (C) Macrophages were infected or not with HIV-1BaL for 90 minutes, washed, and IFNα or IFNγ added once after infection at the indicated concentrations (0.1–10 ng/mL). HIV-1 infection was monitored by p24 levels in the supernatants (day 14 shown; *P < .005) (mean ± SEM).
IFN coordinately enhances macrophage APOBEC3 expression and decreases susceptibility to HIV-1. (A-B) Macrophages (n = 3 donors) were cultured in the presence or absence of 10 ng/mL IFNα or IFNγ for 4 hours or 100 ng/mL LPS for 1 hour and the RNA was processed for microarray analysis. The bar graph shows the fold change for hA3G (A) and hA3A (B) across treatments. (Insets) IFNα dose-response (0, 0.1, 1, 10 ng/mL; 4 hours) induction of hA3G (A) and hA3A (B) by RT-PCR. (C) Macrophages were infected or not with HIV-1BaL for 90 minutes, washed, and IFNα or IFNγ added once after infection at the indicated concentrations (0.1–10 ng/mL). HIV-1 infection was monitored by p24 levels in the supernatants (day 14 shown; *P < .005) (mean ± SEM).
APOBEC3A depletion reverses resistance to HIV-1 in immature monocytic cells
Considering that the enhanced levels of hA3A might be determinants of the relative resistance of monocytes compared with differentiated macrophages to HIV-1, we silenced expression of this member of the cytidine deaminase family using specific siRNA. Transfection of several hA3A-specific siRNA sequences not only significantly decreased hA3A expression but the otherwise HIV-1–resistant monocytes were also converted into more susceptible populations, similar to differentiated macrophages, as measured by nested PCR for viral DNA synthesis (Figure 6A). Parallel monocytes treated with an irrelevant control siRNA, electroporation only, or left untreated exhibited a minimal PCR product consistent with restricted HIV-1 replication in immature monocytes (Figure 6A). This failure of monocytes to resist HIV following depletion of hA3A was sustained through 7 to 14 days, as evident by significantly higher p24 viral antigen in the culture supernatants (Figure 6B; P < .05), comparable to that seen for hA3G-specific siRNA (Figure 6B inset). By Western analysis, hA3A protein levels, which are more evident after IFNα treatment,19 are reduced or absent in siRNA-treated cells (Figure 6C). Diminished hA3A protein following hA3A-specific siRNA, coupled with reduced antiviral function, is consistent with a key role for this member of the cytidine deaminase superfamily in anti–HIV-1 defense. These data newly implicate this host-cell protein in impeding HIV-1 replication and favor the concept that hA3A expression, together with other members of this cytidine deaminase family, is fundamental to the degree of receptiveness of myeloid cells to productive HIV-1 infection. If hA3A represents a defensive maneuver inherent in undifferentiated monocytes, which lose this protective shield during maturation, then evidence that targeting this molecule to embellish host resistance offers a conceptual advance in therapeutic strategies to counterattack HIV-1.
Silencing of APOBEC3A blocks monocyte resistance to HIV. (A) Monocytes were transfected with siRNA sequences specific for hA3A or a control siRNA (siC) or mock transfected (electroporation control [EPC]) before infection with HIV. Two to 3 days after HIV-1 infection, cells were processed and tested for viral DNA synthesis by nested PCR. (B) Transfected monocytes were adhered and infected with HIV-1. Supernatants were harvested every third day for analysis of p24 levels by ELISA (day 13 shown; mean ± SE). Differences between electroporation control (EPC) and siRNA for hA3A were significant (*P < .05). (Inset) Monocytes transfected as described in (A) with siRNA specific for hA3G or control sequences (siC) were infected with HIV-1 and supernatants were tested for HIV p24. (C) Transfected cells were cultured for 48 hours and treated with IFNα (10 ng/mL) overnight before monitoring of hA3A and α tubulin protein levels by Western-blot analysis.
Silencing of APOBEC3A blocks monocyte resistance to HIV. (A) Monocytes were transfected with siRNA sequences specific for hA3A or a control siRNA (siC) or mock transfected (electroporation control [EPC]) before infection with HIV. Two to 3 days after HIV-1 infection, cells were processed and tested for viral DNA synthesis by nested PCR. (B) Transfected monocytes were adhered and infected with HIV-1. Supernatants were harvested every third day for analysis of p24 levels by ELISA (day 13 shown; mean ± SE). Differences between electroporation control (EPC) and siRNA for hA3A were significant (*P < .05). (Inset) Monocytes transfected as described in (A) with siRNA specific for hA3G or control sequences (siC) were infected with HIV-1 and supernatants were tested for HIV p24. (C) Transfected cells were cultured for 48 hours and treated with IFNα (10 ng/mL) overnight before monitoring of hA3A and α tubulin protein levels by Western-blot analysis.
Discussion
For nearly 2 decades, the differential susceptibility of immature monocytes and monocyte-derived differentiated macrophages to HIV-1 has represented a conundrum, especially significant in that identification of potential resistance factors inherent to monocytes might illuminate pathways to block macrophage infection. In an effort to reveal underlying maturation-dependent defense mechanisms, we monitored a battery of molecules associated with macrophage HIV-1 resistance and susceptibility.11 Many of these suspect genes were not differentially expressed during differentiation, with the notable exception of members of the APOBEC cytidine deaminase family that can function as HIV-1 postentry restriction factors. Differentiation of monocytes to DCs and, especially, macrophages with concurrent reduced transcription of APOBEC3 genes parallels their decreasing resistance to viral replication. This viral receptiveness is not dependent on a proliferative or activation response but can be reversed by the antiviral cytokine IFNα and, to a lesser extent, by IFNγ but not by the TLR ligand LPS.19 Since expression of hA3G in a virus-producing cell can lead to abortive retroviral infection following incorporation into progeny virions as they assemble and bud,37,38 reduced hA3G is reasonably linked with failure to suppress infection. The virus itself can also selectively deplete hA3G when HIV-1–encoded Vif targets hA3G for proteasome-mediated destruction through a SOCS-box–mediated interaction with the cullin E3 ubiquitin ligase complex.23,–25,39,40 Vif mutations associated with this SOCS box exhibit less infectivity because hA3G is no longer actively reduced or eliminated.40,–42
The efficiency of the APOBEC innate cellular shield is evident in resting T cells (Chiu et al20 ; G.P., T.W., K.L., W.J., S.W., in preparation) and, as we show, in immature monocytes, which are resistant to HIV-1 and express high levels of low–molecular weight hA3G. However, based on our transcriptome analysis, we identified maturational differences not only in expression of hA3G, an established anti–HIV-1 molecule, but also even more dramatically in hA3A. By multiple parameters, hA3A can be implicated in the innate antiretroviral repertoire of monocytes. hA3A, similar to hA3G, is constitutively high in nonpermissive monocytes, declines during monocyte-to-macrophage differentiation, and low levels are present in macrophages during their most vulnerable period of virus infectibility. Moreover, by silencing hA3A expression in resistant monocytes, these cells convert to a virus-susceptible population. While perhaps surprising that silencing a single member of the APOBEC3 family has such a profound reversal of monocyte resistance to HIV-1, it is possible that hA3A interacts in complex with other members of the cytidine deaminase family, as shown for hA3G and hA3F.43 Although differentiated macrophages lose most of their hA3G and hA3A in parallel with emergent permissiveness to the virus, monocyte-derived DC-SIGN+HLA-DR+CD86+CD11c+ dendritic cells in the immature, nonactivated state retain full or partial APOBEC3 levels. Whether retention of APOBEC3 family proteins is associated with IFNα production during DC maturation44 is currently unclear.
There appears to be a nonequivalency of APOBEC3 family members in mediating antiretroviral activity. APOBEC3F is a cytidine deaminase that has antiviral activity, albeit less active than hA3G,21,22,44,45 whereas hA3B and hA3C were initially reported to be active against SIV not HIV-1.46 However, recent evidence suggests that hA3B can inhibit HIV-1 but, interestingly, is resistant to Vif.47 Nonetheless, in monocytes, hA3A as well as hA3G are constitutively expressed genes that suppress or prevent viral infection. APOBEC3G is associated with an inhibition of reverse transcription in resting T cells,20 and a delayed accumulation of products of late reverse transcription is also characteristic of infection-resistant monocytes,8 consistent with a comparable role of hA3G in these 2 populations. By comparison, the molecular mechanisms by which hA3A inhibits HIV-1 are yet to be elucidated. Whether it triggers cytosine deamination of dinucleotide sequences with unique or overlapping hypermutation patterns,48 is independent of deamination,20,41,49 is targeted by Vif to the proteasome, or is subject to alternative HIV countermeasures must be determined. It is intriguing that in transfected cell lines, hA3A has recently been shown to inhibit adeno-associated virus and retrotransposons50,51 and suggests that hA3A may interfere with HIV infection by a novel pathway.51 While likely not attributable to a single molecular pathway, it is clear that HIV-1–resistant monocytes in which hA3G or hA3A is silenced convert to willing hosts for HIV-1, all other conditions being equal.
Identification of cellular genes that are involved in early events, whether at the level of virion entry, viral DNA synthesis, proviral integration, or production of viral transcripts, directs attention to potential targets for disrupting the viral life cycle.11 In our previous gene screen of macrophages subsequent to HIV-1 infection, we identified a number of candidate genes influenced by viral infection, including cytokines, cell-surface proteins, genes previously linked to HIV-1 replication, and genes not previously associated with HIV-1, including p21,16 which can be explored as candidates for blocking HIV-1 infection. In the case of APOBEC3 proteins, the objective would be to enhance rather than to inhibit their expression. Agonists of these host-cell cytidine deaminases could provide the basis of potent anti–HIV-1 drugs. IFNα, by virtue of its ability to promote hA3G and hA3A expression (Figure 5)19 along with its multiple additional antiviral properties,52 may be considered a natural guardian against HIV-1. In this regard, recent trials administering pegylated IFNα-2b to HIV-1–viremic patients resulted not only in suppression of HIV-1 viral load but also in strikingly augmented APOBEC3 expression in the PBMCs of these treated patients (G.P. et al, manuscript in preparation).
Demonstration that intracellular factors, such as members of the APOBEC3 family, are associated with HIV-1 resistance in less-mature myeloid cells and that repression of these key innate antiretroviral factors during maturation contributes to their receptiveness as viral hosts offers insight into development of therapeutics against HIV-1 infection and AIDS. Originally thought to represent constitutive components of cellular defense readily overcome by HIV-1, our demonstration of the plasticity of this innate intracellular pathway, coupled with its inducibility, supports a potential mechanism to promote viral resistance.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Niki Moutsopoulos and Jennifer Swisher (National Institute of Dental and Craniofacial Research [NIDCR]) for FACS assistance, and Jan Orenstein (George Washington University) for transmission electron microscopy.
This work was supported in part by the Intramural Research Program of the NIH, National Institute of Dental and Craniofacial Research.
Authorship
Contribution: G.P., T.G.-W., S.N., W.J., and K.J.L. designed, performed, and analyzed experiments; Z.G.R. and P.J.M. analyzed Affymetrix data; and S.M.W. designed the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: S. M. Wahl, Building 30, Rm 320, 30 Convent Dr. MSC 4352, NIDCR, NIH, Bethesda, MD 20892-4352; e-mail: smwahl@dir.nidcr.nih.gov.


![Figure 6. Silencing of APOBEC3A blocks monocyte resistance to HIV. (A) Monocytes were transfected with siRNA sequences specific for hA3A or a control siRNA (siC) or mock transfected (electroporation control [EPC]) before infection with HIV. Two to 3 days after HIV-1 infection, cells were processed and tested for viral DNA synthesis by nested PCR. (B) Transfected monocytes were adhered and infected with HIV-1. Supernatants were harvested every third day for analysis of p24 levels by ELISA (day 13 shown; mean ± SE). Differences between electroporation control (EPC) and siRNA for hA3A were significant (*P < .05). (Inset) Monocytes transfected as described in (A) with siRNA specific for hA3G or control sequences (siC) were infected with HIV-1 and supernatants were tested for HIV p24. (C) Transfected cells were cultured for 48 hours and treated with IFNα (10 ng/mL) overnight before monitoring of hA3A and α tubulin protein levels by Western-blot analysis.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/1/10.1182_blood-2006-10-051763/2/m_zh80130702990006.jpeg?Expires=1769087486&Signature=QMUoV9ZRZCqVKuBTzezAZILIk5v6ImfPu1PPKOKsr3RHnNCg3fPzByETfheU-69KBQ8mGIJLP7XRRXCTDA3wyGaC5gKxOegAsK3nDIhgtEQTyHVVcqsA8l3RX4miuy4ck6Vpg2c6sy9ANuIuZWcozIiYQfPanviGESXB-cs0Fo68EiM3SlHGm5OBZyuaGRjl4G-NCPK9gj80uaNl8BSDcKIsRkvaCDVowhsDNTgznxfuj8DwXD3yrqHHZO6u-yCwtSqdDMb4PjUZIfliW1gBirwNY5-JfOqYR23V2TbAYZ4PPhJySzsZXbBY1Ysi4rE8P2JO-2lND425sRTi7sEuyg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)