Abstract
HIV-1 strains use C-C-chemokine receptor 5, CCR5, as a coreceptor for host transmission. Human CCR5 chemokine ligands inhibit binding and infection, whereas CCR5 mutations also inhibit infection by preventing surface expression, resulting in delayed progression to AIDS. Here, we describe a human herpesvirus 6 (HHV-6A) chemokine, U83A, which binds CCR5 with higher affinity than human chemokines, displacing their binding and leading to inhibition of chemotaxis of human leukocytes. Similarly, U83A inhibits infection by HIV-1 strains which use CCR5, but not the CXCR4, coreceptor. Unlike human CCR5 chemokine ligands which induce rapid CCR5 internalization mediated via clathrin, treatment with U83A prevents internalization. A spliced truncated U83A isoform, U83A-N, also binds CCR5 albeit with lower affinity, and this correlates with lower HIV-1 infection inhibition, whereas further truncation abolishes binding and any inhibition. Confocal microscopy confirms CCR5 internalization inhibition by U83A treatment, whereas labeled transferrin uptake shows that endocytosis via clathrin is unaltered. Previous results show that, although U83A-N is an antagonist, U83A is an agonist for CCR1, CCR4, CCR6, and CCR8 present on immune effector and antigen-presenting cells and here also shown for CCR5. Thus, U83A could act as a novel inhibitor of HIV-1 infection while also stimulating local immunity to the virus.
Introduction
Human herpesvirus 6 (HHV-6), like HIV-1, infects and kills CD4+ T lymphocytes. Distinct from HIV-1, it infects up to 95% of populations in infancy, and those infected normally develop effective immunity rather than an AIDS-like illness. Although occasional serious febrile complications are observed during primary infant infections, disease associations are mainly during reactivation of latent infections from immunosuppressive/aberrant conditions, including neuroinflammatory disease in transplant recipients (including solid organ, bone marrow, and hematopoietic stem cell) and disseminated infection in HIV/AIDS.1 Because HHV-6 is a blood-borne virus infection that can be transmitted through epithelial mucosa, an interface with the chemokine system which directs hematopoietic cell traffic is required, affecting virus dissemination and cellular tropism. Chemokines interact with specific receptors present on distinct leukocyte subsets, causing activation and chemotaxis following a chemokine gradient released by infected or damaged cells. They also affect hematopoiesis and antigen presentation in directing antigen-presenting cells to sites of infection or the lymph node.2 Furthermore, chemokine interactions can direct cellular infiltrates into other locations, such as the brain during neuroinflammatory conditions, dependent on local chemokine release and chemokine receptor expression.3 Chemokine receptors CCR5 and CXCR4 are also coreceptors for HIV infectivity, with CCR5 utilization being the main determinant for host transmission.3,4 HHV-6 encodes a chemokine, U83, with distinct forms in 2 strain groups of HHV-6, variant A and B. In infant infections, HHV-6B is dominant in the United States and the United Kingdom, whereas HHV-6A is prevalent in Africa (Zambia).1,5–7 HHV-6A has been anecdotally linked to altered tropism and more severe pathology, with disseminated infections in patients with HIV/AIDS plus fatal neurologic disease in immunosuppressed adult transplant recipients.1 In herpesvirus genome comparisons, U83 is identified as a hypervariable locus among the few HHV-6–specific genes, and, thus, a main candidate for virulence or tropism differences between strains.8
Our previous studies on U83A suggest a foundation for these differences. Although U83B chemokine acts with low potency on CCR2,9 a monocyte receptor, we have shown that U83A acts with high potency as a broad but specific agonist on CCR1, CCR4, CCR6, and CCR8 also binding CCR5 receptors present on monocytic/macrophages, as well as mature T lymphocytes and immature dendritic cells which both have skin-homing properties.10 Thus, U83A of HHV-6A versus U83B of HHV-6B can chemoattract distinct and diverse cellular populations. U83A bound most potently with CCR5, at 0.06 nM, displacing human chemokines, with implications for effects on HIV infection.10 Thus, although both HHV-6A and HHV-6B can infect CD4+ T lymphocytes like HIV,11,12 potential outcomes may be quite distinct, because only U83A efficiently binds the HIV coreceptor, CCR5.10 Here, cell agonism and HIV-1 inhibition by U83A are demonstrated. This is distinct from viral chemokine HIV inhibitors KSHV vMIPII, primarily a broad-spectrum antagonist which inhibits R5 HIV as CCR5 antagonist, and vMIPI, a partial inhibitor on peripheral blood mononuclear cells (PBMCs), by undefined route.13,14 U83A can also have similar activities in that a spliced truncated isoform, U83A-N, produced only early in infection, has antagonist properties.
The human endogenous CCR5 ligands such as CCL3/Mip1α and CCL5/RANTES chemokines are effective inhibitors of HIV-1 infection by preventing entry via the CCR5 coreceptor by R5-using strains. R5 strains appear most important for both vertical and sexual transmission.4 Additionally, CCR5 is viewed as a favorable target for HIV inhibition because naturally occurring deletion mutations (ie, CCR5δ32) are tolerated and give some resistance to infection.15 Further, in some patients with HIV, development of CCR5 antibodies correlate with down-regulation of the CCR5 receptor cell surface expression and inhibition of HIV infection.16 Given drug resistance, new antiviral therapies for other stages of the replicative cycle, including CCR5, are required.17 Furthermore, vaccine development is difficult, because of virus mutability and challenges to stimulate effective immunity. Shown here, U83A may combine properties of an inhibitor via CCR5 with vaccine enhancement via CCR1, CCR4, CCR6, and CCR8 interactions. U83A interactions with HIV-1 are investigated. Earlier, we showed CCR5 binding in transfected COS-7 and leukemic monocytic U937 cells.10 Here, we show CCR5 functional interactions in U373 cells used to assay HIV-1 infection, as well as in human ex vivo PBMCs and characterize HIV-1 inhibition and mechanism of action.
Materials and methods
Blood donation with consent by anonymously coded healthy volunteers from LSHTM is regulated and approved by our institutional review board (London School of Hygiene and Tropical Medicine, University of London).
Cell lines and chemokines
COS-7 cells were obtained from ECACC (Salisbury, United Kingdom). U373-MAGI-CCR5E and MAGI-CXCR4 were from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, donated by Dr Michael Emerman.18 Cell lines were maintained in Dulbecco modified Eagle medium containing 10% fetal bovine serum (FBS), 2 mM L-glutamine, 1 mM sodium pyruvate, 50 U/mL penicillin, 50 μg/mL streptomycin with an additional supplementation of 0.1 mg/mL hygromycin B and 1.0 μg/mL puromycin (Sigma, Poole, United Kingdom) for stably transfected U373-MAGI-CCR5E cell lines. COS-7 cells were transfected with pCDNA3-expressing CCR5 as described.10 Primary human peripheral blood mononuclear cells (PBMCs) were obtained from healthy adult volunteers by centrifugation of EDTA anticoagulated blood over Histopaque-1077 cushion (Sigma-Aldrich, Irvine, United Kingdom). Mononuclear cells were collected from the interphase and washed 3 times in phosphate-buffered saline and used immediately for binding assays or for chemotaxis or HIV-1 inhibition assay after 48 or 72 hours, respectively. Incubation was in low-adherence flasks (Corning, Corning, NY) in RPMI, 5% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 50 U/mL penicillin, 50 μg/mL streptomycin for chemotaxis assay. Chemokines CCL3/MIP-1α and CCL4/MIP-1β (R&D Systems, Abingdon, United Kingdom) were resuspended in PBS 0.1% BSA (fraction V, endotoxin certified; Sigma-Aldrich). Viral chemokines full-length U83A-GS, spliced truncation U83A-N were produced as described previously.10 U83A-Ndel, corresponds to the 17 first N-terminus amino acids of native U83A and was synthesized (Sigma Genosys, Haverhill, United Kingdom).
Receptor binding
Binding assays were performed on U373-MAGI-CCR5E, PBMCs, or transfected COS-7 cells as described.10 Briefly, assays in triplicate contained 2.5 × 106 cells, 166 pM radiolabeled chemokine (specific activity, 2000 Ci/mM (7.4 × 1013 Bq), 125I-labeled CCL3/MIP-1α; Amersham Biosciences, Little Chalfont, United Kingdom), and diluted concentrations of unlabeled competitor chemokines. After a 2-hour incubation on ice, bound chemokine was separated through a phthalate oil cushion, and radioactivity was measured with a gamma counter. Data analyses used Prism 0.1.53 software (GraphPad, San Diego, CA).
Transwell chemotaxis
Migration of PBMCs was evaluated using 96-well microchemotaxis plate (NeuroProbe, Gaithersburg, MD). Migration buffer (HBSS [Invitrogen, Paisley, United Kingdom], 0.1% BSA) supplemented with different concentrations of chemokine was placed into the lower chamber. Cells were resuspended in the same buffer at a concentration of 1 × 107 cells/mL, and a 50-μL cell suspension was placed onto the upper well. After 90 minutes of incubation at 37°C in 5% CO2, cells were carefully wiped off, and the plate was read at 535 nm in a Wallac Victor2 1420 multilabel counter (PerkinElmer, Waltham, MA). Each set was performed in triplicate, and SDs were calculated. Background random migration using only carrier gave the chemotactic index (CI) value of 1. CI values thus represent a fold increase of chemokine directed over the random migration.
Inhibition of HIV entry assay
Stocks of HIV-1 B subtype YU2 (R5 and R3) or NL4-3 (X4) strains were produced by transfecting 293T cells with pYU2 or pNL4-3 plasmid molecular clones, and strains SF2 (R5 and X4) and SF162 (R5) were cultured on PBMCs as described14 (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH from Dr B. Hahn, Dr G. Shaw, and Dr M. Martin). Viral stocks were titrated on U373-MAGI-CCR5E or MAGI-CXCR4 cells, by serial dilution. Specific CCR5 or CXCR4 surface expression was confirmed on the respective cell lines by flow cytometry. Cells were seeded 5 × 104 cells per well of a 48-well plate overnight. The cells were then treated for 30 minutes at 37°C with different concentrations of chemokine or peptide in 75-μL volume. One hundred focus-forming units of HIV virus was added in a volume of 75 μL and incubated for 3 hours at 37°C. The cells were then washed 3 times with PBS and overlaid with 250 μL media containing the chemokine or peptide at the relevant concentration. After 2 days the cells were washed once with 500 μL PBS and fixed with 1% formaldehyde, 0.2% gluteraldehyde in PBS for 5 minutes before being stained with X-Gal staining solution (PBS, 4 mM potassium ferrocyanide, 4 mM potassium ferricyanide, 2 mM MgCl2, and 400 μg/mL X-Gal). Stained foci of infection were counted. Results are the mean of duplicate wells, expressed as percentages of untreated wells.
Receptor internalization and flow cytometry
U373-MAGI-CCR5E cells were suspended at 5 × 106 cells/mL in FACS buffer (PBS, 0.1% BSA). Cells were Fc-blocked by treatment with 1 μg human IgG/105 cells for 15 minutes at room temperature. They were then incubated for various times, as indicated, at 37°C and 5% CO2 in the presence or the absence of CCL3 and/or U83A-GS, U83A-N at various concentrations. Stimulated cells were then incubated at 4°C with FITC-conjugated anti-CCR5 antibody (FAB 182; R&D systems) for 30 minutes. After staining, cells were washed 3 times with ice-cold FACS buffer and suspended at 5 × 105/mL in the same buffer after a 15-minute fixation in 4% PFA. The analysis was done using a FACSCalibur flow cytometer (BD Biosciences, Oxford, United Kingdom). Data were analyzed with CellQuest software (BD Biosciences). Relevant IgG (mouse IgG2B) was used for negative control.
Confocal microscopy
U373-MAGI-CCR5E cells were grown on coverslips for 24 hours then starved in serum-free medium for 30 minutes at 37°C, 5%CO2, and incubated for a further 30 minutes. They were then labeled during 20 minutes at 37°C with 1 μg/mL monoclonal anti–human CCR5-fluorescein antibody (R&D Systems). For antibody feeding, after one washing in prewarmed serum-free medium cells were incubated for a further 10 minutes with buffer containing transferrin-Alexa 594 (594Tf 1/500, 10μg/mL; Molecular Probes, Leiden, The Netherlands) and CCL3 (50 nM), U83A_GS (100 nM) or U83A_N (100 nM) as indicated. They were then washed 3 times in ice-cold PBS containing 0.5% BSA, fixed in 3% paraformaldehyde for 10 minutes. After 3 washings in ice-cold PBS containing 0.5% BSA, free aldehyde groups were quenched with 50 mM NH4Cl in PBS for 10 minutes and permeabilized by 0.05% saponin in 0.5% BSA-PBS (10 minutes). The coverslips were finally washed 3 times and mounted using Vectashield mounting solution (Vector Laboratories, Burlingame, CA) containing DAPI for nucleus detection. Cells were examined using Z-stack sections (at 0.39-μm interval), and pictures were acquired on a Zeiss LSM 510 Axioplan microscope with a Plan-Apochromat 63×/1.4 oil objective by an AxioCam (magnification ×630 under oil immersion; Zeiss, Jena, Germany). Digital images were analyzed with Zeiss LSM Image Browser, version 3.5.0.376 (AxioCam). Fluorochromes were excited at 488 nm for FITC and 543 nm for Alexa Fluor.
Results
CCR5 binding HIV-1–susceptible cells
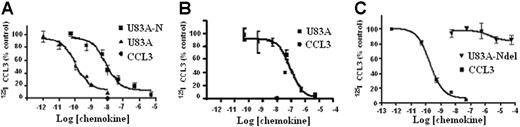
The ability of U83A to bind CCR5 was further tested here on primary cells and cell types used to assay HIV infection to assess their implications for in vivo interactions. First, tested were U373-MAGI-CCR5E, U373 astrocytoma adherent cells used for HIV infection assays. These express the HIV-1 coreceptor CCR518 and not CCR1, CCR2, or CCR8 as confirmed by flow cytometry. In competition binding assays against CCR5 ligand Mip1α/CCL3, U83A showed very high-affinity interactions, IC50 = 1.1 × 10−10 M (Figure 1). This affinity was similar to that shown previously, IC50 = 5.6 × 10−11 M, for COS-CCR5 cells.10 The shorter naturally occurring spliced isoform, U83A-N, was also tested on U373-MAGI-CCR5E and human PBMCs, IC50, 8.3 × 10−9 M and 9.2 × 10−8 M, respectively, similar to previous values obtained on U937 cells expressing CCR1 and CCR5, 5.4 × 10−8 M.10 Thus, U83A-N was also a high-affinity ligand for CCR5 although with reduced potency compared with full-length U83A. The affinity remains in the same range as the human endogenous CCR5 ligands, with full-length U83A one of the highest affinity natural ligands reported. The full-length U83A showed the highest affinity interactions with CCR5 followed by U83A-N, whereas a further truncation, U83A-Ndel, did not bind. This demonstrates the binding specificity is in the N-terminal part of U83A (contained in U83A-N), but that stable interactions are provided by the C-terminal half, also involved in signaling.10
U83A and U83A-N high-affinity binding CCR5 and human chemokine displacement. Competition binding assays on (A) U373-MAGI-CCR5E, (B) PBMCs, and (C) COS-7 CCR5. Data are the percentage of maximum bound iodinated chemokine, determined in absence of cold competitor. (A-B) Total displacement was obtained by 10 nM CCL3 (•); (C) kinetics are shown. Each point represents mean values ± standard deviations of triplicates. IC50 for U373-MAGI-CCR5E cells is as follows: U83A, 1.1 × 10−10 M, U83A-N, 8.3 × 10−9 M; IC50 for PBMCs is as follows: U83A-N, 9.2 × 10−8 M; IC50 for COS-7 CCR5 cells is as follows: CCL3 0.2 × 10−9 M and no displacement by U83A-Ndel.
U83A and U83A-N high-affinity binding CCR5 and human chemokine displacement. Competition binding assays on (A) U373-MAGI-CCR5E, (B) PBMCs, and (C) COS-7 CCR5. Data are the percentage of maximum bound iodinated chemokine, determined in absence of cold competitor. (A-B) Total displacement was obtained by 10 nM CCL3 (•); (C) kinetics are shown. Each point represents mean values ± standard deviations of triplicates. IC50 for U373-MAGI-CCR5E cells is as follows: U83A, 1.1 × 10−10 M, U83A-N, 8.3 × 10−9 M; IC50 for PBMCs is as follows: U83A-N, 9.2 × 10−8 M; IC50 for COS-7 CCR5 cells is as follows: CCL3 0.2 × 10−9 M and no displacement by U83A-Ndel.
Agonist and antagonist activities in CCR5 chemotaxis
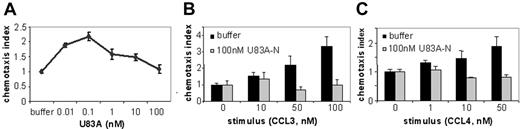
To further investigate actions of U83A on normal function of CCR5 in vivo, we tested agonist and inhibitory effects on CCR5-mediated chemotaxis of ex vivo human donor PBMCs. U83A efficiently mediated chemotaxis on CCR5-expressing PBMCs with a peak activity at 0.1 nM consistent with binding affinity (Figure 2A). To test inhibition of chemotaxis mediated by human chemokine ligands of CCR5, U83A-N was used as this separates binding from the signaling effects of U83A as shown previously for CCR1-, CCR4-, CCR6-, and CCR8-mediated functions.10 U83A-N at 100 nM does not promote chemotaxis (Figure 2B-C) with similar results at 5 to 500 nM. The inhibiting activity of U83A-N was tested against CCL3- and CCL4-induced migrations (Figure 2B-C). CCL3 and CCL4 are both agonists for receptor CCR5; CCL3 also induces CCR1, CCL4 primarily CCR5. Separate migrations of PBMCs from 4 donors were tested. U83A-N was able to efficiently block human chemokine-induced chemotaxis via CCR5. The inhibition of CCL3-mediated chemotaxis via both CCR1 and CCR5 affects more cell types (Figure 2B). However, the inhibition against CCL4 shows U83A can block CCR5 function specifically (Figure 2C). Flow cytometry confirmed expression of both receptors, although CCR5 predominated. Thus, U83A binding acts as an agonist and U83A-N is an antagonist of chemotaxis of CCR5-bearing cells.
U83A agonist and U83A-N antagonist for CCR5-mediated chemotaxis. (A) U83A induced chemotaxis on PBMCs, data shown are representative of 5 independent experiments, with maximum at 0.1 nM dose (P < .01, Dunnett multiple comparison test). U83A-N blocks chemotaxis of ex vivo human PBMCs induced by (B) 50 nM and 100 nM CCL3 or (C) 10 nM and 50 nM CCL4. Data are means ± SDs of triplicates.
U83A agonist and U83A-N antagonist for CCR5-mediated chemotaxis. (A) U83A induced chemotaxis on PBMCs, data shown are representative of 5 independent experiments, with maximum at 0.1 nM dose (P < .01, Dunnett multiple comparison test). U83A-N blocks chemotaxis of ex vivo human PBMCs induced by (B) 50 nM and 100 nM CCL3 or (C) 10 nM and 50 nM CCL4. Data are means ± SDs of triplicates.
Inhibition R5 HIV-1 infection
Having demonstrated efficient functional interactions with cellular CCR5 by the viral chemokines U83A and U83A-N, these were next tested against CCR5 interactions with HIV-1 R5 strains which use CCR5 for binding and cell entry. These were assayed in the human astrocytoma-derived U373-MAGI_CCR5E cells where binding was shown. This is an assay for total virus replication because these cells express β-gal under HIV LTR control, and transactivation by HIV Tat, produced only after integration and virus replication, indicates relative virus production as shown by positive blue foci.18 CD4 and CCR5 are also expressed, allowing infection by most primary HIV strains. An R5 HIV strain, YU2, was tested in inhibition assays. CCR5 human chemokine and HIV inhibitor, RANTES/CCL5, was used as a positive control, with negative control, U83A-Ndel, the truncated peptide which does not bind CCR5 (Figure 1). Figure 3A shows U83A more effective than RANTES/CCL5 in inhibiting HIV infection, with U83A-N at an intermediate potency. The estimated IC50 for the HIV inhibitory effects, of 0.1 nM U83A and 5nM U83A-N, matched the respective affinities for CCR5 and showed up to 50% inhibition at 50 nM. In contrast, U83A-Ndel which does not bind CCR5 had no effect on HIV infection. Furthermore, no inhibitory effect was shown with either U83A-N or CCL5 against infection with CXCR4 using HIV-1 strain on MAGI-CXCR4 cells, demonstrating specificity of U83A-N inhibition for CCR5 using HIV-1 strains (Figure 3B). Up to 60% inhibition was also shown for dual tropic HIV-1 strain SF2 and CCR5-specific SF162 on MAGI-CCR5 cells (Figure 3B-C). For confirmation, HIV effects were also assayed using CEM-NKR-CCR5-luc cells with similar results (not shown). These have the luciferase reporter gene under the transcriptional control of the HIV-2 LTR,19 and inhibition of CCR5-specific HIV-1 strain JRCSF20 was measured by cells responding to Tat expression producing luciferase. Finally, to examine growth inhibition on primary human PBMCs, CCR5-specific HIV-1 strain SF162 was cultured on differentiated PBMCs in the presence of U83A-N or CCL5/RANTES with greater than 95% inhibition demonstrated by U83A-N treatment (Figure 4).
U83A inhibits R5 HIV-1 infection. (A) Using HIV-1 R5 strain on MAGI-CCR5 cells, U83A-N and U83A inhibit HIV-1 foci of infection in the same range of positive control as CCL5/RANTES. The deletion U83A-Ndel had no effect. Results (means ± SDs) are representative of 3 independent experiments. (B) U83A and U83A-N did not inhibit infection by CXCR4 using HIV-1 strain on MAGI-CXCR4 cells. (C) U83A-N and CCL5/RANTES inhibit infection of HIV-1 strain SF2 (dual tropic) on MAGI-CCR5 cells. (D) U83A-N and CCL5/RANTES inhibit infection of HIV-1 strain SF162 (CCR5 specific) on MAGI-CCR5 cells.
U83A inhibits R5 HIV-1 infection. (A) Using HIV-1 R5 strain on MAGI-CCR5 cells, U83A-N and U83A inhibit HIV-1 foci of infection in the same range of positive control as CCL5/RANTES. The deletion U83A-Ndel had no effect. Results (means ± SDs) are representative of 3 independent experiments. (B) U83A and U83A-N did not inhibit infection by CXCR4 using HIV-1 strain on MAGI-CXCR4 cells. (C) U83A-N and CCL5/RANTES inhibit infection of HIV-1 strain SF2 (dual tropic) on MAGI-CCR5 cells. (D) U83A-N and CCL5/RANTES inhibit infection of HIV-1 strain SF162 (CCR5 specific) on MAGI-CCR5 cells.
U83A-N inhibits HIV-1 infection in human PBMC. (A) Flow cytometry for CD14 and CCR5. Cells were cultured for 72 hours allowing monocytic/macrophage differentiation, confirmed by flow cytometry which showed CD14 and CCR5 expression, but not CCR2, CCR3, CCR1, or CCR8. (B) R5 HIV-1 strain SF162 was cultured at an MOI of 0.01, in these cells for 6 days in the presence of U83A-N (gray line) or CCL5/RANTES (black line), and virus was then titrated on MAGI-CCR5 cells as described. Greater than 95% inhibition was demonstrated with U83A-N at 0.1 nM and above. Means ± SDs of duplicates.
U83A-N inhibits HIV-1 infection in human PBMC. (A) Flow cytometry for CD14 and CCR5. Cells were cultured for 72 hours allowing monocytic/macrophage differentiation, confirmed by flow cytometry which showed CD14 and CCR5 expression, but not CCR2, CCR3, CCR1, or CCR8. (B) R5 HIV-1 strain SF162 was cultured at an MOI of 0.01, in these cells for 6 days in the presence of U83A-N (gray line) or CCL5/RANTES (black line), and virus was then titrated on MAGI-CCR5 cells as described. Greater than 95% inhibition was demonstrated with U83A-N at 0.1 nM and above. Means ± SDs of duplicates.
CCR5 internalization inhibition
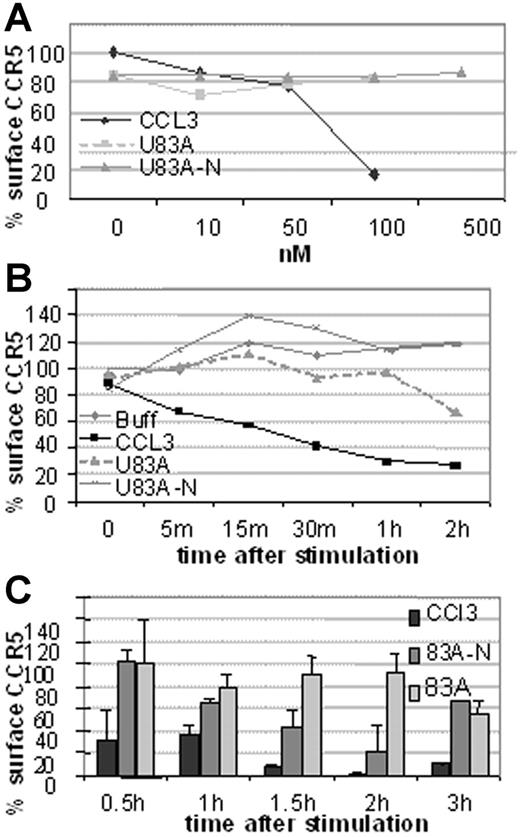
Next investigated was the mechanism of action of HIV-1 inhibition. This could be by blocking binding or by affecting CCR5 cell surface expression. To investigate we used flow cytometry to examine receptor internalization following ligand binding by flow cytometry. First, CCR5 expression on U373-MAGI-CCR5E was monitored before and after 5-minute treatment with various concentrations of human CCR5 chemokine, CCL3, or the viral chemokines, U83A and U83A-N. As shown in Figure 5A, 100 nM CCL3 markedly decreased CCR5 levels, whereas no internalization was shown with the viral chemokines up to 500 nM. After 30-minute exposure to CCL3 most of the CCR5 is internalized with minimal effect by the viral chemokines even up to 2 hours (Figure 5B). In extension to 3 hours there was gradual internalization of CCR5 by U83A with an intermediate effect by U83A-N (Figure 5C). Thus, although human chemokine, CCL3, efficiently and rapidly induced internalization as described previously via a clathrin-mediated pathway,21 U83A and U83A-N showed markedly delayed internalization. Two hours after stimulation 100 nM U83A started to cause receptor internalization (44%), suggesting the mobilization of a different internalization pathway from that induced by CCL3 to internalize CCR5 (43% internalization after a 5-minute incubation).
CCR5 internalization is rapidly induced by CCL3 but not by U83A or U83A-N. Results are representative of 3 sets each. (A) Dose effect of CCL5, U83A, and U83A-N on U373-MAGI-CCR5E CCR5 surface expression after 5 minutes and monitored by flow cytometry. (B) CCR5 internalization kinetics over 2 hours induced by 50 nM CCL5, 100 nM U83A or U83A-N. (C) Quantification of CCR5 internalization induced by 100 nM CCL3 and inhibited by U83A and U83A-N. Mean ± SDs of triplicates.
CCR5 internalization is rapidly induced by CCL3 but not by U83A or U83A-N. Results are representative of 3 sets each. (A) Dose effect of CCL5, U83A, and U83A-N on U373-MAGI-CCR5E CCR5 surface expression after 5 minutes and monitored by flow cytometry. (B) CCR5 internalization kinetics over 2 hours induced by 50 nM CCL5, 100 nM U83A or U83A-N. (C) Quantification of CCR5 internalization induced by 100 nM CCL3 and inhibited by U83A and U83A-N. Mean ± SDs of triplicates.
Inhibition CCR5 recycling by human chemokine
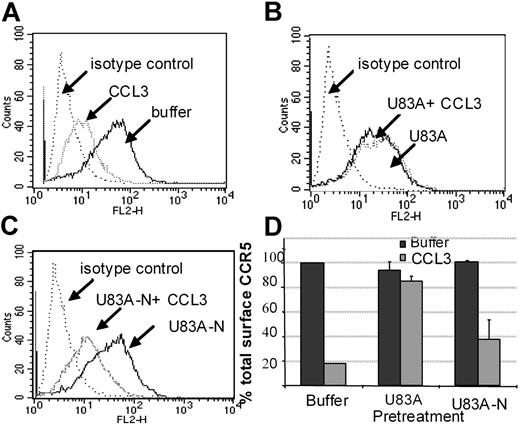
Using flow cytometry, possible blocking of the human agonist induced CCR5 internalization was further tested. Figure 6 shows U83A and U83A-N pretreatment of cells before stimulation by 100 nM CCL3 prevents CCR5 internalization. Inhibition of CCR5 internalization was most efficient with the full-length U83A (10-50 nM) blocking CCL3 (100 nM) induced internalization of CCR5. The spliced U83A-N (100-500 nM) showed 50% inhibition. Thus, U83A and U83A-N displace endogenous human chemokines from binding CCR5, thereby preventing CCR5 internalization and blocking human chemokine-directed chemotaxis as well as preventing utilization of CCR5 as a coreceptor for HIV. The effects correlate with the respective binding affinities.
U83A and U83A-N inhibition of CCR5 internalization induced by CCL3. (A-C) U373-MAGI-CCR5E cells were treated with 100 nM CCL3, 50 nM U83A, 100 nM U83A-N alone or in combination. Surface CCR5 was detected by flow cytometry, before chemokine addition and after a 5-minute chemokine incubation compared with isotype control. (D) Preincubation effects with 50 to 100 nM U83A or 100 to 500 nM U83A-N was followed by stimulation with 100 nM CCL3 (representative of 2 experiments in duplicate; mean ± SDs).
U83A and U83A-N inhibition of CCR5 internalization induced by CCL3. (A-C) U373-MAGI-CCR5E cells were treated with 100 nM CCL3, 50 nM U83A, 100 nM U83A-N alone or in combination. Surface CCR5 was detected by flow cytometry, before chemokine addition and after a 5-minute chemokine incubation compared with isotype control. (D) Preincubation effects with 50 to 100 nM U83A or 100 to 500 nM U83A-N was followed by stimulation with 100 nM CCL3 (representative of 2 experiments in duplicate; mean ± SDs).
Inhibition of CCR5 internalization but not endocytosis
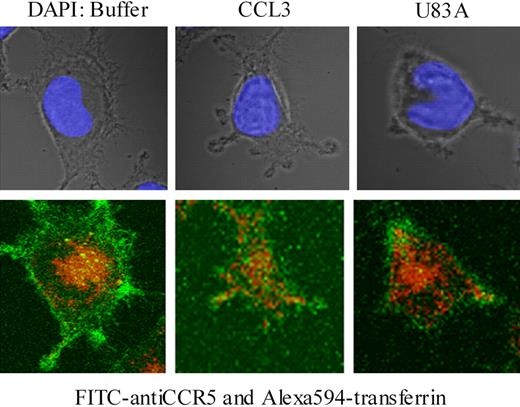
To examine further effects on CCR5 internalization, confocal microscopy was used together with specific antibody “feeding” experiments as described.21 Here, fluorescent-labeled CCR5 antibody (FITC) and transferrin-Alexa 594 were “fed” to the MAGI-CCR5 cells in the presence or absence of human endogenous CCR5 ligand, CCL3, or viral ligand, U83A. Internalization of transferrin-Alexa 594 acted as a positive control for clathrin-mediated endocytosis, where transferrin receptor binding is followed by rapid internalization. In the absence of CCR5 ligand, cells rapidly internalized transferrin (10-20 minutes of incubation), whereas CCR5 remained primarily on the cell surface (Figure 7). With CCL3 treatment CCR5 was rapidly internalized (Figure 7) as previously shown via clathrin-mediated endocytic pathways in Chinese hamster ovary (CHO) or COS-7 cells.21 In contrast, with U83A treatment, CCR5 remained on the cell surface. Although some effects on general morphology and possible “capping” the receptor, there was no internalization of CCR5 even though transferrin was still rapidly endocytosed (Figure 7). Pretreatment of cells with U83A followed by treatment with CCL3 gave similar results (not shown).
U83A blocks clathrin-mediated endocytosis of CCR5 but not transferrin. U373-MAGI-CCR5E cells were grown on coverslips for 24 hours, then exposed to FITC anti-CCR5 (green; bottom) and Alexa Fluor 594-transferrin (red; bottom) for 20 minutes followed by treatment with buffer only, CCR5 ligands 50 nM CCL3 or 100 nM U83A for 10 minutes and mounted with Vectashield mounting solution (DAPI coloration, showing nucleus under incident light in upper panel). Images were taken by a Zeiss 510 laser scanning microscope at × 630 magnification. Fluorochromes were excited at 488 nm for FITC and 543 nm for Alexa Fluor. Serial 0.39-μm confocal slices were captured, and a slice at 3.9 μm is shown from the Z-stack.
U83A blocks clathrin-mediated endocytosis of CCR5 but not transferrin. U373-MAGI-CCR5E cells were grown on coverslips for 24 hours, then exposed to FITC anti-CCR5 (green; bottom) and Alexa Fluor 594-transferrin (red; bottom) for 20 minutes followed by treatment with buffer only, CCR5 ligands 50 nM CCL3 or 100 nM U83A for 10 minutes and mounted with Vectashield mounting solution (DAPI coloration, showing nucleus under incident light in upper panel). Images were taken by a Zeiss 510 laser scanning microscope at × 630 magnification. Fluorochromes were excited at 488 nm for FITC and 543 nm for Alexa Fluor. Serial 0.39-μm confocal slices were captured, and a slice at 3.9 μm is shown from the Z-stack.
Discussion
Overall, these results show U83A as an efficient ligand of CCR5, with highest affinity interactions to date. U83A is an agonist for chemotaxis, whereas the spliced truncated form U83A-N antagonizes human chemokine-induced chemotaxis. These interactions inhibit infectivity of HIV-1 strains which use CCR5 as coreceptor, the primary receptor for HIV-1 transmission. Previously described HIV-inhibitory natural CCR5 antibodies and chemokine ligands appeared to affect receptor internalization,16,21 whereas selected small molecule antagonists affected receptor binding either through direct competition or allosteric effects.15,17,22,23 Modification of CCR5 chemokine ligands can also inhibit internalization.24 The data shown here indicate that viral U83A, unlike human CCR5 chemokine ligands, is not inhibiting HIV-1 infection by receptor internalization, but rather via direct competition or allosteric effects on receptor binding and delaying CCR5 internalization/recycling. CCR3 truncation mutant studies show different human chemokine ligands can mediate altering receptor internalization kinetics.25 This suggests that different signaling pathways may affect receptor recycling. Our recent data on the viral U51 chemokine receptor show different ligands can signal via distinct pathways mediated by different G proteins.26 Some reports suggest distinct G proteins favor cell membrane microdomains such as caveolin in lipid rafts, and this may contribute to the altered pathways shown here and are under further study.
Overall, these results of HHV-6 U83A chemokines on HIV-1 inhibition were similar to those shown for Kaposi sarcoma–associated herpesvirus chemokines KSHV vMIPI and vMIPII although mechanisms are differ. First, KSHV vMIPI at 100 nM was demonstrated to reduce by 80% the infectivity of PBMCs by the CCR5-specific strain SL-2; however, this mechanism remains undefined as vMIPI is CCR8 specific. In the case of KSHV vMIPII, 100 nM caused 50% reduction of HIV-1 infection on the U87/CD4 CCR5 cell line.13,14 Although having similar activities in HIV-1 inhibition, U83A interactions with CCR5 are approximately 50-fold more efficient than those with vMIPII.13,14 Furthermore, vMIPII operates mainly as a broad spectrum chemokine receptor antagonist, whereas U83A is a broad but selective β-chemokine receptor agonist.10 Although the activities of the spliced form U83A-N as CCR5 antagonist and HIV-1 inhibitor resemble vMIPII, both U83A and U83A-N further inhibit CCR5 internalization/recycling as well as leukocyte chemotaxis. Here, the distinction for U83A is the high-affinity interactions with CCR5 as an agonist together with a mechanism which inhibits binding and receptor internalization/recycling. Moreover, because HHV-6 and HIV-1 infect similar cell types, it is more likely that HHV-6 U83A chemokine will compartmentalize in relevant cellular microenvironments in vivo.
There are some studies which indicate in vivo interactions of HHV-6 and HIV. Earlier studies showed HHV-6 viral loads in blood are lowered coincident with the depletion of its target cell, the CD4 T lymphocyte during HIV/AIDS progression.27–29 These results need to be reexamined for HHV-6 strain variants present because only U83 from variant A interacts with CCR5. Thus, for HHV-6A infection, this depletion in the blood may enhance HIV replication in other cell types by removal of a natural, albeit viral, chemokine inhibitor, U83A. In populations where HHV-6A is prevalent (in parts of Africa) and a persistent infection such as other herpesviruses,5 expression of the U83A gene may act as human allelic variation in chemokine or chemokine receptor genes, such as the CCR5δ32 mutation,30 with possible effects on HIV infection and HIV/AIDS progression.
Previously, we further showed U83A agonist activity on CCR1, CCR4, CCR6, and CCR8 present on monocytic/macrophages, effector T lymphocytes, and dendritic cells which can be recruited for virus dissemination plus immune presentation.10 Thus, U83A may also work as a vaccine cellular adjuvant to chemoattract and signal via chemokine receptors on immune effector and antigen-presenting cells. Here, we showed U83A CCR5 agonist activity and HIV-1 infection inhibition. Thus, in a novel combination of properties, U83A could act as an inhibitor and a broad vaccine adjuvant at the same time, combining small molecule drug–like capacity with vaccinelike properties for use in immunizations, or topical microbicides. Given skin homing properties of some U83A-induced chemokine receptors (CCR4, CCR8, CCR6) plus inducible inflammatory receptors (CCR1 and the CCR5 properties shown here), U83A may have particular utility in skin mucosal-directed vaccinations. Thus, U83A may be an effective HIV-1 therapeutic with properties of infection inhibition and immunostimulation. It may also have synergistic effects with existing HIV-1 therapies, including the next generation of small molecule CCR5 inhibitors.
Authorship
Contribution: U.A.G., C.M.P., and J.C. participated in designing the research; J.C. and C.M.P. performed the research; J.C., C.M.P., and U.A.G. controlled and analyzed the data; D.R.D. provided essential reagents; U.A.G. and J.C. wrote the paper; and all authors checked the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ursula A. Gompels, Pathogen Molecular Biology Unit, Department of Infectious & Tropical Diseases, London School of Hygiene & Tropical Medicine, University of London, Keppel St, London WC1E 7HT, United Kingdom; e-mail: ursula.gompels@lshtm.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a project grant from the Biological and Biochemical Sciences Research Council (BBSRC), United Kingdom (U.A.G. and J.C.), and the Health Protection Agency, United Kingdom (C.M.P.).



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal