Abstract
Acquired genomic aberrations have been shown to significantly impact survival in several hematologic malignancies. We analyzed the prognostic value of the most frequent chromosomal changes in a large series of patients with newly diagnosed symptomatic myeloma prospectively enrolled in homogeneous therapeutic trials. All the 1064 patients enrolled in the IFM99 trials conducted by the Intergroupe Francophone du Myélome benefited from an interphase fluorescence in situ hybridization analysis performed on purified bone marrow plasma cells. They were systematically screened for the following genomic aberrations: del(13), t(11;14), t(4;14), hyperdiploidy, MYC translocations, and del(17p). Chromosomal changes were observed in 90% of the patients. The del(13), t(11;14), t(4;14), hyperdiploidy, MYC translocations, and del(17p) were present in 48%, 21%, 14%, 39%, 13%, and 11% of the patients, respectively. After a median follow-up of 41 months, univariate statistical analyses revealed that del(13), t(4;14), nonhyperdiploidy, and del(17p) negatively impacted both the event-free survival and the overall survival, whereas t(11;14) and MYC translocations did not influence the prognosis. Multivariate analyses on 513 patients annotated for all the parameters showed that only t(4;14) and del(17p) retained prognostic value for both the event-free and overall survivals. When compared with the currently used International Staging System, this prognostic model compares favorably. In myeloma, the genomic aberrations t(4;14) and del(17p), together with β2-microglobulin level, are important independent predictors of survival. These findings have implications for the design of risk-adapted treatment strategies.
Introduction
Multiple myeloma is the second most common hematologic cancer, representing 1% of all cancer diagnoses and 2% of all cancer deaths. Despite recent progress in the management of patients, myeloma remains an incurable disease, with a median survival not exceeding 4 years.1 However, this uniform evolution hides a wide heterogeneity in the clinical course; some patients die from disease evolution within a few weeks, whereas others live for more than 10 years. Several prognostic staging systems have been proposed, the most powerful being the recently reported International Staging System (ISS), based on the evaluation of 2 simple biological parameters (ie, the serum levels of β2-microglobulin and albumin).2 Nevertheless, this staging system did not really include the role of (cyto) genetics since very few patients were analyzed for this parameter
Chromosomal abnormalities have been shown to display a major role in disease evolution in several hematologic malignancies. In myeloma, cytogenetics has been hampered by the low proliferative activity of the malignant plasma cells in vitro, and by the frequent low tumor cell infiltrate within the bone marrow specimens. Most large series reported about 30% abnormal karyotypes,3-5 although other techniques not dependent upon obtaining metaphases described genomic aberrations in almost 100% of the cases.6,7 Fluorescence in situ hybridization (FISH) is able to circumvent this pitfall, since it enables the detection of specific chromosomal changes even in noncycling interphase cells. Initial studies using this technique in myeloma demonstrated a high incidence of chromosomal changes,7,8 and suggested that FISH could be used for the assessment of single abnormalities useful for prognostic evaluation.9-13
We designed a study based on interphase FISH for the evaluation of a large series of homogeneously treated patients, using DNA probes specific for the most recurrent chromosomal aberrations observed in myeloma. Our objective was to assess the incidence and clinical relevance of genomic abnormalities in this group of patients treated with high-dose therapy.
Patients, materials, and methods
Approval for this study was obtained from the University Hospitals of Nantes, Toulouse, and Grenoble institutional review boards. Informed consent was provided in accordance with the Declaration of Helsinki.
Patients
Between April 2000 and December 2003, 1064 patients younger than 66 years of age with symptomatic newly diagnosed multiple myeloma were enrolled in the IFM99 therapeutic trials, run by the Intergroupe Francophone du Myélome (IFM; for 81 patients, bone marrow has been analyzed in other laboratories). Briefly, patients received an induction therapy with 4 courses of VAD (vincristine, adriamycin, and dexamethasone), followed by double intensive therapy. The IFM99-02 trial was dedicated for patients with fewer than 2 poor prognosis factors (β2-microglobulin levels above 3 mg/L and del(13) by FISH). After induction, patients received 2 courses of high-dose melphalan (140 mg/m2 and 200 mg/m2), and were then randomized for maintenance therapy—none (arm A), pamidronate (arm B), or pamidronate plus thalidomide (arm C)—until relapse. This trial recruited 780 patients.14 The IFM99-03 trial enrolled 65 patients with 2 poor prognosis factors and with an HLA-identical familial donor.15 After induction, patients received 1 high-dose melphalan course (200 mg/m2), followed by a reduced intensity–conditioned allogeneic transplantation. Finally, the IFM99-04 trial enrolled 219 patients with 2 poor prognosis factors and no HLA-identical familial donor.16 After a similar induction and first high-dose melphalan course, patients received a second melphalan-based intensification (220 mg/m2), and were randomized to receive or not an anti–interleukin-6 (IL-6) antibody during the conditioning regimen. All the patients were analyzed for del(13) by FISH on bone marrow at diagnosis, and 983 of them were referred to the Hematology Laboratory of Nantes and are reported in this study.
Interphase cytogenetic analysis
After overnight shipment, mononuclear cells were separated by gradient-density centrifugation (Ficoll-Hypaque; Eurobio, Les Ulis, France). Plasma cells were then purified using CD138-coated magnetic beads according to the manufacturer's instructions (Miltenyi Biotec, Paris, France), enabling a plasma cell purity higher than 90% (controlled in each patient) as previously described.8 Plasma cells were then analyzed using DNA probes specific for the following chromosomal aberrations: del(13q14) t(11;14)(q13;q32), t(4;14)(p16;q32), MYC rearrangements, hyperdiploidy, and del(17p13). The del(13) was analyzed with a probe specific for the D13S319 locus (purchased from Abbott, Rungis, France). Probes specific for the t(4;14) and t(11;14) translocations were kindly provided by Abbott (Chicago, IL). Hyperdiploidy was assessed using a set of probes specific for chromosomes 5, 9, and 15 (kindly provided by Abbott, Chicago, IL), as previously described (hyperdiploidy if at least 2 probes show extracopies).17 Translocations involving the MYC locus were detected using yeast artificial chromosome (YAC) probes previously described,18 and a commercially available probe, kindly provided by Abbott (Chicago, IL). The del(17p) was assessed using a P53-specific bacterial artificial chromosome (BAC) probe at 17p13 (RPCI-613o12). Probe labeling and FISH procedures have been previously described.8
Statistical analyses
The primary endpoint was the correlation with survival from the time of diagnosis. Kaplan-Meier curves for event-free survival (EFS; defined by the time between diagnosis and the occurrence of progression, relapse, or death) and overall survival (OS) were plotted and compared by the use of the log-rank test. Comparison of frequencies between groups was performed using the chi-square test. Prognostic factors for EFS and OS were determined by means of the Cox proportional hazard model for covariate analysis. As possible prognostic factors, β2-microglobulin levels, albumin levels, hemoglobin levels, platelet counts, isotype, and presence or absence of genomic aberrations (del(13) t(11;14), t(4;14), MYC translocations, hyperdiploidy, and del(17p)) were included in the regression model. For continuous variables, classical cutoffs were selected. To take into account the possible effects of treatment upon prognostic variables, treatment-adjusted statistical analyses were performed. The statistical analyses were performed with the SAS 9.1 software package (SAS Institute Inc, Cary, NC).
Results
Interphase cytogenetic analysis
Since del(13) assessment was required for enrolment in the different trials, patients were primarily analyzed for 13q deletions. Among the 983 bone marrow aspirates received in the lab, 936 were assessable for del(13) (lack of plasma cells or FISH failure in 47 samples). Other probes were analyzed in the following order: t(11;14), t(4;14), hyperdiploidy, MYC, and del(17p). Because of the small number of purified plasma cells in many specimens (median percentage of plasma cells was 6%), these probes have been tested in 746, 716, 657, 571, and 532 patients, respectively (Table 1). del(13) was observed in 449 (48%) patients. The median percentage of plasma cells exhibiting del(13) was 70% (range, 20%-100%). Translocation t(11;14) occurred in 154 (21%) patients, and was associated with del(13) in 60 patients (39% of the t(11;14)-positive patients). Translocation t(4;14) was observed in 100 (14%) patients, and was frequently associated with del(13) (85% of the t(4;14)-positive patients; P < .001). In contrast, t(4;14) and t(11;14) were never associated. Hyperdiploidy was assessed in 256 (39%) patients, and 36% of those presented del(13). A lower incidence of t(11;14) and t(4;14) was also observed in patients with hyperdiploidy (2% and 4.6%, respectively). MYC translocations were observed in 74 (13%) patients. No specific association was observed with other chromosomal abnormalities in these patients with rearranged MYC. Loss of 17p was present in 58 (11%) patients, in a median of 75% of the plasma cells (range, 32%-94%). del(13) was detected in 78% of these patients (P < .001). Translocations t(4;14) and t(11;14) were seen in 11 and 6 patients with del(17p), respectively. No correlation was found with MYC rearrangements or ploidy.
Incidence of chromosomal abnormalities in multiple myeloma
| Genomic aberration . | Incidence, % (no. of patients analyzed for the aberration) . |
|---|---|
| del(13) | 48 (936) |
| t(11;14)(q13;q32) | 21 (746) |
| t(4;14)(p16;q32) | 14 (716) |
| Hyperdiploidy | 39 (657) |
| MYC translocations | 13 (571) |
| del(17p) | 11 (532) |
| Genomic aberration . | Incidence, % (no. of patients analyzed for the aberration) . |
|---|---|
| del(13) | 48 (936) |
| t(11;14)(q13;q32) | 21 (746) |
| t(4;14)(p16;q32) | 14 (716) |
| Hyperdiploidy | 39 (657) |
| MYC translocations | 13 (571) |
| del(17p) | 11 (532) |
Correlation with outcome
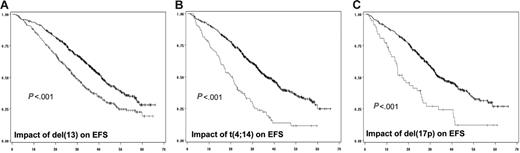
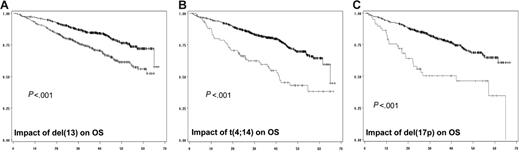
In a first step, we did analyze the prognostic impact of each individual chromosomal aberration on the EFS (Table 2; Figure 1) and OS (Figure 2). When attained, the median survival is given in months from diagnosis. Conversely, if the median survival was not attained, the percentage of patients alive at the median follow-up (ie, 41 months) is given. After a median follow-up of 41 months for surviving patients, 234 of the 936 patients analyzed for chromosomal abnormalities had died. The median EFS for patients with del(13) t(4;14), and del(17p) was 29 months (versus 41 months; P < .001), 20.6 months (versus 36.5 months; P < .001), and 15 months (versus 35 months; P < .001), respectively. Regarding OS, the median was attained for t(4;14) and del(17p) (ie, 41.3 months versus 79% alive at 41 months; P < .001), and 22 months (versus 75% alive at 41 months; P < .001), respectively. For del(13), evaluation at 41 months showed a percentage of surviving patients of 68% (versus 83%; P < .001). Regarding t(11;14), hyperdiploidy, and MYC translocations, no (or marginal) impact on both EFS and OS was observed. For del(13) and del(17p), the prognostic impact was even greater if we split the patients presenting the deletions according to a cutoff of plasma cells presenting the abnormality. Serial analyses showed that the most powerful cutoffs were 74% for del(13) and 60% for del(17p). The median EFSs were 27 months (versus 39 months; P < .001) and 14.6 months (versus 34.7 months; P < .001), respectively, for patients with del(13) of 74% and greater and del(17p) of 60% and greater. Using these cutoffs, 59% of patients with del(13) of 74% and greater were alive at 41 months (versus 80%; P < .001), whereas the median OS was 22.4 months for patients with del(17p) of 60% and greater (versus 75% of patients alive if del(17p) was less than 60%; P < .001).
Prognostic value of chromosomal abnormalities (univariate analysis)
| Genomic aberration . | Impact on EFS, mo*(P) . | Impact on OS†(P) . |
|---|---|---|
| del(13) | 29 vs 41 (< .001) | 68% vs 83% (< .001) |
| t(11;14)(q13;q32) | 35 vs 34 (.2) | 80% vs 74% (.28) |
| t(4;14)(p16;q32) | 20.6 vs 36.5 (< .001) | 41.3 months vs 79% (< .001) |
| Hyperdiploidy | 37 vs 33 (.02) | 82% vs 70% (.006) |
| MYC translocations | 35 vs 37 (.94) | 72% vs 78% (.50) |
| del(17p) | 15 vs 35 (< .001) | 22 months vs 75% (< .001) |
| Genomic aberration . | Impact on EFS, mo*(P) . | Impact on OS†(P) . |
|---|---|---|
| del(13) | 29 vs 41 (< .001) | 68% vs 83% (< .001) |
| t(11;14)(q13;q32) | 35 vs 34 (.2) | 80% vs 74% (.28) |
| t(4;14)(p16;q32) | 20.6 vs 36.5 (< .001) | 41.3 months vs 79% (< .001) |
| Hyperdiploidy | 37 vs 33 (.02) | 82% vs 70% (.006) |
| MYC translocations | 35 vs 37 (.94) | 72% vs 78% (.50) |
| del(17p) | 15 vs 35 (< .001) | 22 months vs 75% (< .001) |
Median EFS for patients presenting the chromosomal abnormality versus that of those who did not present the genomic aberration.
Median OS for patients presenting the chromosomal abnormality versus that of those who did not present the genomic aberration. When the median was not attained, we did calculate the percentage of patients alive at the time of median follow-up (ie, 41 months).
Impact of genomic aberrations on EFS. (A) Kaplan-Meier plot of the impact of del(13) on EFS for the 936 patients analyzed for this abnormality. (B) Impact of t(4;14), analyzed in 716 patients. (C) Value of del(17p) on EFS of 532 patients. The gray curve is for patients presenting the genomic abnormality, whereas the black curve represents the EFS of patients lacking the chromosomal aberration.
Impact of genomic aberrations on EFS. (A) Kaplan-Meier plot of the impact of del(13) on EFS for the 936 patients analyzed for this abnormality. (B) Impact of t(4;14), analyzed in 716 patients. (C) Value of del(17p) on EFS of 532 patients. The gray curve is for patients presenting the genomic abnormality, whereas the black curve represents the EFS of patients lacking the chromosomal aberration.
Impact of genomic aberrations on OS. (A) Kaplan-Meier plot of the impact of del(13) on OS for the 936 patients analyzed for this abnormality. (B) Impact of t(4;14), analyzed in 716 patients. (C) Value of del(17p) on OS of 532 patients. The gray curve is for patients presenting the genomic abnormality, whereas the black curve represents the OS of patients lacking the chromosomal aberration.
Impact of genomic aberrations on OS. (A) Kaplan-Meier plot of the impact of del(13) on OS for the 936 patients analyzed for this abnormality. (B) Impact of t(4;14), analyzed in 716 patients. (C) Value of del(17p) on OS of 532 patients. The gray curve is for patients presenting the genomic abnormality, whereas the black curve represents the OS of patients lacking the chromosomal aberration.
Multivariate analysis
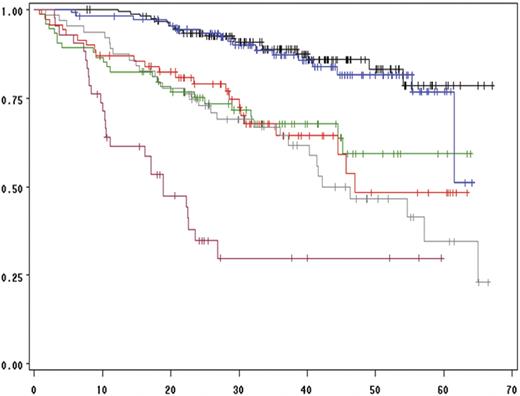
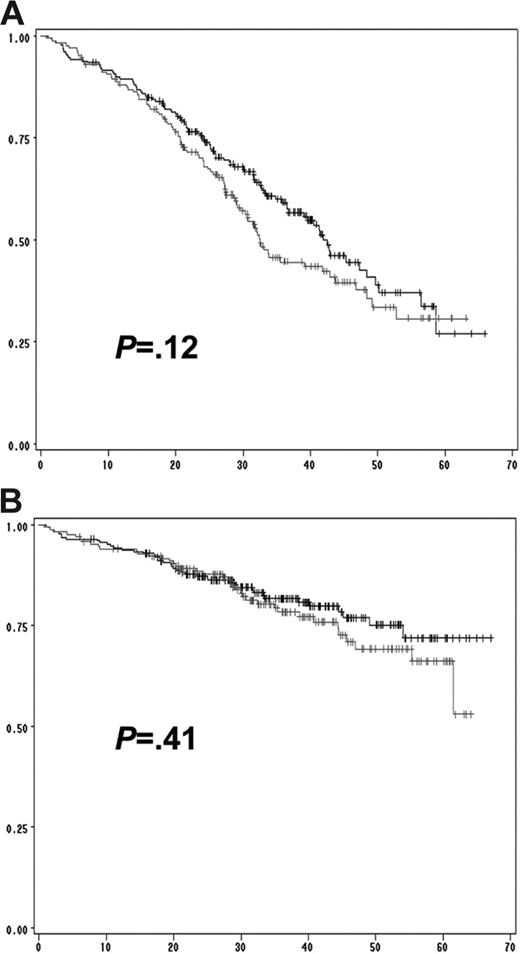
We then performed a multivariate analysis including all the chromosomal aberrations significantly associated with EFS and OS in the univariate analysis (ie, del(13) t(4;14), del(17p), and ploidy) and other parameters shown to be associated with survival in this series: β2-microglobulin level, albumin level, hemoglobin level, and platelet count (Table 3). The analysis was performed on the 513 patients for whom all the parameters were available. Of these, 4 parameters were statistically independent predictors of EFS: t(4;14), del(17p), β2-microglobulin, and hemoglobin levels lower than 100 g/L. A similar analysis for prediction of OS identified 3 factors: t(4;14), del(17p), and β2-microglobulin, with the delineation of 3 groups of patients with highly divergent outcomes (Figure 3). Of note, these analyses showed that the prognostic value of del(13) was almost entirely dependent on the frequent association with t(4;14) and del(17p). In patients lacking these 2 abnormalities, del(13) is not any more significant (Figure 4). Multivariate analyses clearly identified a group of patients with an excellent prognosis (36% of the series): those lacking t(4;14) and del(17p), with a low β2-microglobulin level (expected survival at 4 years = 83%). Conversely, patients presenting either t(4;14) or del(17) and a high β2-microglobulin level have a median OS of only 19 months. Analyses according to treatment randomizations did not modify the results; these 3 parameters retained their independent prognostic significance after treatment-adjusted analyses. We also analyzed the impact of these 2 chromosomal abnormalities in each ISS stage to evaluate the possibility of improving survival prediction. We showed that t(4;14) and/or del(17p) separated 2 groups of patients within each ISS stage (Figures 5–6).
Results of Cox regression analysis of EFS and OS time from diagnosis
| Prognostic parameter . | Hazard ratio for EFS (95% CI) . | P . | Hazard ratio for OS (95% CI) . | P . |
|---|---|---|---|---|
| del(17p) more than 60% | 3.29 (2.23-4.87) | < .001 | 3.93 (2.54-6.08) | < .001 |
| t(4;14) | 2.79 (2.05-3.79) | < .001 | 2.78 (1.90-4.06) | < .001 |
| β2m greater than 4 mg/L | 1.67 (1.28-2.18) | < .001 | 2.83 (2.02-3.97) | < .001 |
| Hb greater than 100 g/L | 1.38 (1.06-1.81) | .017 | — | — |
| Prognostic parameter . | Hazard ratio for EFS (95% CI) . | P . | Hazard ratio for OS (95% CI) . | P . |
|---|---|---|---|---|
| del(17p) more than 60% | 3.29 (2.23-4.87) | < .001 | 3.93 (2.54-6.08) | < .001 |
| t(4;14) | 2.79 (2.05-3.79) | < .001 | 2.78 (1.90-4.06) | < .001 |
| β2m greater than 4 mg/L | 1.67 (1.28-2.18) | < .001 | 2.83 (2.02-3.97) | < .001 |
| Hb greater than 100 g/L | 1.38 (1.06-1.81) | .017 | — | — |
To convert β2-microglobulin level from milligrams per liter to nanomoles per liter, multiply milligrams per liter by 85.
β2m indicates β2-microglobulin level; Hb, hemoglobin level; and —, not significant.
Influence of t(4;14), del(17p), and β2-microglobulin level on overall survival. The black curve is for the 155 patients lacking del(13), t(4;14), and del(17p), and presenting a low β2-microglobulin level (≤ 4 mg/L). The green curve represents the same patients, but with a high β2-microglobulin level (> 4 mg/L; 74 patients). The blue curve depicts the 110 patients lacking t(4;14) and del(17p) with a low β2-microglobulin level, but presenting a del(13). The red curve represents the 69 patients lacking both t(4;14) and del(17p) with a high β2-microglobulin level and with a del(13). The gray curve shows the 63 patients with either a t(4;14) or a del(17p) in more than 60% of their plasma cells, and a low β2-microglobulin level. Finally, the pink curve shows the overall survival of the 42 patients with either a t(4;14) or a del(17p) in more than 60% of their plasma cells, and a high β2-microglobulin level.
Influence of t(4;14), del(17p), and β2-microglobulin level on overall survival. The black curve is for the 155 patients lacking del(13), t(4;14), and del(17p), and presenting a low β2-microglobulin level (≤ 4 mg/L). The green curve represents the same patients, but with a high β2-microglobulin level (> 4 mg/L; 74 patients). The blue curve depicts the 110 patients lacking t(4;14) and del(17p) with a low β2-microglobulin level, but presenting a del(13). The red curve represents the 69 patients lacking both t(4;14) and del(17p) with a high β2-microglobulin level and with a del(13). The gray curve shows the 63 patients with either a t(4;14) or a del(17p) in more than 60% of their plasma cells, and a low β2-microglobulin level. Finally, the pink curve shows the overall survival of the 42 patients with either a t(4;14) or a del(17p) in more than 60% of their plasma cells, and a high β2-microglobulin level.
Prognostic impact of del(13) in patients lacking t(4;14) and del(17p). (A) Prognostic influence of del(13) on EFS in patients presenting neither t(4;14), nor del(17p). (B) Impact of del(13) on OS. No statistically significant difference was observed for both EFS (P = .12) and OS (P = .41). The gray lines represent patients with del(13) but lacking t(4;14) and del(17p); the black lines represent patients lacking all 3 genomic aberrations.
Prognostic impact of del(13) in patients lacking t(4;14) and del(17p). (A) Prognostic influence of del(13) on EFS in patients presenting neither t(4;14), nor del(17p). (B) Impact of del(13) on OS. No statistically significant difference was observed for both EFS (P = .12) and OS (P = .41). The gray lines represent patients with del(13) but lacking t(4;14) and del(17p); the black lines represent patients lacking all 3 genomic aberrations.
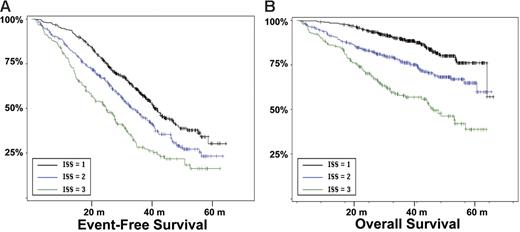
Survival according the ISS stages. (A) EFS according to the ISS stages. (B) OS (in months).
Survival according the ISS stages. (A) EFS according to the ISS stages. (B) OS (in months).
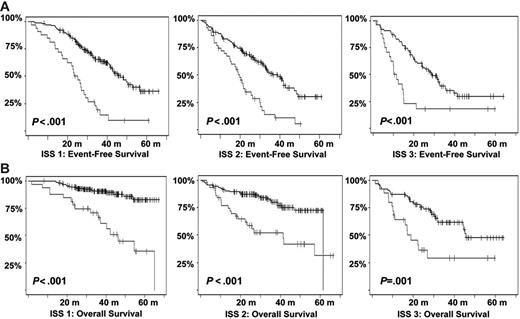
Kaplan-Meier estimates of survival according to the ISS stages and t(4;14) and/or del(17p). (A) EFS among patients in the various ISS stages according to the presence or not of t(4;14) and/or del(17p). (B) OS (in months). Gray lines indicate patients presenting t(4;14) or del(17p); black lines, those lacking the aberrations.
Kaplan-Meier estimates of survival according to the ISS stages and t(4;14) and/or del(17p). (A) EFS among patients in the various ISS stages according to the presence or not of t(4;14) and/or del(17p). (B) OS (in months). Gray lines indicate patients presenting t(4;14) or del(17p); black lines, those lacking the aberrations.
Discussion
We found that even when focusing on a few number of recurrent chromosomal abnormalities, interphase FISH was able to detect genomic changes in almost 90% of the patients with myeloma at diagnosis, about 3 times more frequently than conventional chromosomal banding. This striking difference is most likely due to the low number of plasma cells within the bone marrow specimens sent for laboratory purposes. In this series, the median percentage of plasma cells after mononuclear cell separation was only 6%. In addition, the usually low proliferative activity of malignant plasma cells, and the fact that some of the chromosomal changes are cytogenetically silent at karyotype (like t(4;14)), explain the low informativity of cytogenetics in myeloma. However, the low plasma cell infiltrate present in the samples requires identification of the plasma cells beforehand to perform interphase FISH. Despite these pitfalls, this study showed that genomic information might be obtained in at least 95% of patients with myeloma, even in a multicenter setting.
This study is so far the largest series of patients with newly diagnosed myeloma, analyzed for genomic aberrations, enabling the description of definitive incidences of the most frequent chromosomal abnormalities. The del(13) is the most frequent abnormality (48%), followed by hyperdiploidy (39%), t(11;14) (21%), t(4;14) (14%), MYC translocations (13%), and del(17p) (11%). Moreover, all the patients have been treated with a homogeneous intensive strategy (double transplantation in all cases), which allows for highly valuable prognostic analyses. According to previously reported studies,9-11 del(13) was predictive for both EFS and OS with highly significant P values. However, del(13) was not found to be an independent prognostic factor in the multivariate analysis. Actually, most of the prognostic power of del(13) was related to t(4;14) and del(17p), which are frequently associated with del(13). In patients lacking t(4;14) and del(17p), del(13) was no longer prognostic, whatever the cutoff chosen for its definition (Figure 4).
The analysis of t(4;14) and del(17p) was much more powerful in the prediction of both EFS and OS. Translocation t(4;14) was associated with a median EFS of 20.6 months and a median OS of 41.3 months, which are both highly significantly shorter than those for patients lacking the translocation, in agreement with other smaller series.12,19-24 However, despite its high impact on survival (shown by the multivariate analysis), t(4;14) has to be evaluated in the context of other parameters, and especially β2-microglobulin level. As shown in Figure 3, patients with t(4;14) and a low β2-microglobulin level displayed an outcome close to that of patients lacking the translocation but with a high β2-microglobulin level. Similar conclusions can be drawn for del(17p).12,13,25 Patients presenting the deletion in more than 60% of their plasma cells had a short EFS (14.6 months) and OS (22.4 months), but patients with a low β2-microglobulin level may expect a longer survival (Figure 3). The biological substratum of this major clinical impact is so far unknown for both abnormalities. Translocation t(4;14) is known to deregulate 2 genes, FGFR3 and MMSET.26,27 However, since FGFR3 is not expressed in about one-third of patients with t(4;14),20,21 the target gene is most likely MMSET, whose functions are currently not known. Similarly, the target gene(s) of del(17p) is (are) so far not identified. Even though several authors focused on the P53 gene, formal demonstrations of its deregulation are currently lacking.12,13,25
Other genomic changes have a lower impact on disease evolution. Translocation t(11;14) did not act upon survival, as suggested by some recent studies.28,29 The role of the consequently up-regulated CCND1 gene is not understood so far. Translocations involving MYC did not modify the course of the disease. However, as shown by gene profiling experiments,30 MYC is activated in a large number of patients with myeloma by mechanisms other than translocations, which may have hidden the prognostic impact of these rearrangements. Hyperdiploidy was marginally prognostic in this series. Previous series suggesting a favorable impact of hyperdiploidy on outcome were based on cytogenetics,5,31,32 and thus restricted to patients with an informative karyotype, and were mostly retrospective series including nonhomogeneously treated patients. Evaluation of ploidy by FISH was not dependent upon proliferation and may explore different groups of patients. Furthermore, the assessment of hyperdiploidy using FISH probably underestimates its frequency, and may slightly modify its specific prognostic impact. Nevertheless, because of the marginal impact of hyperdiploidy on survival, this pitfall probably did not introduce a major bias in the analysis. Regarding other potentially prognostic chromosomal aberrations not analyzed in this study, a specific comment is required for t(14;16)(q32;q23),33,34 which has been described as a poor prognosis factor. Because of the scarcity of available plasma cells, we chose to focus our analysis on the most frequent genomic changes, and did not analyze t(14;16), present in less than 5% of the patients.8 Because this translocation is associated with a poor prognosis,12 and is almost constantly associated with del(13), it is highly probable that it would have even “lightened” the prognostic impact of del(13). Finally, we showed that this prognostic model was independent of the treatment, at least in this series of young patients treated with tandem intensification. In particular, the poor prognosis associated with a high β2-microglobulin level, t(4;14), and del(17p) seemed not to be modified by the administration of thalidomide as maintenance therapy. However, because the IFM99-02 trial was dedicated to patients with 0 or 1 poor prognosis factors, these abnormalities were underrepresented in this trial. The analysis could only be performed for del(13), showing that these patients did not benefit from thalidomide maintenance.14 We then analyzed the role of cytogenetic abnormalities according to the ISS. In this classification, only a few patients were analyzed for cytogenetics and/or FISH. We show here that the genetic parameters highly improved the survival prediction power in each ISS stage (Figure 6).
In conclusion, we show that genomic aberrations, evaluated by interphase FISH, play a major role in the evolution of patients with myeloma, extending previous conclusions focused on this topic.35-37 Analysis at diagnosis enables the identification of 3 groups in this large series of patients homogeneously treated by double transplantations: patients who highly benefit from high-dose therapy (patients lacking t(4;14) and del(17p), and who have low β2-microglobulin levels), patients who have a short survival with this type of treatment (patients with either t(4;14) or del(17p), and high β2-microglobulin levels), and an intermediate group. These analyses may have implications for the risk-adapted management of patients with myeloma, at least for the youngest ones. Whether these prognostic parameters are still valid in older patients, or in patients treated with other therapeutic strategies, remain open questions currently under evaluation in other IFM trials.
Authorship
Contribution: H.A.-L. designed and performed the research and wrote the paper; M.A., P.M., and F.G. designed the research and coordinated one of the clinical trials; C.C. performed the statistical analysis; and all other coauthors are members of the IFM board, and participated in the design of the research and in the patients' management.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the IFM appears as Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Correspondence: Hervé Avet-Loiseau, Laboratoire d'Hématologie, Institut de Biologie, 9 quai Moncousu, 44093 Nantes, France; e-mail: herve.avetloiseau@chu-nantes.fr.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We are indebted to Mrs Marine Aliaga, Nadège Gouy, Marie-Christine Boursier, and Karine Pennarun for excellent technical expertise.
This work was supported in part by grants from the Association pour la Recherche sur le Cancer, from the Ligue contre le Cancer (Equipe Labélisée), and from the French Ministry of Health (PHRC 2002).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal