Abstract
The transcription factors Scl and Lmo2 are crucial for development of all blood. An important early requirement for Scl in endothelial development has also been revealed recently in zebrafish embryos, supporting previous findings in scl−/− embryoid bodies. Scl depletion culminates most notably in failure of dorsal aorta formation, potentially revealing a role in the formation of hemogenic endothelium. We now present evidence that the requirements for Lmo2 in zebrafish embryos are essentially the same as for Scl. The expression of important hematopoietic regulators is lost, reduced, or delayed, panendothelial gene expression is down-regulated, and aorta-specific marker expression is lost. The close similarity of the phenotypes for Scl and Lmo2 suggest that they perform these early functions in hemangioblast development within a multiprotein complex, as shown for erythropoiesis. Consistent with this, we find that scl morphants cannot be rescued by a non-Lmo2–binding form of Scl but can be rescued by non-DNA–binding forms, suggesting tethering to target genes through DNA-binding partners linked via Lmo2. Interestingly, unlike other hematopoietic regulators, the Scl/Lmo2 complex does not appear to autoregulate, as neither gene's expression is affected by depletion of the other. Thus, expression of these critical regulators is dependent on continued expression of upstream regulators, which may include cell-extrinsic signals.
Introduction
Lmo2 was first discovered through its homology to the T-cell oncogene lmo1 and has been found to be activated by chromosomal translocations in several T-cell leukemias.1 Like its partner in erythroid cells, scl, during normal development lmo2 is specifically expressed in hematopoietic progenitors, erythroid cells, and megakaryocytes, and in endothelial cells.2,3 Targeted gene ablation in mice revealed an essential role for Lmo2 in primitive hematopoiesis, with lmo2−/− embryos dying at embryonic day (E) 9 to 10 due to severe anemia.2 Lmo2−/− embryonic stem (ES) cells were unable to contribute to any hematopoietic lineage in chimeras, indicating a critical requirement for Lmo2 in definitive hematopoiesis.4 In addition, although lmo2−/− ES cells were initially able to contribute to normal primary capillary plexus formation, they were unable to contribute to the formation of larger vessels, and chimeras with a large contribution of lmo2−/− ES cells exhibited disrupted vascular organization from E9 onwards, indicating a role for Lmo2 in angiogenesis.3
The Lmo2 protein comprises 2 LIM zinc finger–like protein interaction domains and has been found to act as a bridging molecule between the Scl/E47 heterodimer and Gata1/2 in the formation of DNA-binding complexes.5–10 A specific phenylalanine residue in helix 2 of the Scl basic helix-loop-helix (bHLH) domain has been shown to be essential in vitro for hematopoietic activity of Scl. However, rather than being important for heterodimerization with E47, this residue promotes protein interactions with Lmo2.11 Remarkably, introduction of a phenylalanine residue at the corresponding position in the myogenic bHLH, MyoD, was sufficient to confer hematopoietic activity, rescuing hematopoietic development in scl−/− ES cells. The interaction between Scl and Lmo2 is therefore essential for initiation of hematopoiesis in ES cells. However, the in vivo requirement for this interaction has not been formally demonstrated. Nevertheless, the fact that both Scl and Lmo2 are similarly required for primitive and definitive hematopoiesis and for angiogenic remodeling of the vasculature suggests that their activities during these processes may be as partners in a multiprotein complex.
Coinjection of scl and lmo2 mRNA in zebrafish embryos broadens the range of mesodermal tissues that can be converted to a blood and endothelial fate, compared with injection of scl mRNA alone.12 Such gain-of-function experiments suggest that Scl and Lmo2 may drive hematopoietic and endothelial development through specification of the hemangioblast. If true, one would predict that Lmo2 would turn out to be required for endothelial as well as hematopoietic development in loss-of-function experiments, as found for Scl.13
With this in mind, we have examined the role of lmo2 in hematopoietic and endothelial development by knocking down gene expression using antisense morpholinos in zebrafish embryos. In zebrafish, the primitive blood arises from 2 bilateral populations of hemangioblast-like cells within the lateral plate mesoderm of early somite–stage embryos; the anterior lateral-plate mesoderm (ALM), which gives rise to the head vasculature and primitive myeloid cells, and the posterior lateral-plate mesoderm (PLM), which gives rise to the endothelium of the major trunk vessels and a large population of primitive erythrocytes. PLM precursors migrate to the midline, forming the intermediate cell mass (ICM) where primitive erythroid precursors complete their differentiation sandwiched between the forming dorsal aorta (DA) and posterior cardinal vein (PCV). By the end of the first day, a basic vascular loop has been formed and, as the embryonic heart initiates its first contractions, the converged mass of primitive erythrocytes begins to enter circulation. Around this time, runx1 staining reveals a row of single hematopoietic stem cells (HSCs) that remain associated with the DA.14
Our findings essentially recapitulate those of Scl-depleted zebrafish embryos: primitive and definitive hematopoiesis were lost, and endothelial development was severely disrupted, culminating in loss of the DA. Comparison of the earliest defects in gene expression in lmo2 morphants with those of scl morphants did not reveal any major differences, implicating Lmo2 as a partner to Scl during this early stage of its activity in hematopoietic and endothelial development. Consistent with this, we show that Lmo2 binding is essential but DNA binding is dispensable for rescue of hematopoiesis and normal endothelial gene expression in scl morphants. This implies the presence of additional DNA-binding components of the Scl/Lmo2 multiprotein complex.
Materials and methods
Morpholinos
Splice morpholinos (MOs) are a particularly powerful method of target gene knockdown because efficacy of splicing inhibition can be directly assessed by reverse transcriptase–polymerase chain reaction (RT-PCR) analysis of the splice products generated.15 Lmo2 functions as a bridging molecule in multiprotein complexes, mediating protein-protein interactions via 2 zinc finger–like LIM domains.16 To ensure disruption of these domains, upstream splice sites (ie, the exon 4 splice donor or exon 5 splice acceptor) would have to be targeted (Figure 1A). However, polymorphisms in the intronic sequence at the exon 4 splice donor, and the possibility that in-frame splicing between exons 4 and 6 might occur if the exon 5 splice acceptor was targeted, producing a novel protein with one LIM domain and potentially abnormal function, ruled out either sequence as a good candidate for effective splice MO-mediated gene knockdown. It was therefore not practical to use a splice-blocking approach to knock down lmo2 expression in zebrafish. We therefore used antisense MOs designed to block lmo2 translation.
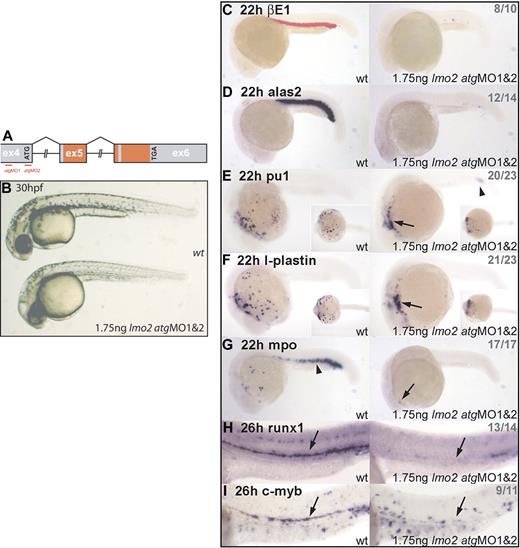
Lmo2 morphants exhibit normal gross morphology but lack hematopoietic gene expression. (A) Genomic structure of the lmo2 gene. Exons (ex) depicted as boxes; atg start and tga stop codons are indicated; intron sizes not to scale; red-shaded regions encode the 2 LIM zinc finger–like domains (the first is split across exons 5 and 6); atgMO binding sites are shown (red lines). (B) Live 30-hpf embryos; lateral views; anterior, left. Lmo2 morphants developed without nonspecific defects. (C-G) Whole-mount embryos; lateral views; anterior, left; inset images in panels E and F show ventral view. (H-I) Whole-mount embryos; lateral view; close-up of trunk/tail region; anterior, left. Numbers of embryos represented in gray. (C-D) Erythroid, βE1, and alas2 expression is abolished in all morphants at 22 hpf. (E-G) In 22-hpf morphants, expression of primitive myeloid genes pu.1, l-plastin, and mpo was severely reduced. Remaining cells were restricted to the heart region (arrows) and, in the case of pu.1, the posterior ICM (arrowhead). (H-I) Expression of HSC markers runx1 and c-myb was significantly down-regulated in the ventral DA of Lmo2-depleted embryos (arrows).
Lmo2 morphants exhibit normal gross morphology but lack hematopoietic gene expression. (A) Genomic structure of the lmo2 gene. Exons (ex) depicted as boxes; atg start and tga stop codons are indicated; intron sizes not to scale; red-shaded regions encode the 2 LIM zinc finger–like domains (the first is split across exons 5 and 6); atgMO binding sites are shown (red lines). (B) Live 30-hpf embryos; lateral views; anterior, left. Lmo2 morphants developed without nonspecific defects. (C-G) Whole-mount embryos; lateral views; anterior, left; inset images in panels E and F show ventral view. (H-I) Whole-mount embryos; lateral view; close-up of trunk/tail region; anterior, left. Numbers of embryos represented in gray. (C-D) Erythroid, βE1, and alas2 expression is abolished in all morphants at 22 hpf. (E-G) In 22-hpf morphants, expression of primitive myeloid genes pu.1, l-plastin, and mpo was severely reduced. Remaining cells were restricted to the heart region (arrows) and, in the case of pu.1, the posterior ICM (arrowhead). (H-I) Expression of HSC markers runx1 and c-myb was significantly down-regulated in the ventral DA of Lmo2-depleted embryos (arrows).
Antisense MOs were obtained from GeneTools (Philomath, OR) with sequences scl spliceMO 5′-AATGCTCTTACCATCGTTGATTTCA-3′, lmo2 atgMO1 5′-CAGTGGCGTTGTTCAATTTCTCCGG-3′, and lmo2 atgMO2 5′-GTAGAAGCCATTTTCAATATGATTC-3′. 10 μg/μL and 2.5 μg/μL stock solutions in dH2O of scl spliceMO and lmo2 atgMO1 plus MO2, respectively, were prepared and injected into 1- to 16-cell–stage embryos at the yolk/blastomere boundary. Quantities of MOs injected were titrated to appropriate effective nontoxic levels (ie, 6.5 ng scl spliceMO and 1.75 ng lmo2 atgMO1 and MO2).
mRNA for injection
Murine lmo2 and zebrafish scl full-length mRNAs were generated as previously described.12 mSCL MH2 and mSCL MH2F constructs11 were subcloned by PCR and recombined into a pBUT3 gateway destination vector in front of and in frame with an HA tag. Truncated Scl constructs were made by amplifying sequence encoding Scl bHLH and HLH domains from pFC617 using forward primers 5′-CTCCTCTAGAGCCACCATGGATCCTAGCAATCGAGTCAAGCGC-3′ and 5′-CTCCTCTAGAGCCACCATGGAGCGCTGGCGACAGCAGAATG-3′, respectively, and reverse primer 5′-CTCCCTCGAGTGGCCCGGGCGGGAGCTTCACC-3′. Products were cloned into pBUT2-MT.17 mRNAs were transcribed from linearized templates using mMessage mMachine (Ambion, Huntingdon, United Kingdom). mRNAs were injected, either alone or in combination with scl MO, into one blastomere of embryos at the 1/2- and occasionally 4-cell stage.
Western analysis
Whole 6- to 10-somite (s)–stage embryos (15 per lane) were homogenized by vortexing with glass beads (Sigma, Poole, United Kingdom) in 2 × RSB, run on a 4% to 12% SDS–polyacrylamide gel, and immunoblotted. Membranes were probed with rat anti-HA (1:1000; Roche, Welwyn Garden City, United Kingdom) followed by antirat (1:2000; Merck Biosciences, Nottingham, United Kingdom) antibodies, or by mouse anti–β-actin (Clone AC 15; Sigma) followed by antimouse (1:2000; Amersham, Buckinghamshire, United Kingdom) antibodies. Signal was detected by enhanced chemiluminescence (ECL; Amersham).
Whole-mount in situ hybridization
Breeding zebrafish were maintained and embryos were raised and staged according to Westerfield.18 Whole-mount in situ hybridization on zebrafish embryos was carried out as previously described.19 Antisense RNAs for in situ hybridization were transcribed from linearized templates using Promega's T3, T7, and SP6 RNA polymerases in the presence of digoxigenin (DIG)–labeled nucleotides (Roche, Burgess Hill, United Kingdom). DIG antibody–alkaline phosphatase conjugate was detected using BM-Purple (Roche) or Fast Red (Sigma). Msr is a novel zebrafish vein–specific marker (IMAGE clone 6 795 34747 ; corresponds to predicted full-length zebrafish clone XM_682265, and shares approximately 65% protein sequence identity with Xenopus Msr20 and approximately 52% with mouse Msr, which shows vein-specific expression in the retinal vasculature21 ).
Image acquisition and processing
Embryos were visualized using a Nikon SMZ 1500 zoom stereomicroscope (Nikon, Melville, NY) equipped with a Nikon 1.0 × WD 54 objective lens with an adjustable internal lens ranging from 0.75 × to 11.25 × magnification. Embryos were viewed at 9 to 11.25 × magnification, depending on size. Images were obtained using a Nikon DXM 1200 digital camera and Nikon ACT-1 software (version 2.12) and processed with Adobe Photoshop CS software, version 8.0 (Adobe Systems, San Jose, CA).
Results
Lmo2-depleted zebrafish embryos fail to develop primitive or definitive blood, phenocopying the lmo2−/− mouse
We used antisense MOs designed to block lmo2 translation (“Materials and methods”). The most effective strategy, as assessed by effect on downstream gene expression and incidence of nonspecific defects, entailed injection of 2 MOs, the first overlapping the lmo2 atg, and the second binding within the 5′ untranslated region (UTR) approximately 300 bp upstream of the atg (Figure 1A). Embryos injected with 1.75 ng of each of these MOs developed essentially normally and without nonspecific defects (Figure 1B).
Although lmo2 morphant embryos appeared grossly normal (Figure 1B), primitive erythrocytes were not observed, either in circulation or accumulated elsewhere. Therefore, Lmo2 depletion in zebrafish embryos recapitulates the phenotype of the lmo2−/− mouse with respect to the loss of primitive blood.2 However, unlike the mouse, and although they eventually develop pericardial edemas, the morphant embryos were able to survive for several days in the absence of blood circulation. Expression of the primitive erythroid genes beta embryonic globin 1 (βE1)22 and aminolevulinate synthase 2 (alas2),23 at 22 hours after fertilization (hpf), was lost in all but a few cells in the ICM (Figure 1C-D). Similarly, expression of pu.1,24 l-plastin,25 and myeloperoxidase (mpo)26 in primitive myeloid cells was down-regulated in 22-hpf morphants (Figure 1E-G). The nonmotile cells remaining under the heart and in the posterior ICM are likely to be improperly programmed myeloid precursors (Figure 1E-G, arrows; Figure 1E, arrowhead). The mpo expression marking erythroid precursors (Figure 1G, arrowhead) was lost.
Expression of runx1 and c-myb at 26 hpf in the region of the ventral DA, which marks the emerging HSCs, was significantly depleted in morphant embryos (Figure 1H-I, arrows). The loss of expression of these definitive HSC markers in the absence of Lmo2 indicates a failure of definitive hematopoiesis in these embryos, a conclusion also drawn from studies of mouse chimeras where lmo2−/− ES cells were found to be unable to contribute to any definitive hematopoietic lineage.4 However, because the absence of circulation in zebrafish embryos does not confer immediate lethality as it does in the mouse, the failure of DA-associated HSCs to emerge can be observed in Lmo2-depleted embryos as opposed to chimeras.
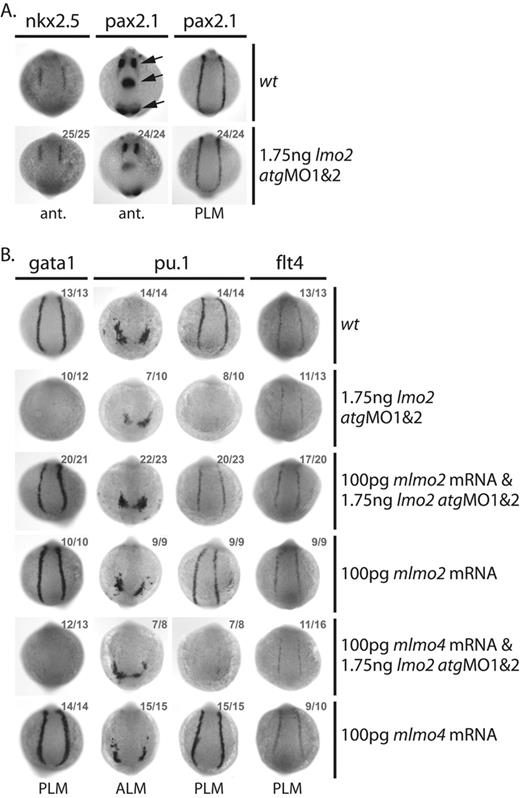
Several observations indicate that the phenotypes generated by MO injection were specific to knockdown of lmo2. First, morphant embryos developed normally without nonspecific defects, either morphologically (Figure 1B) or in terms of gene expression: nonhematopoietic genes nkx2.527 in the precardiac mesoderm and pax2.128 in pronephric duct progenitors, tissues that neighbor the developing blood and endothelium, were unperturbed in morphants at the 10s stage (Figure 2A). Similarly, pax2.1 expression in the optic stalk, midbrain/hindbrain boundary region, and otic vesicle was also unaffected by Lmo2 depletion at 10s (Figure 2A, arrows). Secondly, MO-induced defects in hematopoietic and endothelial gene expression were rescued by coinjection with a murine lmo2 mRNA: the lost expression of gata129 and pu.1 in the PLM, and the reduction in expression of pu.1 in the ALM and flt430 in the PLM of 10s embryos was restored to normal levels (Figure 2B). Coinjection of the lmo2 atgMOs with mRNA encoding an alternative member of the Lmo family, murine lmo4, did not restore normal gata1, pu.1, or flt4 expression, confirming specificity of the rescue (Figure 2B).12
Lmo2 atgMO-mediated knockdown of Lmo2 expression is highly specific. (A-B) Whole-mount 10s embryos; views as indicated; dorsal, top; numbers of embryos represented in gray. (A) Expression of the nonhematopoietic genes nkx2.5 in the precardiac lateral plate mesoderm and pax2.1 in the optic stalk, midbrain/hindbrain boundary region, otic vesicle (arrows, bottom to top), and progenitors of the pronephric duct, immediately adjacent to the primitive erythroid and endothelial precursors in the PLM, was unaffected in morphants. (B) Coinjection of murine lmo2, but not lmo4, mRNA with the lmo2 atgMOs rescued early gata1, pu.1, and flt4 expression levels.
Lmo2 atgMO-mediated knockdown of Lmo2 expression is highly specific. (A-B) Whole-mount 10s embryos; views as indicated; dorsal, top; numbers of embryos represented in gray. (A) Expression of the nonhematopoietic genes nkx2.5 in the precardiac lateral plate mesoderm and pax2.1 in the optic stalk, midbrain/hindbrain boundary region, otic vesicle (arrows, bottom to top), and progenitors of the pronephric duct, immediately adjacent to the primitive erythroid and endothelial precursors in the PLM, was unaffected in morphants. (B) Coinjection of murine lmo2, but not lmo4, mRNA with the lmo2 atgMOs rescued early gata1, pu.1, and flt4 expression levels.
The position of Lmo2 in the regulatory cascade of hematopoietic genes in the PLM (primitive erythroid and HSCs)
Precisely where in the regulatory networks leading to blood formation Lmo2 acts was ascertained by analyzing the earliest traceable defects in gene expression in morphant embryos. Lmo2 expression in the PLM initiates between the 2s and 3s stages, approximately 20 minutes after that of scl (Figure 3I). Genes expressed in the PLM could be divided into 4 categories with respect to their requirement for Lmo2.
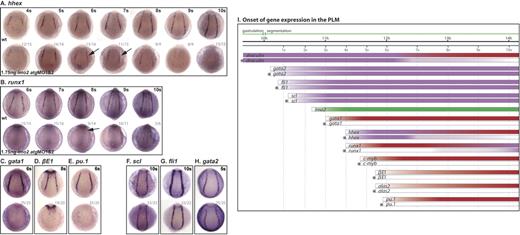
Lmo2 is a critical regulator of PLM development. (A-H) Whole-mount embryos; posterior views; dorsal, top; numbers of embryos represented in gray. (A) Hhex expression initiated as normal, appeared reduced at 6s/7s (arrows) and was lost by 10s. (B) Runx1 was at first dependent on Scl, initiated weakly in the PLM of morphants at 8s (arrow), and increased toward the 10s stage. (C-E) Gata1, βE1, and pu.1 expression was not observed in the PLM of morphant embryos. (F-H) Scl, fli1 and gata2 expression in the PLM was unaffected in 10s morphants. (I) Data from in situ analyses are summarized schematically. Bars represent the changing expression of genes over time. *Gene expression in morphants. Purple indicates Lmo2 independence; red, Lmo2 dependence; and green, lmo2 expression.
Lmo2 is a critical regulator of PLM development. (A-H) Whole-mount embryos; posterior views; dorsal, top; numbers of embryos represented in gray. (A) Hhex expression initiated as normal, appeared reduced at 6s/7s (arrows) and was lost by 10s. (B) Runx1 was at first dependent on Scl, initiated weakly in the PLM of morphants at 8s (arrow), and increased toward the 10s stage. (C-E) Gata1, βE1, and pu.1 expression was not observed in the PLM of morphant embryos. (F-H) Scl, fli1 and gata2 expression in the PLM was unaffected in 10s morphants. (I) Data from in situ analyses are summarized schematically. Bars represent the changing expression of genes over time. *Gene expression in morphants. Purple indicates Lmo2 independence; red, Lmo2 dependence; and green, lmo2 expression.
Dependent: for initiation and maintenance.
Expression of the erythroid terminal differentiation genes, βE1 globin and alas2, and the regulators, gata1, pu.1, and c-myb, was not initiated in lmo2 morphants (Figure 3C-E, I; data not shown). Lmo2 is therefore essential, either directly or indirectly, for the initiation and maintenance of expression of these genes.
Dependent: for timing of initiation.
Runx1 expression in the PLM of lmo2 morphants started at the 8s stage as opposed to 4s in wild-type embryos, and gradually increased (Figure 3B [arrow], I). This suggests that Lmo2 is required for the normal initiation of runx1 expression in the PLM, although its expression is subsequently activated and strengthened by other factors. This delay was also seen in scl morphants, albeit not to the same extent as it is in lmo2 morphants (runx1 expression initiated at 6s as opposed to 8s).13
Dependent: for maintenance.
Early broad expression of the zinc-finger transcription factor draculin,25 which initiates prior to that of scl and lmo2, must be independent of both. However, reduction of draculin expression in lmo2 morphants (ie, Lmo2 dependence), was evident from the 7s stage and increased until, at 10s, draculin expression was lost in all but the most posterior PLM cells around the tailbud (data not shown; Figure 3I). This region of the PLM is both Scl and Lmo2 negative at this time (data not shown), and therefore draculin expression would not be expected to be Scl or Lmo2 dependent. This category also includes hhex,31 normally expressed in the PLM from around 4s. In lmo2 morphants, hhex expression initiated normally but became reduced from around the 6s/7s stage and was essentially lost by 10s (Figure 3A [arrows], I). This indicates that while initiation of hhex expression is Lmo2 independent, like draculin, maintenance of hhex in the PLM from around 6s/7s onwards requires Lmo2.
Independent.
Genes in this category include putative hemangioblast markers scl, gata2,29 and fli1,30 which are expressed in the PLM before lmo2, and must therefore be initiated independently of this transcription factor. The expression of these genes in lmo2 morphants was unaffected at 10s and earlier (Figure 3F-I; data not shown). Thus, a precursor population, expressing hemangioblast genes, was still present in the PLM, although it was unable to elaborate the blood program in the absence of Lmo2.
The position of Lmo2 in the regulatory cascade of hematopoietic genes in the ALM (primitive myeloid)
As in the PLM, lmo2 is expressed in the ALM from 2s to 3s (Figure 4G). The consequences of Lmo2 depletion for the expression of hematopoietic genes in this region fell into the same 4 categories.
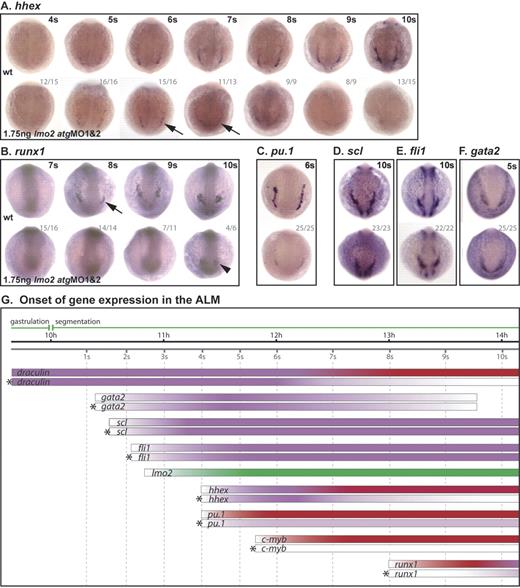
Lmo2 is a critical regulator of ALM development. (A-F) Whole-mount embryos; posterior views; dorsal, top; numbers of embryos represented in gray. (A) Although staining is generally weak in these embryos, hhex expression initiated as normal, appeared reduced at 6s/7s (arrows) and was lost by 10s. (B) Runx1 was at first dependent on Scl, initiating only weakly in the ALM of morphants at 10s (arrowhead). Pu.1 expression in the ALM of morphant embryos was substantially reduced (C), while scl, fli1, and gata2 expression was unaffected (D-F). (G) Data from in situ analyses are summarized schematically as previously described for Figure 3.
Lmo2 is a critical regulator of ALM development. (A-F) Whole-mount embryos; posterior views; dorsal, top; numbers of embryos represented in gray. (A) Although staining is generally weak in these embryos, hhex expression initiated as normal, appeared reduced at 6s/7s (arrows) and was lost by 10s. (B) Runx1 was at first dependent on Scl, initiating only weakly in the ALM of morphants at 10s (arrowhead). Pu.1 expression in the ALM of morphant embryos was substantially reduced (C), while scl, fli1, and gata2 expression was unaffected (D-F). (G) Data from in situ analyses are summarized schematically as previously described for Figure 3.
Dependent: for initiation and maintenance.
Pu.1 and c-myb expression was substantially reduced or lost in the ALM of lmo2 morphants (Figure 4C, G; data not shown).
Thus, these regulators of myeloid differentiation are dependent, either directly or indirectly, on Lmo2 for their expression.
Dependent: for timing of initiation.
Initiation of runx1 expression in the ALM was delayed in MO-injected embryos, from 8s (Figure 4B [arrow], G) to 10s (Figure 4B [arrowhead], G), indicating a requirement for Lmo2 for the correct initiation of runx1 expression in the ALM. In this instance, the delay in ALM runx1 expression was the same as previously observed in scl morphants,13 indicating an identical requirement for lmo2 expression.
Dependent: for maintenance.
As in the PLM, the broad expression of draculin in the ALM region before lmo2 is switched on was, as expected, controlled independently of Lmo2. However, by 7s, when draculin expression is becoming restricted to the ALM, its expression was substantially reduced in morphants, and was lost by 10s (data not shown; Figure 4G). In addition, the profile of hhex expression in the ALM of lmo2 morphants followed that of the PLM with normal expression at 4s that decayed by 7s and was absent by 10s (Figure 4A [arrows], G). Therefore, as in the PLM, the expression of draculin and hhex in the ALM becomes Lmo2 dependent at 7s.
Independent.
In lmo2 morphants, expression of scl, fli1, and gata2 in the ALM initiated normally (Figure 4D-G). Persistence of expression of these early hemangioblast genes indicates that precursor cells were not lost, but their elaboration of the myeloid program was compromised in the absence of Lmo2.
Endothelial gene expression is reduced in Lmo2-depleted embryos
The expression of endothelial markers was significantly down-regulated following injection of the lmo2 atgMOs. While fli1 expression in the PLM of 10s stage morphants was normal (Figure 3G), flk132 and flt4 expression was significantly lower (Figure 5A-B), indicating an important early role for Lmo2 in establishing the program of endothelial development.
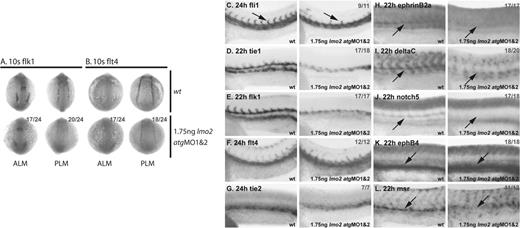
Endothelial development is severely disrupted in Lmo2-depleted embryos. (A-B) Whole-mount 10s embryos; views as indicated; dorsal, top. (C-L) Whole-mount embryos; lateral view; close-up of trunk/tail region; anterior, left. Numbers of embryos represented in gray. (A-B) At 10s, flk1 and flt4 expression in the ALM and PLM was substantially reduced in morphant embryos. (C) In 24-hpf morphants, fli1 expression was reduced in the trunk region and sprouting of primary ISVs was mostly absent (arrows). (D-E) Tie1 and flk1 expression was significantly reduced in the trunk in 22-hpf morphants. (F-G) However, flt4 and tie2 expression at 24hpf was not as significantly down-regulated in morphant embryos. (H-J; arrows) Expression of ephrinB2a (H) deltaC (I), and notch5 (J) in the DA was lost in 22-hpf morphants. (K-L; arrows) Although expression of the vein-specific marker ephB4 (K) was not down-regulated, msr expression in the PCV was lost (L).
Endothelial development is severely disrupted in Lmo2-depleted embryos. (A-B) Whole-mount 10s embryos; views as indicated; dorsal, top. (C-L) Whole-mount embryos; lateral view; close-up of trunk/tail region; anterior, left. Numbers of embryos represented in gray. (A-B) At 10s, flk1 and flt4 expression in the ALM and PLM was substantially reduced in morphant embryos. (C) In 24-hpf morphants, fli1 expression was reduced in the trunk region and sprouting of primary ISVs was mostly absent (arrows). (D-E) Tie1 and flk1 expression was significantly reduced in the trunk in 22-hpf morphants. (F-G) However, flt4 and tie2 expression at 24hpf was not as significantly down-regulated in morphant embryos. (H-J; arrows) Expression of ephrinB2a (H) deltaC (I), and notch5 (J) in the DA was lost in 22-hpf morphants. (K-L; arrows) Although expression of the vein-specific marker ephB4 (K) was not down-regulated, msr expression in the PCV was lost (L).
At 24 hpf, when endothelial precursors have migrated to the midline to form the axial vessels, expression of fli1 was also reduced, and intersomitic vessels (ISVs) failed to sprout normally (Figure 5C). At 22 hpf, expression of tie133 normally delineates the 2 axial vessels, the DA and PCV, separated by a tie1− mass of differentiating primitive erythrocytes that have not yet entered circulation. However, staining in lmo2 morphant embryos revealed fewer cells, and these were restricted to the PCV and expressing tie1 only weakly (Figure 5D). The expression of flk1, which is usually stronger in the DA than the PCV at this time, was similarly restricted to the PCV in lmo2 morphants (Figure 5E). However, the expression of flt4 and tie2,33 which is becoming restricted to the PCV by 24 hpf, was not as significantly reduced in morphants (Figure 5F-G). Furthermore, the expression of artery-specific markers ephrinB2a,34 deltaC,34 and notch534 in the DA was lost (Figure 5H-J), while vein-specific ephB434 expression in the PCV was not down-regulated (Figure 5K). Together, these data suggest that the DA is more severely affected than the PCV by the depletion of Lmo2. However, although ephB4 expression was not down-regulated, PCV development was not entirely normal as residual tie1 and flk1 expression in the PCV was reduced (Figure 5D-E). Furthermore, expression of a novel zebrafish vein–specific marker, msr (“Materials and methods”), was also lost (Figure 5L). Therefore, DA development was severely disrupted, and, while PCV endothelial cells were present, their development was also abnormal. This indicates a fundamental requirement for Lmo2 in endothelial development from the earliest stages.
Lmo2 but not DNA binding by Scl is required for specification of the primitive erythroid, myeloid and endothelial lineages
Lmo2 has been shown to interact with Scl in a multiprotein complex during erythropoiesis.5,6,8–11 From the data presented thus far, with 1 minor exception, the molecular phenotype of the lmo2 morphant is indistinguishable from that of the scl morphant,13 suggesting that the early functions of these 2 proteins may also be effected in a multiprotein complex. To test this prediction, we analyzed the requirement for Lmo2 and DNA binding by Scl in the specification of the primitive erythroid, myeloid, and endothelial lineages. Earlier studies, in scl−/− ES cells, demonstrated that the phenylalanine residue in the second helix of Scl, necessary for interaction with Lmo2, is required to rescue hematopoiesis.11 DNA binding, however, is dispensable.35 In neither case was endothelial development examined. DNA-binding mutants were also shown to restore both primitive hematopoietic and endothelial gene expression in the zebrafish mutant cloche, which lacks scl expression. However, as these mutants were capable of inducing the expression of endogenous wild-type scl, it was impossible to assess the true Scl DNA-binding requirement for specification of these lineages. This problem is avoided in scl morphants, because although endogenous scl expression may be induced, wild-type Scl protein cannot be translated due to aberrant splicing of the scl pre-mRNA.
As shown previously,13 coinjection of wild-type, correctly spliced scl mRNA rescued primitive erythroid (gata1), myeloid (pu.1), and endothelial (flt4) gene expression in scl morphants (Figure 6). However, truncated scl constructs consisting of either the bHLH region or the HLH region alone were capable of similar rescues (Figure 6). In addition, the constitutive expression of the truncated Scl proteins in these embryos, just as for wild-type Scl, caused ectopic endothelial gene expression in the paraxial mesoderm.17 We therefore conclude that the ability of Scl to direct the differentiation of mesodermal precursors toward a hemangioblast cell fate does not require direct DNA binding by Scl. To determine whether Scl is interacting with Lmo2 when carrying out this function, we made use of the mouse scl constructs used in the ES cell rescues.11 We first tested Scl mutated in the critical phenylalanine residue for the ability to rescue scl morphant zebrafish embryos. Unfortunately, this protein was not stable in developing embryos and so the failure to rescue could not be assigned to the failure to interact with Lmo2 (data not shown). We therefore tested mSCL MH2, in which the second helix of the mouse Scl protein is substituted with that of the nonhematopoietic bHLH MyoD. While this protein is still able to dimerize with E2A, it is unable to interact with Lmo2 and cannot rescue in vitro hematopoiesis from scl−/− ES cells. In contrast, mSCL MH2F includes a further modification: a single amino acid substitution which reintroduces the critical phenylalanine residue of the Scl second helix, restoring the ability of the construct to interact with Lmo2 and therefore rescue hematopoiesis from scl−/− ES cells. When mSCL MH2 and MH2F RNAs were tested for their abilities to rescue scl morphant zebrafish embryos, MH2F but not MH2 was able to rescue gata1, pu.1, and flt4 expression, representing primitive erythroid, myeloid, and endothelial lineages (Figure 7A). Western analysis showed that similar amounts of protein had been synthesized in the injected embryos (Figure 7B). We therefore conclude that Scl and Lmo2 act together in a multiprotein complex to induce hemangioblasts in zebrafish embryos.
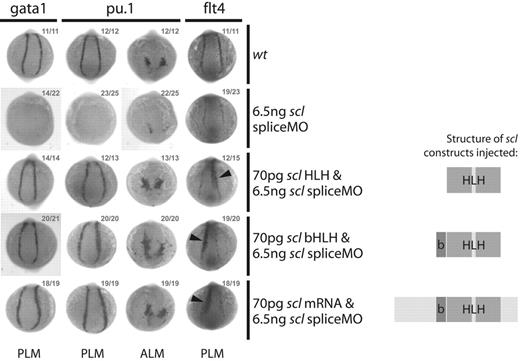
Scl DNA binding is not required for early erythroid, myeloid, or endothelial gene expression. Whole-mount 10s embryos; posterior/anterior views as indicated; dorsal, top; numbers of embryos represented in gray. Coinjection of scl expression constructs containing only the bHLH or HLH domains with the scl spliceMO was sufficient to rescue early gata1 and pu.1 expression in the PLM and the PLM and ALM, respectively. Expression of the endothelial gene flt4 in the PLM could also be restored to normal levels, and ectopic expression of full-length scl or either construct resulted in the expansion of flt4 expression into the presomitic mesoderm (arrowheads).12,17 Structures of Scl constructs used in each case are illustrated.
Scl DNA binding is not required for early erythroid, myeloid, or endothelial gene expression. Whole-mount 10s embryos; posterior/anterior views as indicated; dorsal, top; numbers of embryos represented in gray. Coinjection of scl expression constructs containing only the bHLH or HLH domains with the scl spliceMO was sufficient to rescue early gata1 and pu.1 expression in the PLM and the PLM and ALM, respectively. Expression of the endothelial gene flt4 in the PLM could also be restored to normal levels, and ectopic expression of full-length scl or either construct resulted in the expansion of flt4 expression into the presomitic mesoderm (arrowheads).12,17 Structures of Scl constructs used in each case are illustrated.
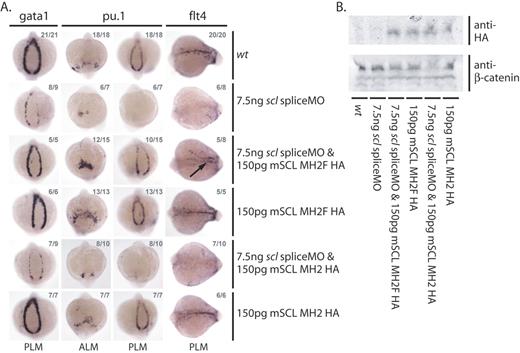
Lmo2 binding is required for early erythroid, myeloid, and endothelial gene expression. (A) Erythroid (gata1 and pu.1 in the PLM), myeloid (pu.1 in the ALM), and endothelial (flt4) gene expression defects in scl morphants can only be rescued by coinjection with an mRNA encoding a bHLH construct which includes the phenylalanine residue of the Scl second helix essential for the interaction between Scl and Lmo2. Whole-mount 15s embryos; views as indicated; numbers of embryos represented in gray. Arrow indicates rescued endothelial gene expression in the position of the dorsal aorta. (B) Western analysis shows that both HA-tagged constructs are expressed in injected whole-embryo extracts.
Lmo2 binding is required for early erythroid, myeloid, and endothelial gene expression. (A) Erythroid (gata1 and pu.1 in the PLM), myeloid (pu.1 in the ALM), and endothelial (flt4) gene expression defects in scl morphants can only be rescued by coinjection with an mRNA encoding a bHLH construct which includes the phenylalanine residue of the Scl second helix essential for the interaction between Scl and Lmo2. Whole-mount 15s embryos; views as indicated; numbers of embryos represented in gray. Arrow indicates rescued endothelial gene expression in the position of the dorsal aorta. (B) Western analysis shows that both HA-tagged constructs are expressed in injected whole-embryo extracts.
Discussion
Lmo2 is essential for hematopoiesis
MO-mediated knockdown of lmo2 expression in zebrafish effectively recapitulated the gross hematopoietic defects of the lmo2−/− mouse. Morphant embryos developed without primitive or definitive blood: primitive erythroid gene expression was essentially lost, primitive myeloid gene expression was significantly reduced, and the expression of genes associated with definitive hematopoiesis in DA-associated HSCs was mostly absent.
Just as lmo2-null mouse embryos display the same gross hematopoietic defects as those with targeted deletions in scl,2,4 the effects on hematopoietic gene expression seen following injection of the lmo2 atgMOs were essentially the same as those previously obtained in scl morphants.13 Expression of fli1 and gata2, which initiates prior to that of both scl and lmo2 and was normal in both morphants, is therefore Scl and Lmo2 independent. The direct or indirect requirements for Lmo2 for expression of all other hematopoietic and endothelial genes analyzed were essentially the same as the requirements for Scl,13 strongly suggesting that Scl and Lmo2 act together within a multiprotein complex during development and differentiation of the hemangioblast. The only observed exception to this was a subtle difference in the control of runx1 expression in the PLM. Runx1 expression, in the absence of Scl, began to recover at an earlier timepoint than in the absence of Lmo2. This is likely to reflect a genuine difference between control by Scl and Lmo2 rather than a difference in penetrance of MOs, as the delay in runx1 expression in the ALM was the same in both morphants. It is possible that Lmo2 may interact and collaborate with as yet unknown factors that rescue expression of runx1 in the PLM in the absence of Scl, enabling expression of runx1 at an earlier timepoint than is possible in the absence of Lmo2.
In our previous study we used the length of time between the first appearance of scl expression and that of its target genes to determine the likelihood of those targets being directly activated by Scl.13 Candidate direct targets included gata1 and runx1 in the PLM and pu.1 in the ALM. As lmo2 expression initiates approximately 20 minutes later than that of scl in both the PLM and ALM, there would be even less time between the assembly of a fully functional Scl/Lmo2 complex and activation of target genes for the generation of intermediates. Lmo2 expression initiates approximately 20 minutes before that of gata1 in the PLM, 30 minutes before that of pu.1 in the ALM, and 30 minutes before that of runx1 in the PLM. This shorter period therefore increases the likelihood that expression of gata1, pu.1, and runx1 is directly activated by the Scl/Lmo2 multiprotein complex (Figure 8). It is also possible that early PLM expression of c-myb, a proto-oncogene believed to play a role in controlling the proliferation and differentiation of HSCs,36 is activated directly by the Scl/Lmo2 complex as its expression initiates around 40 minutes after that of lmo2 (Figure 8). This is the first time that a direct role for the Scl/Lmo2 complex in regulation of c-myb expression has been suggested.
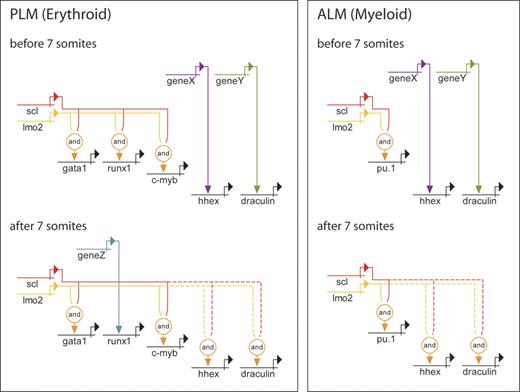
Genetic regulatory networks controlling early PLM and ALM development. Direct relationships, illustrated by continuous lines, are defined by 3 criteria: (1) target gene expression is affected by perturbation of activator; (2) target gene and activator are coexpressed; (3) target gene promoter/enhancer sequences contain binding sites for activators, or length of time between perturbation of activator and effect on target gene is probably insufficient to allow for synthesis of intermediates. Where criteria 1 and 2 are met but 3 is unknown, the relationship is described as indirect and depicted by a dashed line. PLM (erythroid): the data show that initiation of gata1 and c-myb expression is both Scl and Lmo2 dependent. Initially, runx1 expression is dependent on Scl and Lmo2, while hhex and draculin are Scl and Lmo2 independent. After 7s, hhex and draculin become dependent on Scl and Lmo2, and runx1 expression gradually becomes Scl and Lmo2 independent. ALM (myeloid): pu.1 expression is initially dependent on Scl and Lmo2, while hhex and draculin are independent. After 7s, hhex and draculin become Scl and Lmo2 dependent. Unknown activators of Scl- and Lmo2-independent genes are depicted as genes X, Y, and Z. It is possible that X and Y represent the same gene due to similarities in timing of involvement.
Genetic regulatory networks controlling early PLM and ALM development. Direct relationships, illustrated by continuous lines, are defined by 3 criteria: (1) target gene expression is affected by perturbation of activator; (2) target gene and activator are coexpressed; (3) target gene promoter/enhancer sequences contain binding sites for activators, or length of time between perturbation of activator and effect on target gene is probably insufficient to allow for synthesis of intermediates. Where criteria 1 and 2 are met but 3 is unknown, the relationship is described as indirect and depicted by a dashed line. PLM (erythroid): the data show that initiation of gata1 and c-myb expression is both Scl and Lmo2 dependent. Initially, runx1 expression is dependent on Scl and Lmo2, while hhex and draculin are Scl and Lmo2 independent. After 7s, hhex and draculin become dependent on Scl and Lmo2, and runx1 expression gradually becomes Scl and Lmo2 independent. ALM (myeloid): pu.1 expression is initially dependent on Scl and Lmo2, while hhex and draculin are independent. After 7s, hhex and draculin become Scl and Lmo2 dependent. Unknown activators of Scl- and Lmo2-independent genes are depicted as genes X, Y, and Z. It is possible that X and Y represent the same gene due to similarities in timing of involvement.
Again, just as in scl morphants,13 hhex and draculin expression in both the PLM and ALM was initially Lmo2 independent, but from the 7s stage onwards became progressively reduced and therefore increasingly dependent on Lmo2 for maintenance (Figure 8). The delay in onset of runx1 expression in both lmo2 and scl morphants13 indicates that it is initially dependent on activation by the Scl/Lmo2 multiprotein complex, but is subsequently driven by alternative factors (Figure 8).
To conclude, the defects in gene expression in the PLM and ALM in both morphants imply critical early roles for Lmo2 and Scl within a multiprotein complex that functions at or near the top of the hierarchy of transcription factors governing hematopoietic development in both populations.
Lmo2 and endothelial development
The endothelial defects generated on injection of the lmo2 atgMOs were the same as those seen in scl morphants with respect to the genes monitored.13 Early expression of flk1 and flt4 was reduced in endothelial precursors, indicating an early requirement for Lmo2 in the endothelial program. Later, panendothelial gene expression in the trunk region was reduced, ISV formation was mostly absent, and DA-specific gene expression was lost. These observations support a role for Lmo2, alongside Scl, in vascular endothelial growth factor (VEGF) signaling through maintenance of flk1 expression. This early dependence on the Scl/Lmo2 multiprotein complex for correct flk1 expression, and the resulting severe early endothelial defects, may be peculiar to zebrafish where scl and lmo2 expression initiates before that of flk1. However, in mouse embryos, although flk1 is expressed prior to and independently of scl, there is evidence to suggest a role for Scl, and presumably therefore the Scl/Lmo2 multiprotein complex, in maintenance of flk1 expression.37–39 In doing so, the Scl/Lmo2 complex may still play a role in mice in the development of hemogenic endothelium at the time of HSC emergence in the aorta-gonad-mesonephros (AGM). Consistent with this notion, expression of both genes is elevated in the DA at this time.40
As in scl morphants, ephB4 expression in the PCV in lmo2 morphants was normal, while msr expression was lost, suggesting that PCV development is also compromised when Lmo2 or Scl is depleted. As PCV development is essentially normal when VEGF signaling alone is disrupted, there must be an extra requirement for the Scl/Lmo2 complex during venous endothelial development over and above its role in VEGF signaling. This comparatively lesser and VEGF-independent role in endothelial development may be analogous to the late angiogenic defects observed in scl−/− and lmo2−/− mouse embryos.3,41–43
Lmo2 and Scl are obligate partners
The close molecular similarity of the morphant phenotypes for lmo2 and scl raised the possibility that they might be working together in a complex, as has been shown during erythropoiesis.5,6,8–11 Consistent with this suggestion, we found that Scl's function could not be carried out by a mutant version unable to bind Lmo2 although a non-DNA binding mutant was still active, implying a critical requirement for the interaction between Scl and Lmo2 and the presence of cofactors that tether them to the DNA, allowing activation of target genes. Lmo2 has been shown to bind Scl via specific residues in the second helix, bridging an interaction between the Scl/E47 heterodimer and either GATA1 or GATA2.11 It seems, therefore, that the presence of Lmo2 is essential for activation of Scl target genes in the absence of Scl DNA binding to mediate the interaction between Scl and the DNA-binding subunits of the complex. However, in addition, MO-mediated knockdown of lmo2 expression in zebrafish embryos strongly suggests that this interaction is essential for activation of Scl target genes even when Scl DNA-binding activity is intact. The DNA-binding mutant Scl data recapitulate previous studies, both in vitro in ES cells and in vivo in zebrafish embryos, in which Scl, but not direct DNA binding by Scl, was shown to be required for the specification of the primitive hematopoietic lineages.35,44 Also, for the first time, we show that Scl's role in normal endothelial development, potentially at the level of the hemangioblast, does not require DNA binding. The data available, therefore, strongly suggest that Scl and Lmo2 are obligate partners within a multiprotein complex that drives development and differentiation of the hemangioblast.
Is positive feedback required for maintenance of scl and lmo2 expression?
Positive feedback loops and autoregulation are common features of genetic regulatory networks, important for stabilizing expression of key regulators and driving the forward momentum of differentiation (Howard and Davidson45 and references therein). Indeed, such relationships are frequently observed in the regulatory networks that drive hematopoiesis, with gata1, gata2, fli1, pu.1, and c-myb all using autoregulation to maintain their expression (Swiers et al46 and references therein). However, identifying autoregulation in MO studies can be misleading: target gene RNAs are likely to be degraded either because they are defective or because they are not on polysomes. However, since the evidence strongly suggests that Scl and Lmo2 activate target genes as obligate partners, disruption of that partnership through MO-mediated knockdown of one component will indirectly reveal the requirement for positive feedback in maintaining gene expression of the other. Thus, at least until the 10s stage, scl expression was unperturbed in lmo2 morphants and must therefore be Lmo2 independent, just as lmo2 expression in scl morphants was normal and therefore Scl independent. Since loss of either component appears to be sufficient to abolish the activity of the entire complex, it seems unlikely that positive feedback by the Scl/Lmo2 complex is involved in maintaining the levels of Scl and Lmo2, at least during early development and differentiation of the hemangioblast.
Authorship
Conflict-of-interest statement: The authors declare no competing financial interests.
Correspondence: Roger Patient, Weatherall Institute of Molecular Medicine, Oxford University, John Radcliffe Hospital, Headington, Oxford, OX3 9DS United Kingdom; e-mail: roger.patient@imm.ox.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We would like to thank C. Porcher for mSCL MH2 and MH2F constructs, and J. Campos-Ortega, Y. Cheng, K. Crosier, M. Fishman, M. Hammerschmidt, P. Ingham, G. Lieschke, K. Peters, A. Rodaway, D. Stainier, S. Wilson, and L. Zon for probes.
Supported by the Medical Research Council (MRC) (R.P. and M.G.), the Biotechnology and Biological Sciences Research Council (BBSRC) (L.J.P.), and National Institutes of Health (NIH) grants PO1-CA-65493 (C.M.V. and C.E.E.) and GM63904 (S.C.E.).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal