Abstract
Data on 23 patients with low-grade non-Hodgkin lymphomas (NHLs), 4 mantle (MT), 4 marginal zone (MZ), and 15 follicular (FL), were analyzed and compared with 10 high-risk (HR) B-cell chronic lymphocytic leukemias (B-CLLs) with lymph node involvement and 4 diffuse large-cell lymphomas (DLCLs). A significant increase in circulating Vδ1 T lymphocytes producing interleukin-4 (IL-4) was found in patients with FL, MT, and MZ NHL, at variance with DLCL and HR B-CLL. IL-4 was also detectable in the sera and lymph nodes of the same patients. In 19 of the 23 patients with NHL with increased circulating Vδ1 T lymphocytes, B cells expressing the UL-16–binding proteins (ULBPs) ULBP2 or ULBP3 or both were found in peripheral blood, bone marrow, or lymph nodes. Of note, in HR B-CLL or in DLCL, where leukemic cells were negative for ULBPs, no Vδ1 T-cell increase was found. Moreover, Vδ1 T lymphocytes from patients with FL NHL proliferate in response to ULBP2+ and ULBP3+ lymphoma cells. Finally, patients with high expression of ULBPs, increased circulating Vδ1 T lymphocytes, and high levels of serum IL-4 showed stable disease in a 1-year follow-up in contrast to patients with low circulating Vδ1 T cells and undetectable IL-4 or ULBPs.
Introduction
Low-grade non-Hodgkin lymphomas (NHLs) and chronic B-cell lymphocytic leukemia (B-CLL) with lymph node involvement frequently relapse or become resistant to chemotherapy.1,2 In particular, indolent follicular lymphoma (FL) is the second most frequent lymphoid malignancy after diffuse large-cell lymphoma (DLCL). The disease course is often characterized by shortening of disease-free periods after each treatment or transformation to aggressive lymphoma.2 Nevertheless, there is increasing evidence for an immune surveillance against cancer, including hematologic malignancies, making of interest the possibility of manipulating the immune system to eliminate neoplastic cells. In particular, it has been reported that mice deficient in innate effector cells, such as natural killer (NK) cells or γδ T cells, show a significant increased incidence of tumors.3,4 Furthermore, several studies have demonstrated a role for human γδ T lymphocytes in recognition of transformed cells.5–7 Among these lymphocytes 2 main subsets are known: the circulating Vδ2 subset is capable of killing myeloma and Burkitt lymphoma cells,8,9 whereas the Vδ1 subset, which is mainly located in the mucosal-associated lymphoid tissue, is implicated in the defense against epithelial cancers.5,10,11
It has been reported that in some hematologic malignancies circulating γδ T lymphocytes are increased; Vδ1 T cells have been found with high frequency in acute leukemias and low-risk B-CLL,12–14 whereas an expansion of Vδ2 T lymphocytes has been described in multiple myeloma and low-grade NHL after therapy with aminobiphosphonates.9,15 Indeed, the Vδ2 subset recognizes phosphate-containing nonpeptide antigens, without the need of antigen processing and presentation by conventional MHC molecules.16–18 In turn, Vδ1 T cells can bind to the human MHC class I-related molecules MIC-A and MIC-B expressed by epithelial tumors19,20 and by some leukemic cell lines.21 More recently, the MIC-A–related UL-16–binding protein (ULBP) 1, 2, 3, 4 molecules have been found to be recognized by γδ T cells through interaction with the NKG2D receptor.21–23 In mice, γδ T lymphocytes were shown to kill tumor cells through the recognition of the murine retinoic acid early inducible (RAE-1) protein, an MHC-related molecule like the human MIC-A.24
We have recently reported that circulating Vδ1 T lymphocytes are increased in nonprogessor, low-risk B-CLL patients showing a detectable expression of ULBP3 on leukemic cells; these lymphocytes exert antitumor activity in vitro to autologous leukemic B cells.14
In this paper, we show that in patients with low-grade lymphomas expressing ULBP2 or ULBP3 or both, circulating Vδ1 T lymphocytes producing interleukin 4 (IL-4) were increased, in contrast to high-risk (HR) B-CLL or DLCL, which were negative for ULBP2 and ULBP3. Furthermore, Vδ1 T lymphocytes proliferated in vitro in response to lymphoma cells expressing ULBP2 and ULBP3. Finally, patients with high expression of ULBPs on their B cells, increased circulating Vδ1 T lymphocytes, and high levels of serum IL-4 showed stable disease in a 1-year follow-up in contrast to patients with low IL-4 levels, reduced numbers of circulating γδ T cells, and undetectable expression of ULBPs.
Patients, materials, and methods
Patients
Twenty-three patients with low-grade NHL, 4 mantle (MT), 4 marginal zone (MZ), and 15 follicular (FL) lymphomas, and 10 patients with HR B-CLL with lymph node involvement were consecutively enrolled in this study and compared to 4 patients with DLCL. Bone marrow (BM) and peripheral blood (PB) or, in some cases, lymph node (LN) biopsy specimens were obtained under conventional diagnostic procedures, after patients provided verbal informed consent at the moment of the procedure, at the Clinical Hematology Division (Department of Hematology and Oncology, University of Genoa, Genoa, Italy). NHLs were diagnosed according to the Revised European American Classification of Lymphoma Neoplasms (REAL)25 ; B-CLL patients met the diagnostic criteria of the National Cancer Institute Working Group (NCI-WG)26 and were staged according to the Rai modified criteria.27 Their clinical features are summarized in Table 1. One-year follow-up was available for 20 of the NHL patients enrolled in this study. Stable disease (SD) was defined as no significant reduction or measurable change in lymphoma manifestations. Progressive disease (PD) meant an increase of frequency and severity of disease-associated symptoms, occurrence of new nodal or extranodal lesions, or any combination. Fifteen healthy subjects, matched for sex and age, were also studied.
Characteristics of NHL and HR B-CLL patients
| Patient no. . | Sex . | Age, y . | Year* . | Diagnosis† (stage) . | Prior therapy‡ . | Off therapy,§ mo . | Chromosomeabnormality‖ . | Lymphocytes,¶ ×109/L(no. of CD3+ cells) . |
|---|---|---|---|---|---|---|---|---|
| 01 | M | 82 | 2003 | FL (IIIB) | GEOX | 12 | t(14;18) | 1.7 (1.1) |
| 02 | M | 60 | 2002 | FL (IVA) | None | — | None | 2.4 (1.9) |
| 03 | F | 76 | 2002 | FL (II) | (VACOP-B) | 24 | t(14;18) | 2.1 (1.4) |
| 04 | F | 56 | 1999 | FL (IB) | Flu/Ctx/Nov | 24 | None | 2.2 (1.7) |
| 05 | F | 64 | 2002 | FL (IVA) | None | — | None | 2.9 (2.7) |
| 06 | M | 57 | 2000 | FL (IIB) | (FEN) | — | t(14;18); t(8;14) | 1.8 (2.2) |
| 07 | F | 50 | 2004 | FL (IVB) | None | — | None | 1.6 (1.3) |
| 08 | M | 52 | 2004 | FL (II) | VP | 6 | None | 4.3 (3.2) |
| 09 | F | 70 | 2004 | FL (IIIs) | None | — | None | 3.2 (2.3) |
| 10 | F | 64 | 2002 | FL (IVA) | Flu/Ctx/Nov | 36 | t(14;18) | 1.8 (1.0) |
| 11 | F | 57 | 1995 | FL (III) | MACOP-B | 48 | None | 3.8 (2.6) |
| 12 | M | 42 | 2002 | FL (III) | (MACOP-B) | — | None | 4.8 (2.6) |
| 13 | F | 58 | 2004 | FL (II) | (IEV) | — | None | 3.9 (2.2) |
| 14 | M | 53 | 2004 | FL (IVA) | None | — | None | 3.4 (2.3) |
| 15 | F | 60 | 2003 | FL (I) | None | — | None | 2.3 (1.7) |
| 16 | M | 58 | 2004 | MT (IVA) | (FEN + MT) | — | t(14;18) | 2.9 (2.0) |
| 17 | F | 56 | 2004 | MT (II) | None | — | None | 3.1 (2.2) |
| 18 | M | 55 | 2004 | MT (II) | None | — | None | 3.2 (2.1) |
| 19 | M | 58 | 2003 | MT (II) | (IEV) | — | t(14;18) | 2.9 (1.9) |
| 20 | F | 62 | 2004 | MZ (II) | None | — | None | 3.6 (2.8) |
| 21 | F | 70 | 1997 | MZ (IVA) | (GEOX) | — | None | 2.6 (1.6) |
| 22 | F | 60 | 2004 | MZ (IVA) | (IEV) | — | t(14;18) | 3.2 (2.2) |
| 23 | M | 58 | 2003 | MZ (I) | None | — | None | 3.4 (2.4) |
| 24 | M | 62 | 2001 | DLCL | VACOP-B | 36 | None | 3.8 (2.6) |
| 25 | F | 49 | 2004 | DLCL | FEN + MT | 14 | t(14;18) | 2.7 (2.1) |
| 26 | F | 71 | 2004 | DLCL | VACOP-B + MT | 6 | None | 2.5 (1.7) |
| 27 | M | 57 | 2004 | DLCL | (VACOP-B) | — | t(14;18); t(3;14) | 1.9 (1.2) |
| 28 | M | 60 | 1998 | B-CLL (HR) | None | — | ND | 10.9 (1.9) |
| 29 | F | 64 | 1999 | B-CLL (HR) | None | — | ND | 9.8 (1.8) |
| 30 | F | 65 | 2002 | B-CLL (HR) | None | — | ND | 33.4 (4.6) |
| 31 | M | 61 | 2001 | B-CLL (HR) | Flu/Ctx | 10 | ND | 18.9 (1.8) |
| 32 | F | 56 | 1999 | B-CLL (HR) | None | — | ND | 41.5 (3.6) |
| 33 | M | 60 | 2001 | B-CLL (HR) | Flu | 12 | ND | 46.8 (4.1) |
| 34 | M | 65 | 2000 | B-CLL (HR) | None | — | ND | 68.5 (3.6) |
| 35 | M | 50 | 1999 | B-CLL (HR) | None | — | ND | 40.2 (2.8) |
| 36 | F | 59 | 2003 | B-CLL (HR) | None | — | ND | 15.6 (2.9) |
| 37 | M | 62 | 2003 | B-CLL (HR) | None | — | ND | 28.8 (5.9) |
| Patient no. . | Sex . | Age, y . | Year* . | Diagnosis† (stage) . | Prior therapy‡ . | Off therapy,§ mo . | Chromosomeabnormality‖ . | Lymphocytes,¶ ×109/L(no. of CD3+ cells) . |
|---|---|---|---|---|---|---|---|---|
| 01 | M | 82 | 2003 | FL (IIIB) | GEOX | 12 | t(14;18) | 1.7 (1.1) |
| 02 | M | 60 | 2002 | FL (IVA) | None | — | None | 2.4 (1.9) |
| 03 | F | 76 | 2002 | FL (II) | (VACOP-B) | 24 | t(14;18) | 2.1 (1.4) |
| 04 | F | 56 | 1999 | FL (IB) | Flu/Ctx/Nov | 24 | None | 2.2 (1.7) |
| 05 | F | 64 | 2002 | FL (IVA) | None | — | None | 2.9 (2.7) |
| 06 | M | 57 | 2000 | FL (IIB) | (FEN) | — | t(14;18); t(8;14) | 1.8 (2.2) |
| 07 | F | 50 | 2004 | FL (IVB) | None | — | None | 1.6 (1.3) |
| 08 | M | 52 | 2004 | FL (II) | VP | 6 | None | 4.3 (3.2) |
| 09 | F | 70 | 2004 | FL (IIIs) | None | — | None | 3.2 (2.3) |
| 10 | F | 64 | 2002 | FL (IVA) | Flu/Ctx/Nov | 36 | t(14;18) | 1.8 (1.0) |
| 11 | F | 57 | 1995 | FL (III) | MACOP-B | 48 | None | 3.8 (2.6) |
| 12 | M | 42 | 2002 | FL (III) | (MACOP-B) | — | None | 4.8 (2.6) |
| 13 | F | 58 | 2004 | FL (II) | (IEV) | — | None | 3.9 (2.2) |
| 14 | M | 53 | 2004 | FL (IVA) | None | — | None | 3.4 (2.3) |
| 15 | F | 60 | 2003 | FL (I) | None | — | None | 2.3 (1.7) |
| 16 | M | 58 | 2004 | MT (IVA) | (FEN + MT) | — | t(14;18) | 2.9 (2.0) |
| 17 | F | 56 | 2004 | MT (II) | None | — | None | 3.1 (2.2) |
| 18 | M | 55 | 2004 | MT (II) | None | — | None | 3.2 (2.1) |
| 19 | M | 58 | 2003 | MT (II) | (IEV) | — | t(14;18) | 2.9 (1.9) |
| 20 | F | 62 | 2004 | MZ (II) | None | — | None | 3.6 (2.8) |
| 21 | F | 70 | 1997 | MZ (IVA) | (GEOX) | — | None | 2.6 (1.6) |
| 22 | F | 60 | 2004 | MZ (IVA) | (IEV) | — | t(14;18) | 3.2 (2.2) |
| 23 | M | 58 | 2003 | MZ (I) | None | — | None | 3.4 (2.4) |
| 24 | M | 62 | 2001 | DLCL | VACOP-B | 36 | None | 3.8 (2.6) |
| 25 | F | 49 | 2004 | DLCL | FEN + MT | 14 | t(14;18) | 2.7 (2.1) |
| 26 | F | 71 | 2004 | DLCL | VACOP-B + MT | 6 | None | 2.5 (1.7) |
| 27 | M | 57 | 2004 | DLCL | (VACOP-B) | — | t(14;18); t(3;14) | 1.9 (1.2) |
| 28 | M | 60 | 1998 | B-CLL (HR) | None | — | ND | 10.9 (1.9) |
| 29 | F | 64 | 1999 | B-CLL (HR) | None | — | ND | 9.8 (1.8) |
| 30 | F | 65 | 2002 | B-CLL (HR) | None | — | ND | 33.4 (4.6) |
| 31 | M | 61 | 2001 | B-CLL (HR) | Flu/Ctx | 10 | ND | 18.9 (1.8) |
| 32 | F | 56 | 1999 | B-CLL (HR) | None | — | ND | 41.5 (3.6) |
| 33 | M | 60 | 2001 | B-CLL (HR) | Flu | 12 | ND | 46.8 (4.1) |
| 34 | M | 65 | 2000 | B-CLL (HR) | None | — | ND | 68.5 (3.6) |
| 35 | M | 50 | 1999 | B-CLL (HR) | None | — | ND | 40.2 (2.8) |
| 36 | F | 59 | 2003 | B-CLL (HR) | None | — | ND | 15.6 (2.9) |
| 37 | M | 62 | 2003 | B-CLL (HR) | None | — | ND | 28.8 (5.9) |
GEOX indicates gemcitabine, oxaliplatin, dexamethasone; VACOP-B, etoposide, doxorubicin, cyclophosphamide, vincristine, prednisone, bleomycin; Flu/Ctx/Nov, fludarabine/cyclophosphamide/novobiocin; FEN, fludarabine, etoposide, mitoxantrone; VP, vincristine, prednisone; MACOP-B, methotrexate, doxorubicin, cyclophosphamide, vincristine, prednisone, bleomycin; IEV, ifosfamide, etoposide, vincristine; MT, methotrexate; ND, not determined; and —, no therapy.
Year of diagnosis and staging.
Diagnosis and stage defined according to the REAL classification25 and the Rai modified classification.27
Chemotherapy. Therapy started after this study is indicated in the parentheses.
Time from the last cycle of chemotherapy and the time of this study.
Translocations: t(14;18), Bcl-2; t(3;14) Bcl-6; t(8;14) c-myc.28
Lymphocyte count at the time of the study.
Monoclonal antibodies and reagents
The anti-Vδ1 monoclonal antibody (mAb) A13 and anti-Vδ2 mAb (BB3 or γδ123R3 clones, both IgG1) were prepared as described.14 The anti–αβ TCR BMA031 (IgG1) and anti-Vδ1 mAb MCA2080 (IgG1) were obtained from Serotec (Cergy Saint-Christophe, France). The fluorescein isothiocyanate (FITC)–conjugated anti-CD5 (IgG1), the phycoerythrin (PE)–conjugated anti-CD23 (IgG1), the PE–anti-CD20 (IgG2a), the PE–anti-IL-4, the allophycocyanin (APC)–conjugated anti-CD3, and the purified anti-CD3 (IgG1) mAbs were from BD PharMingen Europe (Milan, Italy). The purified anti–IL-4 MAB304 and the anti-NKG2D (MAB139) mAb (both IgG1) were from R&D Systems (Milan, Italy) and the anti–HLA class-I W6/32 (IgG2a)–producing hybridoma was purchased from the American Type Culture Collection (Manassas, VA), and the mAb was purified by affinity chromatography. The anti–MIC-A mAbs AMO1 and BAMO3 were from Immatics Biotechnologies (Tubingen, Germany) and the anti-ULPB mAbs (anti–ULBP2 M311 and anti–ULBP3 M551) were kindly provided by Amgen (Seattle, WA, MTA no. 200309766-001). Phytohemagglutinin (PHA), pokeweed mitogen (PWM), all-trans-retinoic acid (ATRA), and murine immunoglobulins were from Sigma Chemicals (St Louis, MO) and recombinant IL-2 was from PeproTech (London, United Kingdom).
Proliferation of γδ T-cell populations
Mononuclear cells were isolated from peripheral blood (PBMCs) or BM (BMMCs) or LN (LNMCs) of patients with NHL or B-CLL and healthy donors (HDs) by Ficoll-Hypaque gradient. Cells were seeded at 105 cells/well in 96-well U-bottomed microplates (Greiner Labortecknic, Nurtingen, Germany) and cultured in RPMI 1640 medium supplemented with 200 mM l-glutamine 10% FCS, 1 μg/mL PHA, 25 U/mL rIL-2, and 105 irradiated (3000 rad) C1R or H9 lymphoma cell lines as described.14 Autologous B cells were obtained from BM by negative depletion with anti-CD14–, anti-CD4–, and anti-CD8–coated magnetic microbeads (StemCell Technologies, Vancouver, BC, Canada) and were more than 98% pure as assessed by CD20 staining. The majority of B cells in FL lymphomas did not express CD5 and were weakly CD23+ or CD23−, MT and MZ lymphomas were CD5+ and CD23+, respectively, whereas CLL were CD5+CD23+ as described25 (not shown). This phenotype, together with morphology, was also used to identify neoplastic B cells in PB, BM, or LNs.
Experiments with Vδ1 or Vδ2 T cells purified by negative immunodepletion with magnetic microbeads (StemCell Technologies) as described,14 were also performed, adding the purified populations (2 × 104 cells/well) to autologous irradiated (3000 rad) PBMCs in the absence or presence of irradiated H9 cells (5 × 103 cells/well). Proliferation of γδ T cells to the ULBP2+ ULBP3+ H9 or the C1R cell line that do not express at the cell-surface ULBP2 and ULBP3 was evaluated by counting the number of A13+MCA2080+ (Vδ1) or BB3+ (Vδ1) cells at different times (1, 3, 5, 7, and 10 days) of culture. At day 3, 25 U/mL rIL-2 was added to the cultures. In some experiments, PBMCs, BMMCs, or LNMCs were labeled with carboxy-fluorescein diacetate succinimidyl ester (CFSE; Sigma Chemical) at the beginning of culture; at the indicated time points, cells were harvested, stained with APC-anti-CD3 and PE-A13 or with APC-anti-CD3 and PE-BB3 and analyzed by flow cytometry as described.14 Data were analyzed using the ModFit LT version 3.0 computer program (Verity Software House, Topsham, ME).
Immunofluorescence and cytofluorometric analysis
Single or double immunofluorescence on PBMCs or BMMCs or LNMCs of patients and PBMCs from HDs was performed with the various FITC-, PE-, or APC-conjugated mAbs or with mAbs labeled with the fluorochromes Alexa Fluor-488 or -594 contained in the Zenon Tricolor Labeling Kit for mouse IgG1, IgG2a, or IgG2b (Molecular Probes Europe, Leiden, The Netherlands). Immunofluorescence staining of cultured cells was performed as described elsewhere.14 Control aliquots were stained with Alexa Fluor-labeled isotype-matched irrelevant mAbs. The cytoplasmic content of IL-4 was evaluated after cell fixation (3% paraformaldehyde in PBS, 5 minutes at 4°C) and permeabilization (0.1% Nonidet-P40 in PBS, 5 minutes at 4°C), with the specific PE–anti-IL-4 mAb (BD PharMingen). Samples were run on a flow cytometer (FACScan, Becton Dickinson, Heidelberg, Germany) equipped with an argon ion laser. Data were analyzed using CellQuest computer program and are expressed as log red fluorescence intensity (arbitrary units [au]) versus log green fluorescence intensity (au) or versus number of cells or as mean fluorescence intensity (MFI, au). Calibration was assessed with CALIBRITE particles (Becton Dickinson) using the AutoCOMP computer program.
Evaluation of mRNA for ULBPs
Total RNA was prepared from B cells purified from PB or BM or LN using TRIpure (Sigma Chemical) and reverse transcribed using the reverse transcription kit from Amplimedical (Bioline Division, Milan, Italy). In some experiments, RNA was extracted by B cells pretreated with 10 μg/mL ATRA for 1 to 12 hours. The resulting cDNA was amplified by polymerase chain reaction (PCR) with specific primer pairs in 32 cycles at 95°C for 1 minute and 60°C and 72°C for 1 minute. Oligonucleotide sequences (forward; reverse) were: β-actin: 5′-CATACTCCTGCTTGCTGATCC-3′, 5′-ACTCCATCATGAAGTGTGACG-3′; ULBP2: 5′-TTACTTCTCAATGGGAGACTGT-3′, 5′-TGTGCCTGAGGACATGGCGA-3′; ULBP3: 5′-CCTGATGCACAGGAAGAAGAG-3′, 5′-TATGGCTTTGGGTTGAGCTAAG-3′. Amplicons were examined on 2% agarose gel for correct size.13 Images were acquired by Chemi 550 (AlphaInnotech, San Leonardo, CA) and analyzed by Gel Pro Analyzer 3.1 (Media Cybernetics, Silver Spring, MD). Results are expressed as pixel number × 10−3 au.
Measurement of Th1/Th2 cytokines in the sera of NHL patients
Human IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, TNF-α, TNF-β, and IFN-γ levels were measured in the sera of patients with NHL using the FlowCytomix Multiplex kit (Bender MedSystem, Vienna, Austria). The assay is based on a mixture of beads of different size, coated with capture antibodies specific for each cytokine to be analyzed, and a biotin-conjugated second antibody mixture to detect the cytokine of interest. After 2 hours of incubation at room temperature with diluted sera (1:4 dilution), streptavidin-PE was added for 1 hour at room temperature. Samples were then run on a flow cytometer (FACScan, Becton Dickinson), analyzed with BMS FlowCytomix software (Bender MedSystem), and referred to a standard curve. Results are expressed as picogram per milliliter for each cytokine.
Immunohistochemistry
Snap-frozen LN samples, embedded in OCT compound, from 6 FL NHL patients and 3 normal LNs, obtained under diagnostic procedures, were analyzed for the expression of ULBPs in situ; immunohistochemistry was performed on cryosections obtained from patients where frozen samples were available, because ULBPs mAbs are not functional in paraffin-embedded tissues. Thin tissue sections (5 μm) were cut on slides and endogenous peroxidase was quenched with 1.5% hydrogen peroxidase in methanol for 10 minutes. The anti-ULBP2, anti-ULBP3, anti–IL-4, and anti-Vδ1 mAbs were added at 5 μg/mL concentration, and an isotypic unrelated antibody was used as negative control (DakoCytomation, Glostrup, Denmark). A polymeric 2-step method (Supersensitive IHC Detection System, Biogenex, San Ramon, CA) was used as a revelation system, according to the manufacturer's instructions, using 3,3′-diaminobenzidine as chromogen. Then, the slides were counterstained with hematoxylin, cover-slipped, and analyzed under an IX70 microscope (Olympus Biosystem, Planegg, Germany) equipped with a charged coupled device (CCD) camera (Camedia 4040Zoom, Olympus).
Statistical analysis
Data are presented as mean ± SD. Statistical analysis was performed using the Student t test. P values below .05 were considered statistically significant.
Results
Expression of ULBPs in NHL
Expression of ULBPs has been described in acute myeloid leukemias and low-risk B-CLL where it is thought to support one of the arms of natural immunity, represented by γδ T lymphocytes13,14 ; however, it is not known whether these molecules are present and play a role also in low-grade NHL cells. BM- or PB-purified B cells from 23 patients with low-grade lymphomas, 4 MT, 4 MZ, and 15 FL, were analyzed for the expression of ULBPs and compared to 10 HR B-CLL with LN involvement (Table 1). The majority of B cells in FL lymphomas did not express CD5 and were weakly CD23+ or CD23−, MT and MZ lymphomas were CD5+ and CD23+, respectively, whereas CLLs were CD5+CD23+ as described25 (not shown). This phenotype, together with morphology, was also used to identify neoplastic B cells in PB, BM, or LNs.
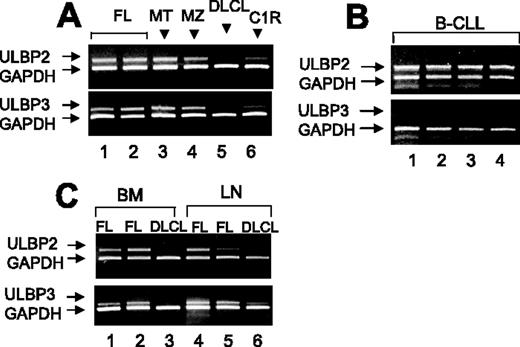
As shown in Figure 1, ULBP2 mRNA was transcribed in purified PB B cells from all MT or MZ, in 14 of 15 FL lymphomas (Figure 1A), and in 8 of 10 B-CLLs (Figure 1 B). ULBP3 mRNA was found in all MZ, 1 MT, and 12 of 15 FL lymphomas (Figure 1A) and not in HR B-CLL (Figure 1B). Furthermore, in 4 FL (Figure 1C shows 2 representative cases), 2 MT and 2 MZ lymphomas (not shown), also purified B cells from BM and LNs were analyzed and both ULBP2 and ULBP3 mRNA were found (Figure 1C); on the other hand, no ULBP2 and ULBP3 mRNA transcription was detected in B cells from PB (Figure 1A) BM, or LNs (panel C) of DLCLs.
Transcription of ULBP2 and ULBP3 in B cells from NHL patients. Total RNA from B cells purified from PB (A-B) or BM or LNs (C) was reverse transcribed and amplified by PCR with specific primers for ULBPs compared to GAPDH. (A-C) ULBP2 and ULBP3 mRNA expression in purified PB B cells from (A) MT or MZ (1 representative case of 4 studied) or FL lymphomas (2 representative cases of 15 analyzed) or DLCL (1 representative case of 4), (B) B-CLL (4 representative cases of 10), and (C) BM- or LN-derived B cells from 2 FL lymphomas and one DLCL.
Transcription of ULBP2 and ULBP3 in B cells from NHL patients. Total RNA from B cells purified from PB (A-B) or BM or LNs (C) was reverse transcribed and amplified by PCR with specific primers for ULBPs compared to GAPDH. (A-C) ULBP2 and ULBP3 mRNA expression in purified PB B cells from (A) MT or MZ (1 representative case of 4 studied) or FL lymphomas (2 representative cases of 15 analyzed) or DLCL (1 representative case of 4), (B) B-CLL (4 representative cases of 10), and (C) BM- or LN-derived B cells from 2 FL lymphomas and one DLCL.
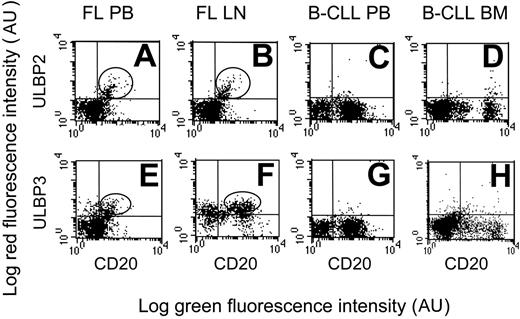
Interestingly, ULPB2 was expressed at the cell surface of PB- and LN-purified (Figure 2A-B, one representative case) or BM-purified (not shown) B lymphocytes in NHL lymphomas (14 of 15 FL and all MT or MZ lymphomas); likewise, ULPB3 was expressed in all MZ and in 10 of 15 FL lymphomas (Figure 2E-F, one representative case). On the other hand, neither DLCL (not shown) nor HR B-CLL expressed ULBP2 (Figure 2C-D, one representative case) or ULBP3 (Figure 2G-H, one representative case) as surface proteins.
Expression of ULBP2 and ULBP3 in NHL B cells. ULPB2 (A-D) or ULBP3 (E-H) expression at the cell surface of PB or LN B lymphocytes in FL NHL (A-B, E-F, 1 representative case of 15 studied) or of PB or BM B lymphocytes in B-CLL (C-D, G-H, 1 representative case of 10 studied), as indicated. B lymphocytes are identified by FITC-conjugated anti-CD20 mAb. Expression of ULBP2 and ULBP3 was assessed by indirect immunofluorescence, with the specific mAbs followed by isotype-specific PE-GAM immunoglobulin and FACS analysis. Results are expressed as log green fluorescence intensity (au) versus log red fluorescence intensity (au).
Expression of ULBP2 and ULBP3 in NHL B cells. ULPB2 (A-D) or ULBP3 (E-H) expression at the cell surface of PB or LN B lymphocytes in FL NHL (A-B, E-F, 1 representative case of 15 studied) or of PB or BM B lymphocytes in B-CLL (C-D, G-H, 1 representative case of 10 studied), as indicated. B lymphocytes are identified by FITC-conjugated anti-CD20 mAb. Expression of ULBP2 and ULBP3 was assessed by indirect immunofluorescence, with the specific mAbs followed by isotype-specific PE-GAM immunoglobulin and FACS analysis. Results are expressed as log green fluorescence intensity (au) versus log red fluorescence intensity (au).
Expansion of Vδ1 T lymphocytes from NHL patients with ULBP2 expression on B cells
Cytofluorimetric analysis of ex vivo isolated PBMCs showed that in NHL expressing ULBP2 or ULBP3 or both (19 of 23) circulating Vδ1, but not Vδ2, T lymphocytes were strongly increased compared to HDs (absolute number/μL blood: 107.0 ± 35.1 versus 15.2 ± 1.1); conversely in DLCL, as well as in HR B-CLL, where neoplastic cells were ULBP−, no Vδ1 T-cell increase was found (not shown). Thus, a close correlation between ULBP expression and number of circulating Vδ1 T cells exists in low-grade lymphomas.
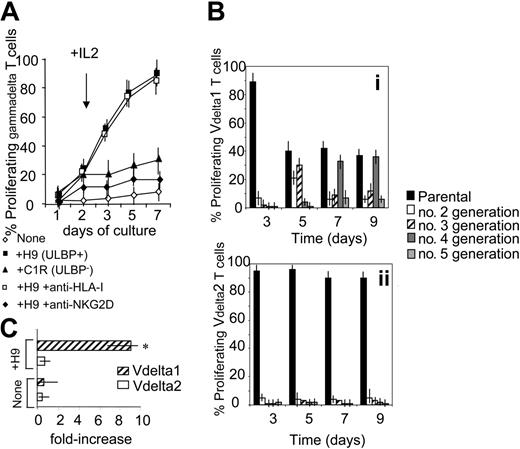
To determine whether the expression of ULBPs on tumor cells might contribute to Vδ1 T-cell expansion, PBMCs from FL NHL patients were stained with CFSE and cocultured with irradiated ULBPs+ H9 or ULBP− C1R lymphoma cells. At day 2, 25 U/mL rIL-2 was added (arrow in Figure 3A). In some experiments 5 μg/mL anti–HLA-I or anti-NKG2D mAb was added during the culture, to define the role of these receptors in γδ T-cell growth. At days 1, 5, and 7, cells were recovered and stained with PE-conjugated anti-γδ mAb and samples were run on a Fluorescence-Activated Cell Sorter (FACS) (Becton Dickinson, Bedford, MA) gated on γδ T cells. Proliferating γδ T cells were identified as red cells with decreased green fluorescence intensity. Of note, a strong proliferation of γδ T lymphocytes (about 80%, n = 10) was observed in all the patients after 7 days of culture (Figure 3A). These cells were all NKG2D+ and mostly (> 90%) Vδ1 (not shown). This proliferation was inhibited by anti-NKG2D, but not by anti–HLA-I, mAbs (Figure 3A). On the other hand, little proliferation was evident on coculture with C1R lymphoma cells that are ULBP2− and ULBP3− (Figure 3A). These data would suggest that interaction between NKG2D on Vδ1 T lymphocytes and ULBPs on neoplastic B cells is needed to induce T-cell proliferation and that in patients with NHL the expansion of Vδ1 T cells might be due to signals delivered by autologous neoplastic B cells.
γδ T lymphocytes from NHL patients proliferate in response to lymphoma cells. (A) PBMCs from FL NHL patients were stained with CFSE and cocultured with ULBP+ H9 or ULBP− C1R lymphoma cells. At day 2, 25 U/mL rIL-2 was added (arrow). In some experiments 5 μg/mL anti–HLA-I or anti-NKG2D mAb was added at the onset of the culture. At days 1, 5, and 7, cells were recovered and stained with PE-conjugated anti-γδ mAb and samples were run on a FACS gated on γδ T cells. Results are expressed as percentage of proliferating γδ T cells, identified as red cells with decreased green fluorescence intensity; mean ± SD of experiments performed with PBMCs from 10 FL NHL patients. (B) Proliferating Vδ1 (i) or Vδ2 (ii) T cells from 10 FL lymphoma patients, stained with CFSE, cultured as in panel A, and identified with PE-conjugated anti-Vδ1 or anti-Vδ2 mAb as red cells with decreased green fluorescence intensity. Data were analyzed with ModFit LT computer program and results expressed as percentage of Vδ1 or Vδ2 cells in second, third, fourth, or fifth generation, compared to the parental nonproliferating population, at different time points, as indicated. (C) Vδ1 or Vδ2 T cells were purified by negative immunodepletion with magnetic microbeads as described14 and cultured with irradiated PBMCs alone (none) or with H9 lymphoma cells (+H9). After 7 days cells were recovered, counted, checked for Vδ1 or Vδ2 expression, and the results expressed as mean fold-increase in cell number ± SD of experiments performed with cells obtained from 6 FL patients. Asterisk indicates P < .01 versus Vδ2.
γδ T lymphocytes from NHL patients proliferate in response to lymphoma cells. (A) PBMCs from FL NHL patients were stained with CFSE and cocultured with ULBP+ H9 or ULBP− C1R lymphoma cells. At day 2, 25 U/mL rIL-2 was added (arrow). In some experiments 5 μg/mL anti–HLA-I or anti-NKG2D mAb was added at the onset of the culture. At days 1, 5, and 7, cells were recovered and stained with PE-conjugated anti-γδ mAb and samples were run on a FACS gated on γδ T cells. Results are expressed as percentage of proliferating γδ T cells, identified as red cells with decreased green fluorescence intensity; mean ± SD of experiments performed with PBMCs from 10 FL NHL patients. (B) Proliferating Vδ1 (i) or Vδ2 (ii) T cells from 10 FL lymphoma patients, stained with CFSE, cultured as in panel A, and identified with PE-conjugated anti-Vδ1 or anti-Vδ2 mAb as red cells with decreased green fluorescence intensity. Data were analyzed with ModFit LT computer program and results expressed as percentage of Vδ1 or Vδ2 cells in second, third, fourth, or fifth generation, compared to the parental nonproliferating population, at different time points, as indicated. (C) Vδ1 or Vδ2 T cells were purified by negative immunodepletion with magnetic microbeads as described14 and cultured with irradiated PBMCs alone (none) or with H9 lymphoma cells (+H9). After 7 days cells were recovered, counted, checked for Vδ1 or Vδ2 expression, and the results expressed as mean fold-increase in cell number ± SD of experiments performed with cells obtained from 6 FL patients. Asterisk indicates P < .01 versus Vδ2.
Gating on γδ T-cell subsets showed that proliferation of Vδ1 T cells in response to H9 lymphoma cells was stronger than that of Vδ2 T lymphocytes; indeed, analysis with a ModFit LT computer program revealed that about 60% of Vδ1 T cells were in second or third generation on day 5, and in fourth or fifth generation on day 7 or 9 (Figure 3Bi). Conversely, the percentage of Vδ2 T cells in second, third, fourth, or fifth generation was almost negligible compared to the parental nonproliferating populations, at the different time points studied (Figure 3Bii). This was also confirmed by experiments performed with purified Vδ1 or Vδ2 T cells added to irradiated PBMCs alone or with H9 lymphoma cells. After 7 days cells were recovered, counted, and checked for Vδ1 or Vδ2 expression and the results showed a 10-fold increase in Vδ1, but not Vδ2, cell number (Figure 3C). This would further support that the former γδ T-cell subset is preferentially involved in the response to ULBP antigens and might also explain why in NHL patients circulating Vδ1 T cells, rather than Vδ2, are increased.
Production of IL-4 by Vδ1 T lymphocytes from NHL patients
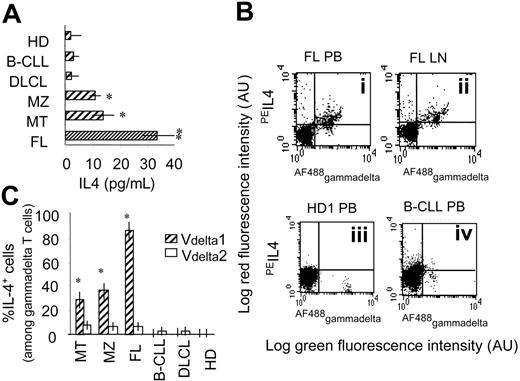
It has been reported that the pattern of cytokine production plays a role in the control of NHL; in particular high levels of IL-4 are related to a good prognosis.29 Thus, we measured IL-4 in the serum of NHL patients using a cytofluorimetric assay that evaluates a panel of Th1/Th2 cytokines, including IL-1β, IL-2, IL-6, IL-8, IL-10, TNF-α, TNF-β, and IFN-γ. We found that only IL-4 is detectable in the serum of patients with FL lymphoma, and to a lesser extent in MT and MZ, but not in DLCL nor in HR B-CLL (Figure 4A). To determine the relative contribution of γδ T cells to IL-4 production, PBMCs were separated from 6 HDs, 10 of 15 FL, 8 of 10 B-CLL, and 4 of 4 DLCL patients (Figure 4 shows 1 representative FL in panel Bi-ii, 1 HD in panel Biii, and 1 B-CLL in panel Biv), and stained with antibodies to identify γδ lymphocytes and cytoplasmic IL-4. In 4 cases of FL, BMMCs were also analyzed. As shown in Figure 4B, γδ T lymphocytes isolated from PB or BM of FL NHL patients were already positive for IL-4 (Figures 4Bi-ii,C) at variance with γδ T lymphocytes from HDs (Figures 4Biii,C), B-CLL patients (Figures 4Biv,C) or DLCL patients (Figure 4C). Interestingly, the majority of ex vivo isolated Vδ1 T cells were IL-4+, both in FL and in MZ or MT NHL patients, at variance with Vδ2 T lymphocytes (Figure 4C); these data would suggest that the Vδ1 T-cell subset in NHL received a stimulus in vivo to produce IL-4.
γδ T lymphocytes from NHL patients produce IL-4. (A) Human IL-4 measured in the sera of HD and patients using the FlowCytomix Multiplex kit. Samples were then run on a flow cytometer (FACScan), analyzed with BMS FlowCytomix software, and referred to a standard curve. Results are expressed as picograms per milliliter and are the mean ± SD from 15 FL, 4 MT, 4 MZ lymphomas, 4 DLCL, 10 B-CLL patients, and 6 HDs. *P < .01 versus HD or B-CLL or DLCL; **P < .01 versus HDs or B-CLL or DLCL, or MZ or MT. (B-C) PBMCs were separated from HDs and FL, B-CLL, and DLCL patients and stained with Alexa Fluor-488–conjugated anti-γδ (B) or anti-Vδ1 or anti-Vδ2 (C) mAb and, after fixation and permeabilization, cytoplasmic staining with PE–anti-IL-4 was performed. (B) Top dot plots are one representative FL staining of PB (i) or LN cells (ii); bottom dot plots are one representative HD (iii) and one representative B-CLL (iv). Results are expressed as log red fluorescence intensity versus log green fluorescence intensity (au). (C) Percentage of IL-4+ cells, among Vδ1 or Vδ2 T cells, calculated as percentage of red cells (PE–anti-IL-4 mAb) after gating on green cells detected by immunofluorescence staining with specific anti-Vδ1 or anti-Vδ2 mAbs. Results are the mean ± SD from 15 FL, 4 MT, 4 MZ lymphomas, 4 DLCL, and 10 B-CLL patients, and 6 HDs. *P < .01 versus Vδ2.
γδ T lymphocytes from NHL patients produce IL-4. (A) Human IL-4 measured in the sera of HD and patients using the FlowCytomix Multiplex kit. Samples were then run on a flow cytometer (FACScan), analyzed with BMS FlowCytomix software, and referred to a standard curve. Results are expressed as picograms per milliliter and are the mean ± SD from 15 FL, 4 MT, 4 MZ lymphomas, 4 DLCL, 10 B-CLL patients, and 6 HDs. *P < .01 versus HD or B-CLL or DLCL; **P < .01 versus HDs or B-CLL or DLCL, or MZ or MT. (B-C) PBMCs were separated from HDs and FL, B-CLL, and DLCL patients and stained with Alexa Fluor-488–conjugated anti-γδ (B) or anti-Vδ1 or anti-Vδ2 (C) mAb and, after fixation and permeabilization, cytoplasmic staining with PE–anti-IL-4 was performed. (B) Top dot plots are one representative FL staining of PB (i) or LN cells (ii); bottom dot plots are one representative HD (iii) and one representative B-CLL (iv). Results are expressed as log red fluorescence intensity versus log green fluorescence intensity (au). (C) Percentage of IL-4+ cells, among Vδ1 or Vδ2 T cells, calculated as percentage of red cells (PE–anti-IL-4 mAb) after gating on green cells detected by immunofluorescence staining with specific anti-Vδ1 or anti-Vδ2 mAbs. Results are the mean ± SD from 15 FL, 4 MT, 4 MZ lymphomas, 4 DLCL, and 10 B-CLL patients, and 6 HDs. *P < .01 versus Vδ2.
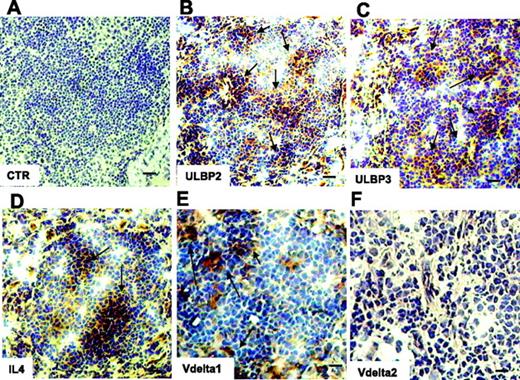
This was further supported by the finding that IL-4 was detected by immunostaining in serial sections of the LNs from 4 NHL of 6 analyzed (Figure 5D); interestingly, in the same specimens ULBP2 and ULBP3 expression was detected as well (Figure 5B-C), at variance with the 2 patients who were negative for IL-4 expression and with normal LNs (not shown). Interestingly, LNs expressing ULBPs and IL-4 were infiltrated by Vδ1 (Figure 5E) but not by Vδ2 T lymphocytes (Figure 5F). Of note, no Vδ1 nor Vδ2 T cells were found in the IL-4− LNs (not shown).
Expression of ULBP2, ULBP3, and IL-4 and localization of Vδ1 T cells in NHL in situ. Thin sections (5 μm) from frozen LNs of 6 FL NHL patients were immunostained with anti-ULBP2 (B), anti-ULBP3 (C), anti–IL-4 (D), or anti-Vδ1 (E) or anti-Vδ2 (F) mAbs, all at 5 μg/mL, followed by the peroxidase revelation system and counterstained with hematoxylin. Panel A is a negative control, an isotypic unrelated antibody (CTR). Arrows in panels B-E indicate positive cells. Slides were cover-slipped and analyzed under a IX70 microscope, equipped with a CCD camera, at ×20 magnification (A-D, bar represents 40 μm), or at ×40 magnification (E-F, bar represents 20 μm). Images were captured with Olympus Plan APO 20×/0.40 NA (A-D) or Olympus UAPO 40×/1.35 NA oil objectives. The total magnification was 200× or 400× according to the 20× or 40× objective used. Images were taken with CELL Olympic Biosystem imaging software version 2.0.
Expression of ULBP2, ULBP3, and IL-4 and localization of Vδ1 T cells in NHL in situ. Thin sections (5 μm) from frozen LNs of 6 FL NHL patients were immunostained with anti-ULBP2 (B), anti-ULBP3 (C), anti–IL-4 (D), or anti-Vδ1 (E) or anti-Vδ2 (F) mAbs, all at 5 μg/mL, followed by the peroxidase revelation system and counterstained with hematoxylin. Panel A is a negative control, an isotypic unrelated antibody (CTR). Arrows in panels B-E indicate positive cells. Slides were cover-slipped and analyzed under a IX70 microscope, equipped with a CCD camera, at ×20 magnification (A-D, bar represents 40 μm), or at ×40 magnification (E-F, bar represents 20 μm). Images were captured with Olympus Plan APO 20×/0.40 NA (A-D) or Olympus UAPO 40×/1.35 NA oil objectives. The total magnification was 200× or 400× according to the 20× or 40× objective used. Images were taken with CELL Olympic Biosystem imaging software version 2.0.
Relationship between ULBPs expression, peripheral γδ T cells, and disease progression
One-year follow-up was available for 20 of the NHL patients analyzed in this study. Interestingly, patients with high number of Vδ1 T lymphocytes (Table 2, group 1: 114.0 ± 35.1 absolute number of cells/μL blood) showed stable disease, they did not display BCl2 or BCl6 gene translocation, and ULBP2 or ULBP3 was expressed on the surface of B cells; furthermore, these data closely correlate with the presence of high levels of IL-4 (Table 2, group 1: 18.0 ± 11.7 pg/mL) in the serum of these patients. In contrast, in 6 NHL patients (3 FL, 1 MZ, 2 MT), who had low numbers of circulating Vδ1T cells (Table 2, group 2: 8.6 ± 6.7; group 1 versus group 2: P < .01) and low levels of serum IL-4 (Table 2, group 2: 2.9 ± 2.0, group 1 versus group 2: P < .01), the disease progressed over 1 year. In these patients ULBP2 or ULBP3 was not expressed at the surface of leukemic cells and they displayed BCl2 or BCl6 gene translocation (Table 2), which are poor prognostic markers.28
Characteristics of low-grade NHL patients: follow-up at 1 year
| Patient no.* . | Stage atdiagnosis† . | One-y follow-up‡(stage in 2005) . | Vδ1§ . | ULBP2/3mRNA‖ . | Surface¶ULBP2/3 . | Serum IL-4level,# pg/mL . | Chromosomeabnormalities** . |
|---|---|---|---|---|---|---|---|
| Group 1 | |||||||
| 2 | FL IVA | SD | 90.6 | +/+ | +/+ | 23.7 | — |
| 4 | FL IB | SD | 100.3 | −/+ | −/+ | 30.7 | — |
| 5 | FL IVA | SD | 101.6 | +/− | +/− | 18.1 | — |
| 7 | FL IVB | SD | 97.8 | −/+ | −/+ | 15.3 | — |
| 8 | FL II | SD | 220.5 | +/+ | +/+ | 8.9 | — |
| 9 | FL IIIs | SD | 100.1 | +/+ | +/+ | 13.7 | — |
| 11 | FL III | SD | 124.6 | +/+ | +/+ | 54.5 | — |
| 13 | FL III | SD | 58.4 | −/+ | −/+ | 7.9 | — |
| 14 | FL IVA | SD | 110.4 | +/+ | +/+ | 12.0 | — |
| 15 | FL I | SD | 132.6 | +/− | +/− | 15.6 | — |
| 17 | MT II | SD | 106.8 | +/+ | +/− | 18.6 | — |
| 18 | MT II | SD | 99.6 | +/+ | −/+ | 14.6 | — |
| 20 | MZ II | SD | 112.1 | +/− | +/− | 9.8 | — |
| 23 | MZ I | SD | 141.4 | +/+ | +/+ | 9.6 | — |
| Group 2 | |||||||
| 1 | FL IIIB | PD (IV) | 9.9 | −/+ | −/− | 2.2 | t(14;18) |
| 3 | FL II | PD (III) | 12.1 | −/+ | −/− | 6.7 | t(14;18) |
| 6 | FL IIB | PD (III) | 10.8 | +/− | −/− | 2.2 | t(14;18); t(8;14) |
| 10 | FL IVA | SD | 18.9 | +/− | +/− | 2.5 | t(14;18) |
| 19 | MT II | PD (III) | 0.1 | −/+ | −/− | 0.0 | t(14;18) |
| 22 | MZ IVA | PD (IVB) | 0.1 | +/− | +/− | 3.9 | t(14;18) |
| Patient no.* . | Stage atdiagnosis† . | One-y follow-up‡(stage in 2005) . | Vδ1§ . | ULBP2/3mRNA‖ . | Surface¶ULBP2/3 . | Serum IL-4level,# pg/mL . | Chromosomeabnormalities** . |
|---|---|---|---|---|---|---|---|
| Group 1 | |||||||
| 2 | FL IVA | SD | 90.6 | +/+ | +/+ | 23.7 | — |
| 4 | FL IB | SD | 100.3 | −/+ | −/+ | 30.7 | — |
| 5 | FL IVA | SD | 101.6 | +/− | +/− | 18.1 | — |
| 7 | FL IVB | SD | 97.8 | −/+ | −/+ | 15.3 | — |
| 8 | FL II | SD | 220.5 | +/+ | +/+ | 8.9 | — |
| 9 | FL IIIs | SD | 100.1 | +/+ | +/+ | 13.7 | — |
| 11 | FL III | SD | 124.6 | +/+ | +/+ | 54.5 | — |
| 13 | FL III | SD | 58.4 | −/+ | −/+ | 7.9 | — |
| 14 | FL IVA | SD | 110.4 | +/+ | +/+ | 12.0 | — |
| 15 | FL I | SD | 132.6 | +/− | +/− | 15.6 | — |
| 17 | MT II | SD | 106.8 | +/+ | +/− | 18.6 | — |
| 18 | MT II | SD | 99.6 | +/+ | −/+ | 14.6 | — |
| 20 | MZ II | SD | 112.1 | +/− | +/− | 9.8 | — |
| 23 | MZ I | SD | 141.4 | +/+ | +/+ | 9.6 | — |
| Group 2 | |||||||
| 1 | FL IIIB | PD (IV) | 9.9 | −/+ | −/− | 2.2 | t(14;18) |
| 3 | FL II | PD (III) | 12.1 | −/+ | −/− | 6.7 | t(14;18) |
| 6 | FL IIB | PD (III) | 10.8 | +/− | −/− | 2.2 | t(14;18); t(8;14) |
| 10 | FL IVA | SD | 18.9 | +/− | +/− | 2.5 | t(14;18) |
| 19 | MT II | PD (III) | 0.1 | −/+ | −/− | 0.0 | t(14;18) |
| 22 | MZ IVA | PD (IVB) | 0.1 | +/− | +/− | 3.9 | t(14;18) |
SD indicates stable disease; PD, progressive disease; and —, no abnormalities.
The number is referred to patients listed in Table 1.
Diagnosis and stage are defined according to the REAL classification.25
Stage after 1 year.
Absolute number of peripheral-blood Vδ1 T lymphocytes/μL at the beginning of the study (early 2004); these numbers did not significantly change after 1 year (2005). Mean ± SD: group 1, 114.0 ± 35.1; group 2, 8.6 ± 6.7; group 1 versus group 2, P < .01. Number of Vδ1 T cells in healthy donors, 15 ± 1.
ULBP2 or ULBP3 mRNA expression evaluated by PCR; +, positive; −, negative.
ULBP2 or ULBP3 surface expression evaluated by immunofluorescence and cytofluorimetric analysis with the specific anti-ULBP2 mAb labeled with Alexa Fluor 594; results are expressed as positive (+), that is. MFI >25 (au); negative (−), that is MFI <20; MFI of Alexa Fluor-labeled isotype-matched irrelevant mAb was 15 au.
IL-4 was measured by enzyme-linked immunosorbent assay in the sera of NHL patients in 2004; results are expressed as picograms per milliliter. Mean ± SD: group 1, 18.0 ± 11.7; group 2, 2.9 ± 2.0; group 1 versus group 2; P < .01. IL-4 in the serum of healthy donors was undetectable.
Translocations: t(14;18), Bcl-2; t(3;14) Bcl-6; t(8;14) c-myc.28
Discussion
In the present work, we show that γδ T cells are likely to provide a contribution to the antitumor response against low-grade NHL, possibly recognizing ULBPs on lymphoma cells. This study started from a number of observations. It has been reported that circulating Vδ2 T lymphocytes are able to kill lymphoma cells,4,8 and we and others have shown that the Vδ1 subset, mainly resident in the mucosal tissue, is effective against acute myeloid leukemias and B-CLL.12–14 The increase in circulating Vδ1 T lymphocytes, reported in acute myeloid leukemias, has been taken to suggest that Vδ1 T cells may either exert an antitumor effect by themselves, or can be used to potentiate antitumor therapies.12,13 Moreover, we reported that in B-CLL patients with increased numbers of Vδ1 T cells, neoplastic B cells showed good prognostic markers such as mutated IgVH genes and the lack of CD38 antigen.14,30 In contrast, one third of B-CLL patients who did not have increased circulating Vδ1 T cells showed disease progression.14
Possible targets for effector γδ T cells are the ULBPs and the nonconventional MHC-related molecules MIC-A and MIC-B that can be expressed by hematologic neoplasias.14,21–23 We here provide evidence that Vδ1 T lymphocytes may be involved also in the antitumor response against low-grade NHL. First, Vδ1 T-cell increase was found in the majority of FL lymphoma patients, but not in DLCL nor in HR B-CLL. Second, Vδ1 T cells from patients with low-grade NHL proliferate in response to lymphoma B cells and this proliferation is related to the expression of ULBPs. Indeed, ULBP2 or ULBP3 is expressed on B cells of FL, MT, and MZ NHL, but not on DLCL or HR B-CLL. It is of note that the Vδ1 T-cell subset not only is significantly expanded in vivo, but also appears to proliferate in vitro more efficiently than the Vδ2 subpopulation in response to ULBP+ lymphoma cells; this is conceivably due to the fact that Vδ2 T cells recognize phosphate antigens rather than ULBPs.8,9 Moreover, NKG2D is involved in the interaction between Vδ1 T lymphocytes and lymphoma cells. Of note, ex vivo isolated Vδ1 T cells from FL NHL patients expressed intracytoplasmic IL-4; in the same patients, high levels of serum IL-4 were detected. Interestingly, IL-4 has been reported as a favorable prognostic marker in NHL.29 We propose that the role of Vδ1 T lymphocytes, which usually represent a resident lymphocyte population, is played at the tumor site, such as LNs or BM in the case of low-grade NHL (mainly FL lymphomas) and low-risk B-CLL. In these sites, an efficient expression of ULBPs and MIC-A might occur, in agreement with that observed in solid tumors.19,31 Along this line, histochemical analysis on LN or BM specimens from 4 FL lymphomas and 2 low-risk B-CLL patients, provide evidence for the expression of ULBP2 or ULBP3 on neoplastic B lymphocytes. Thus, the increased levels of circulating Vδ1 T cells would be a consequence of a response that had occurred in affected tissues.
In high-risk B-CLL or DLCL, B cells may lack, or express at low levels, the relevant antigens recognized by γδ T lymphocytes, and this possibly prevents or impairs their elimination, thus contributing to the more aggressive course of these diseases. On the other hand, in FL lymphomas B cells might be more susceptible to the microenvironment where they reside, capable of inducing a strong and stable expression of ULBPs.
More importantly, we found that in most low-risk B-CLL patients14 and in low-grade NHL (this work), the expression of ULBP2 or ULBP3 on the surface of neoplastic B cells and an increased number of circulating Vδ1 T lymphocytes correlate with favorable prognostic factors and are associated with disease stability. On the contrary, in patients with low percentages of Vδ1 T lymphocytes and ULBP− B cells, the malignant cells also present unfavorable prognostic markers and the disease progressed over 1 year. Likewise, in aggressive NHL, such as DLCL, or in HR B-CLL, no expression of ULBPs or expansion of peripheral γδ T lymphocytes was observed. Finally, it is of note that in FL lymphomas high serum levels of IL-4 are found, compared to those detectable in HDs or in patients with B-CLL or DLCL, which is considered a favorable prognostic factor in NHL, possibly due to a differentiation action of this cytokine.32,33 Also noteworthy is the fact that IL-4 could be detected in LNs from FLs expressing ULBPs and infiltrated by Vδ1 T lymphocytes.
In conclusion, our results point to a role for Vδ1 T lymphocytes in the antitumor defense against low-grade lymphomas and highlight a potential therapeutic use for this T-cell subset. The use of γδ T cells in immunotherapy has been proposed in humans15,34 ; their function might be potentiated by the use of drugs able to induce or up-regulate the molecules recognized by either Vδ1 or Vδ2 T-cell subsets, such as ATRA or biphosphonates.9,35,14 Along this line, we have previously reported that ATRA up-regulates the expression of ULBP3 on B-CLL14 ; accordingly, preliminary experiments on 2 DLCLs showed that ATRA can induce the expression of ULBP2/3 on lymphoma B cells as well (not shown). Interestingly, it has been reported that ATRA has a negative effect on B-cell growth in NHL,32 thus further supporting the use of this drug in NHL therapy.
Authorship
Contribution: S.C. and A.B. performed research; P.D. performed histochemistry; J.L.R. and M.G. analyzed clinical and experimental data; and A.P. and M.R.Z. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alessandro Poggi, Laboratory of Experimental Oncology D, National Institute for Cancer Research, L.go R.Benzi 10, I-16132 Genoa, Italy; e-mail: alessandro.poggi@istge.it or zocchi.maria@hsr.it.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was partially supported by the Italian Association for Cancer Research (AIRC; A.P. and M.R.Z.). We thank Amgen (M.T.A. no. 200309766-001) for kindly providing the anti-ULBPs antibodies.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal