Abstract
Adult bone marrow (BM) contains hematopoietic stem cells (HSCs) as well as a nonhematopoietic, stromal cell population. Within this stromal population are mesenchymal stem cells (MSCs), which not only support hematopoiesis but also differentiate into multiple lineages, including fat, bone, and cartilage. Because of this multipotentiality, the MSC is an attractive candidate for clinical applications to repair or regenerate damaged tissues of mesenchymal origin. However, research progress has been hampered by the limited existing knowledge of the biology of these cells, particularly by the lack of a suitable marker for their prospective isolation. Here, we report that SSEA-4, an early embryonic glycolipid antigen commonly used as a marker for undifferentiated pluripotent human embryonic stem cells and cleavage to blastocyst stage embryos, also identifies the adult mesenchymal stem cell population.
Introduction
Stem cells, which are responsible for maintenance and repair of most adult tissues, are distinguished by their potential for both self-renewal and multilineage differentiation. These properties make stem cells an attractive alternative to treat various degenerative disorders. In adults, a well-known source of stem cells is the bone marrow (BM) compartment, which contains hematopoietic stem cells (HSCs) as well as a nonhematopoietic, stromal cell population. The later, identified as marrow stromal cells or mesenchymal stem cells (MSCs), not only support hematopoiesis but also differentiate along various mesodermal lineages to generate osteoblasts, chondrocytes, and adipocytes.1-3 Because of these properties, MSCs have been exploited for their potential to repair or regenerate damaged tissues of mesenchymal origin, including bone fracture healing,4 tendon repair,5 cartilage regeneration,6,7 and support of engraftment after chemotherapy.8
One group has also identified a subpopulation of bone marrow–derived cells, termed multipotent adult progenitor cells (MAPCs), which possess longer proliferative capacity and broader differentiation potential than MSCs,9,10 including cells of all 3 germ layers in vitro, as well as the ability to generate chimeric mice when injected into blastocysts.10 To date, the relation of these cells to MSCs is not known. It is possible that MAPCs arise in vitro through genetic or epigenetic changes.
MSCs were first identified more than 20 years ago by Friedenstein et al,11-13 who isolated these cells by simply exploiting their ability to adhere to tissue culture plastic. Despite the enormous number of studies that have focused on the biology of MSCs in the past decade, the method for their derivation has not changed much from their initial identification.1-3,14,15 The caveat of such a method is heterogeneity of the resulting adherent cells. It is known that hematopoietic progenitors can persist for a number of weeks in such cultures, with MSCs comprising a minority of the cell population.15 Unlike their well-characterized neighbors, the HSCs, the study of which was revolutionized by methods for prospective isolation,16-18 a good marker for the purification of the MSCs is lacking; without prospective isolation and purification it is very difficult to ascribe any attribute with certainty to the MSC. Once established in culture, these cells have been reported to express a variety of cell-lineage–specific antigens, including, among others, adhesion molecules, integrins, and growth factor receptors2 ; however the expression of these markers varies depending on the culture conditions used.19,20 Moreover, the phenotype of an established population of homogenous MSCs is obtained after weeks in culture, when multilineage potential may be already somewhat compromised. Thus, such criteria do not necessarily identify MSCs in their native state; therefore, little progress has been made in their prospective purification.
Here, we demonstrate that SSEA-4, a stage-specific embryonic antigen previously thought to mark specifically human embryonic stem cells and very early cleavage to blastocyst stage embryos, also identifies an adult mesenchymal stem cell population. We further show that this marker can be used for the prospective isolation of MSCs from whole human bone marrow aspirates.
Materials and methods
Animals
Rosa 26-lacZ transgenic and Balb-c mice (both from Jackson Laboratories, Bar Harbor, ME) were used as BM donors. Rag2−/−γc−/− immunodeficient mice (Taconic Laboratories, Germantown, NY) were used as recipients for the in vivo bone assay. Animal care and all procedures were performed according to University of Texas (UT) Southwestern institutional guidelines.
Generation of immortalized mesenchymal stem cell lines
Whole BM was harvested from the femurs and tibias of male Rosa 26-lacZ transgenic mice after being killed by CO2 inhalation. Cells were plated at 2.4 × 106/cm2 in the following medium: low-glucose Dulbecco modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 40% MCDB-201 (Sigma, St Louis, MO), 10% FBS (Invitrogen), 1 × insulin-transferrin-selenium (ITS; Sigma), 1 × linoleic acid-bovine serum albumin (Sigma), 10−8 M dexamethasone (Sigma), 10−4 M ascorbic acid 2-phosphate (Sigma), 50 U/mL penicillin/streptomycin, and the following growth factors: 10 ng/mL hPDGF-BB (R&D Systems, Minneapolis, MN), 10 ng/mL mEGF (Sigma), and 1000 U/mL mLIF (Chemicon, Temecula, CA). After 3 days of incubation in a humidified incubator at 37°C with 5% CO2, the nonadherent hematopoietic cells were discarded. The adherent cells were passaged after a further 2 days of culture in the same medium. Cells were fed or passaged every 2 days to be kept subconfluent.
BM for mouse primary MSC cultures
Whole BM was flushed from the femurs and tibias of male Balb-c mice with Iscove modified Dulbecco medium (IMDM; Invitrogen), supplemented with 5% FBS. In some experiments, to obtain the bone-associated cells, femurs and tibias were cut into small pieces and crushed using a mortar and pestle. Bone fragments were treated with 0.2% collagenase (Sigma), gently agitated for 1.5 hours at 37°C. After the incubation period, the bone-associated cells were combined with the flushed out cells forming a single-cell suspension. Cells were plated in the same medium described for the generation of immortalized MSC lines, at a density of 1.9 × 106 cells/cm2. After 72 hours, medium and any cells in suspension were aspirated and replaced with fresh medium. Cultures were passaged every 4 days when subconfluent (60%-70%) at a cell density of 1.3 to 2 × 104 cells/cm2.
BM for human primary MSC cultures
Bone marrow samples were obtained from volunteer donors with their informed consent, under a protocol approved by the Institutional Review Board of the UT Southwestern Medical Center. In some instances, bone marrow was purchased from Cambrex (Walkersville, MD). Human MSCs were isolated from marrow of donors with a modification of Pittenger protocol.2 Briefly, cells were separated from 20 to 30 mL marrow diluted with 4 volumes of Minimum Essential Medium Alpha Medium (α-MEM; Invitrogen) containing 10% FBS by centrifugation at 900g for 15 minutes. Cells were washed with phosphate-buffered saline (PBS) once. Cells were plated at 2 × 106 cells/cm2 in expansion medium (low-glucose) DMEM supplemented with 40% MCDB-201, 10% FBS, 1 × ITS, 1 × linoleic acid–bovine serum albumin, 10−8 M dexamethasone, 10−4 M ascorbic acid 2-phosphate, 50 U/mL penicillin/streptomycin, 10 ng/mL hPDGF-BB, and 10 ng/mL hEGF. After 72 hours of culture, suspended cells were removed and replaced with fresh media. Medium was changed every 3 days thereafter. MSCs grew as colonies which were detached with 0.1% trypsin-EDTA and subcultured at a density of 4 to 8 × 103 cells/cm2. Standardized preparations of hMSCs obtained from the Tulane Center for Gene Therapy were cultured at a cell density of 100 cells/cm2 in α-MEM containing 16.5% FBS, as recommended.
Flow cytometry and sorting of MSCs
For SSEA-4, SSEA-3, and SSEA-1 antigens, supernatants from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City) were used. The SSEA-4 mouse monoclonal antibody recognizes the SSEA-4 antigen expressed on human and mouse cells.21 For secondary staining to this antibody, we used fluorescein-5-isothiocyanate (FITC)–conjugated goat antimouse. As isotype control, a matching FITC-conjugated mouse IgG3 antibody (Pharmingen, San Diego, CA) was used. For secondary staining to SSEA-1 and SSEA-3, we used FITC-conjugated goat anti–mouse Ig and PE-conjugated goat anti–rat Ig, respectively (Pharmingen). As matching isotype controls, FITC-conjugated mouse and rat IgM antibodies were used (for SSEA-1 and SSEA-3, respectively). In the mouse double-staining experiment, a PE-conjugated anti–SSEA-4 (R&D Systems) and an APC-conjugated anti-CD45 were used.
For the other mouse surface markers, we used PE-conjugated antimouse antibodies to CD45, Sca-1, c-Kit, and Flk-1 (Pharmingen).
FITC-conjugated antihuman antibody to CD105 (SH2, Endoglin; Serotec, Raleigh, NC), CD45 as well as PE-conjugated antihuman CD44, CD73 (SH3), and CD90 (Thy-1) (Pharmingen) were applied for characterization of human MSCs.
Adherent cells were collected after a short incubation with trypsin. Cells were washed twice, first with IMDM 10% FBS and then with blocking buffer (PBS 1% FBS), suspended in the same buffer containing 0.25 μg/106 cells of Fc block (Pharmingen) and placed on ice for 5 minutes. Antibody was added at 1 μg/106 cells and incubated at 4°C for 30 minutes before washing with blocking buffer, except for staining with the supernatants from the hybridoma bank (SSEA-1, SSEA-3, and SSEA-4), whereby cells were resuspended in 100 μL supernatant and incubation lasted 40 minutes. When secondary staining was required, cells were counterstained with appropriate secondary antibody for 20 minutes at 4°C, followed by washing with blocking buffer. Stained cells were analyzed on a FACS Aria cell-sorting system (Becton Dickinson, San Jose, CA) after addition of propidium iodide (Pharmingen) to exclude dead cells. In the sorting experiments, MSCs were purified based on the expression of SSEA-4 (positives and negatives). For clonal analysis, SSEA-4+ cells were FACS-deposited into single wells of a 96-well dish. Wells with single MSC colonies were harvested and expanded into clonal cell lines. These clones were tested individually for multipotentiality by further subculture in adipogenic, osteogenic, and chondrogenic media.
Induction protocols
For fat differentiation, cells were plated in the presence of α-MEM medium containing horse serum (12.5%; Invitrogen), FBS (12.5%), penicillin/streptomycin (50 U/mL), 2-mercaptoethanol (10−4 M; Sigma), and hydrocortisone (10−4 M; StemCell Technologies, Vancouver, BC, Canada). Characterization of fat was performed by Oil Red O staining. For osteogenic differentiation, cells were cultured in the presence of DMEM medium with fetal calf serum (%), penicillin/streptomycin (50 U/mL), ascorbate (50 μmol/L; Sigma), dexamethasone (0.1 μmol/L; Sigma), β-glycerolphosphate (10 mmol/L; Sigma). Characterization was performed by Alizarin Red S staining, which detects calcium deposition. For chondrogenesis, cells were cultured as pellets at 3 × 105 cells/mL in serum-free chemically defined medium consisting of DMEM (high-glucose), 1 × insulin-transferrin-selenium, 1 × linoleic acid–bovine serum albumin, ascorbate 2-phosphate (50 μg/mL), dexamethasone (100 nM), and 10 ng/mL hTGF-β1 (R&D Systems). Media was changed every 2 days.
All of the bright-field images were taken on an Olympus IX70 microscope (Olympus Optical, Tokyo, Japan) with a Nikon DXM1200 digital camera (Nikon, Tokyo, Japan) and Nikon ACT-1 software. The images from immunofluorescent staining were captured with an Olympus BX50 microscope, an Olympus U-CMAD digital camera, and MetaVue 5.0 software.
In vivo assay for bone formation
MSC cultures initiated with sorted SSEA4+ cells from human BM were transplanted subcutaneously into immunodeficient mice on a hydroxyapatite-collagen scaffold (Collagraft, NeuColl; Zimmer, Warsaw, IN).22 We used cells at passage 5 or 8 (2 independent donors). Briefly, cells were resuspended in 5 mL expansion medium and mixed with scaffold particles in a 50-mL conical tube at a density of 10 × 106 cells/cm2 and incubated for 90 minutes at 37°C with slow shaking (60 rpm). Scaffolds loaded with the cells were immediately implanted into subcutaneous pockets on the dorsal surface of 10- to 12-week-old male Rag2−/−γc−/− mice (n = 14). Mice were anesthetized with a combination of intraperitoneal ketamine (140 mg/kg) and xylazine (7 mg/kg). A total of 12 implants were prepared from each donor. Each mouse received 3 to 5 implants, always including one scaffold without cells as a negative control.
Implants were recovered after 4 to 8 weeks, fixed in 4% buffered paraformaldehyde for 2 days, followed by 1 day in 20% sucrose, and then frozen in OCT compound. Serial cryosections of entire implants were analyzed for bone formation. To evaluate tissue morphology, sections were stained by Hematoxylin and Eosin as well as in 1% Toluidine Blue. Mineral deposition was detected by von Kossa (3% AgNO3 and contrasting with Nuclear fast red) and Alizarin Red S staining. Expression of osteocalcin was analyzed with a specific goat anti–human osteocalcin antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Tissue sections were permeabilized with 0.3% triton for 30 minutes, blocked with 10% donkey serum, and incubated with primary antibody diluted (1:500) in 2% donkey serum/PBS at 4°C overnight. Cy3 donkey anti–goat IgG (1:800) was used as a secondary antibody, and DAPI (4,6-diamidino-2-phenylindole) was used to counterstain nuclei in the cells.
Reverse transcriptase–polymerase chain reaction (RT-PCR) analysis
Total RNA was isolated using Trizol (Invitrogen) as recommended by the manufacturer. First-strand cDNA was produced using Superscript II reverse transcriptase (Invitrogen) with Oligo dT. Five percent of first-strand reaction was used for each ensuing PCR reaction. Primer sequences and PCR conditions are described in Tables 1 and 2 (mouse and human, respectively).
Reverse transcription–polymerase chain reaction mouse primer sequences information
| . | Primer pair sequence . | . | . | |
|---|---|---|---|---|
| Primer . | Forward . | Reverse . | Annealing temperature, °C . | Product size, bp . |
| β-actin | GTGGGGCGCCCCAGGCACCA | CTCCTTAATGTCACGCACGATTTC | 50 | 520 |
| PPAR-γ2 | GGAGATTCTCCTGTTGACCCAG | GGCACTCAATGGCCATGAG | 65 | 403 |
| C/EBP-α | GATAAAGCCAAACAACGCAACG | CTAGAGATCCAGCGACCCGAA | 50 | 253 |
| LPL | TTAACTACCCCCTAGACAACGTCCA | AAGAGATGAATGGAGCGCTCG | 50 | 388 |
| Adipsin | AGACCCCTACCCTTGCAATACG | TGTTACCATTTGTGATGTTTTCGATC | 50 | 376 |
| OPN | TGGAGATCGAATTCTGCTTG | TCAAGTGCTTGAGGGCATAC | 50 | 720 |
| Cbfa-1 | AGTAGCCAGGTTCAACGATCTGA | GGACCGTCCACTGTCACTTTAATA | 50 | 137 |
| Coll II | GGCAACAGCAGGTTCACATA | CCACACCAAATTCCTGTTCA | 55 | 166 |
| Agg | CTAAGTTCCAGGGTCACTGTTACCG | TCCTCTCCGGTGGCAAAGAAGTTG | 59 | 270 |
| Coll X | AAGGAGTGCCTGGACACAAT | GTCGTAATGCTGCTGCCTAT | 55 | 461 |
| . | Primer pair sequence . | . | . | |
|---|---|---|---|---|
| Primer . | Forward . | Reverse . | Annealing temperature, °C . | Product size, bp . |
| β-actin | GTGGGGCGCCCCAGGCACCA | CTCCTTAATGTCACGCACGATTTC | 50 | 520 |
| PPAR-γ2 | GGAGATTCTCCTGTTGACCCAG | GGCACTCAATGGCCATGAG | 65 | 403 |
| C/EBP-α | GATAAAGCCAAACAACGCAACG | CTAGAGATCCAGCGACCCGAA | 50 | 253 |
| LPL | TTAACTACCCCCTAGACAACGTCCA | AAGAGATGAATGGAGCGCTCG | 50 | 388 |
| Adipsin | AGACCCCTACCCTTGCAATACG | TGTTACCATTTGTGATGTTTTCGATC | 50 | 376 |
| OPN | TGGAGATCGAATTCTGCTTG | TCAAGTGCTTGAGGGCATAC | 50 | 720 |
| Cbfa-1 | AGTAGCCAGGTTCAACGATCTGA | GGACCGTCCACTGTCACTTTAATA | 50 | 137 |
| Coll II | GGCAACAGCAGGTTCACATA | CCACACCAAATTCCTGTTCA | 55 | 166 |
| Agg | CTAAGTTCCAGGGTCACTGTTACCG | TCCTCTCCGGTGGCAAAGAAGTTG | 59 | 270 |
| Coll X | AAGGAGTGCCTGGACACAAT | GTCGTAATGCTGCTGCCTAT | 55 | 461 |
PPAR-γ2 indicates peroxisome proliferator-activated receptor γ 2; C/EBP-α, CCAAT/enhancer-binding protein-α; LPL, lipoprotein lipase; OPN, osteopontin; Cbfa-1, core-binding factor-1, Coll II, collagen II; Agg, aggrecan; Coll X, collagen X.
Reverse transcription–polymerase chain reaction human primer sequences information
| . | Primer pair sequence . | . | . | . | |
|---|---|---|---|---|---|
| Primer . | Forward . | Reverse . | Annealing temperature, °C . | Product size, bp . | |
| β-actin | ACCATGGATGATGATATCGCC | GTGCCAGATTTTCTCCATGTC | 55 | 244 | |
| PPAR-γ2 | GCTGTTATGGGTGAAACTCT | ATAAGGTGGAGATGCAGGCT | 55 | 351 | |
| LPL | GTCCGTGGCTACCTGTCAT | AGCCCTTTCTCAAAGGCTTC | 55 | 717 | |
| Adipsin | GGTCACCCAAGCAACAAAGT | CCTCCTGCGTTCAAGTCATC | 55 | 271 | |
| OPN | CACATCGGAATGCTCATTGC | ATCACCTGTGCCATACCAGT | 50 | 663 | |
| Cbfa-1 | ACTGGGCCCTTTTTCAGA | GCGGAAGCATTCTGGAA | 50 | 317 | |
| ALP | CTGGTAGGCGATGTCCTTA | ACGTGGCTAAGAATGTCATC | 50 | 475 | |
| OC | CTCACACTCCTCGCCCTA | CCTGAAAGCCGATGTGGT | 50 | 262 | |
| Coll II | TTTCCCAGGTCAAGATGGTC | TCACCTGGTTTTCCACCTTC | 54 | 498 | |
| Aggrecan | TGAGGAGGGCTGGAACAAGTACC | GGAGGTGGTAATTGCAGGGAACA | 60 | 350 | |
| Coll X | GCCCAAGAGGTGCCCCTGGAATAC | CCTGAGAAAGAGGAGTGGACATAC | 57 | 703 | |
| . | Primer pair sequence . | . | . | . | |
|---|---|---|---|---|---|
| Primer . | Forward . | Reverse . | Annealing temperature, °C . | Product size, bp . | |
| β-actin | ACCATGGATGATGATATCGCC | GTGCCAGATTTTCTCCATGTC | 55 | 244 | |
| PPAR-γ2 | GCTGTTATGGGTGAAACTCT | ATAAGGTGGAGATGCAGGCT | 55 | 351 | |
| LPL | GTCCGTGGCTACCTGTCAT | AGCCCTTTCTCAAAGGCTTC | 55 | 717 | |
| Adipsin | GGTCACCCAAGCAACAAAGT | CCTCCTGCGTTCAAGTCATC | 55 | 271 | |
| OPN | CACATCGGAATGCTCATTGC | ATCACCTGTGCCATACCAGT | 50 | 663 | |
| Cbfa-1 | ACTGGGCCCTTTTTCAGA | GCGGAAGCATTCTGGAA | 50 | 317 | |
| ALP | CTGGTAGGCGATGTCCTTA | ACGTGGCTAAGAATGTCATC | 50 | 475 | |
| OC | CTCACACTCCTCGCCCTA | CCTGAAAGCCGATGTGGT | 50 | 262 | |
| Coll II | TTTCCCAGGTCAAGATGGTC | TCACCTGGTTTTCCACCTTC | 54 | 498 | |
| Aggrecan | TGAGGAGGGCTGGAACAAGTACC | GGAGGTGGTAATTGCAGGGAACA | 60 | 350 | |
| Coll X | GCCCAAGAGGTGCCCCTGGAATAC | CCTGAGAAAGAGGAGTGGACATAC | 57 | 703 | |
ALP indicates alkaline phosphatase; OC, osteocalcin.
Results
Clonal mesenchymal stem cell lines from mouse bone marrow
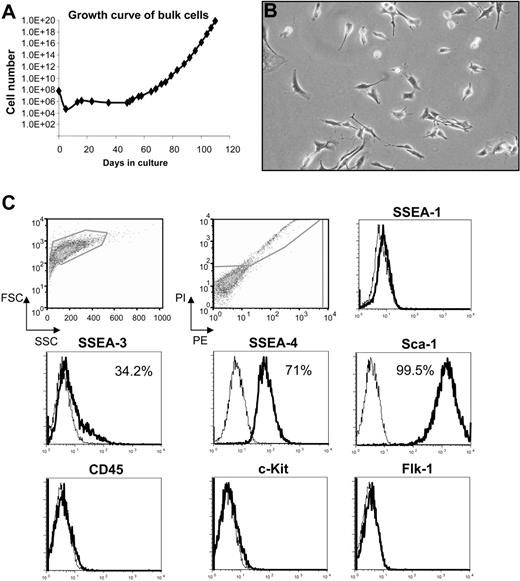
To study the early mesenchymal population present in adult BM, we cultured mouse BM cells obtained from adult Rosa26-lacZ transgenic mice in growth medium described for isolation of MAPCs.9,10 We observed a delayed outgrowth of exponentially growing populations of cells with spindle-shaped morphology, indicative of MSCs23 (Figure 1A-B). These cells doubled every 31.5 hours and were maintained in culture for more than 100 days with no sign of senescence or differentiation. MAPCs have been reported to express certain pluripotency markers characteristic of murine embryonic stem (ES) cells, including the transcription factor Oct-4.10 To determine the relationship between these immortalized cell lines and MAPCs, we tested our cells for these markers as well as other stem cell and lineage-specific antigens. Flow cytometry analysis revealed that large numbers of cells expressed high levels of Sca-1 (99.4%), SSEA-4 (71%), and SSEA-3 (34.2%), but all cells were negative for c-Kit, CD45, Flk-1, and SSEA-1 (Figure 1C). Using real-time PCR, we were unable to detect Oct-4 expression (data not shown). These results demonstrate major differences between these cells and MAPCs, which were reported to be SSEA-1+Sca-1lowFlk-1lowCD45−c-Kit−.10 Furthermore, these cells were single-cell cloned without difficulty at 30 population doublings, by fluorescence-activated cell sorting (FACS).
Characterization of primitive adherent BM cells. (A) Growth curve of cells derived from bone marrow (BM) of Rosa 26-LacZ mice; (B) their morphology under an inverted microscope (original magnification, 10×/0.3 NA Ph1), and (C) expression of surface markers at 39 cell doublings (≅ 100 days in culture). Histogram profiles are gated on forward and side scatters, and PI-negative (live) cells are shown in the first 2 panels. Plots show isotype control staining profile (thin line) versus specific antibody staining profile (thick line). Percentages represent the fraction of cells that express a given surface antigen.
Characterization of primitive adherent BM cells. (A) Growth curve of cells derived from bone marrow (BM) of Rosa 26-LacZ mice; (B) their morphology under an inverted microscope (original magnification, 10×/0.3 NA Ph1), and (C) expression of surface markers at 39 cell doublings (≅ 100 days in culture). Histogram profiles are gated on forward and side scatters, and PI-negative (live) cells are shown in the first 2 panels. Plots show isotype control staining profile (thin line) versus specific antibody staining profile (thick line). Percentages represent the fraction of cells that express a given surface antigen.
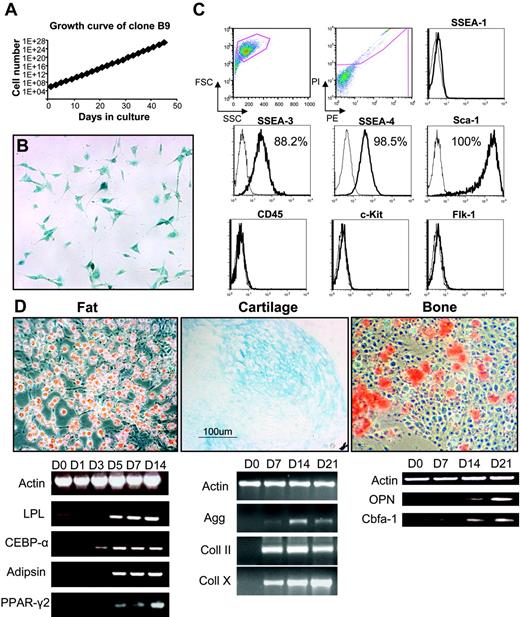
A representative clone, B9, was chosen for further characterization. The doubling time for this clone was 14.8 hours (Figure 2A), and, despite the extended time in culture, it had not changed morphology and still stained positive for LacZ (Figure 2B), although karyotype analysis revealed various abnormalities, including trisomy 1 and 15; monosomy 12, 14, and 17; and translocations (10 > 8 and 9 > 5). Cell-surface marker analyses (Figure 2C) demonstrated high levels of expression for Sca-1 (100%), SSEA-3 (88.2%), and SSEA-4 (98.5%). As seen with the bulk (uncloned) population, clone B9 was negative for CD45, Flk-1, SSEA-1 (Figure 2C), and Oct-4 (data not shown). Analysis of 4 other clones revealed a similar profile, lack of CD45 and nearly 100% expression of Sca-1 and SSEA-4 (data not shown). Despite their karyotypic abnormalities, these cells retained multilineage differentiation potential (Figure 2D). When grown confluent or in media containing hydrocortisone, B9 cells (as well as other clones) differentiated robustly into adipocytes, as demonstrated by Oil Red O staining and expression of fat-related genes, which are all present after 5 days of induction (Figure 2D). When exposed to β-glycerolphosphate, ascorbate, and dexamethasone, calcium deposition was visible after 2 weeks in culture, which coincided with osteopontin expression (Figure 2D), indicative of osteoblast differentiation. In the presence of TGF-β, cells differentiated into cartilage as evidenced by Alcian Blue staining and expression of cartilage-related genes (Figure 2D). We failed to observe blood or endothelial development in vitro or in vivo when cells were transplanted into immunodeficient nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (data not shown). These results suggest that these clones represent immortalized mesenchymal stem cell lines. The fact that they express very high levels of SSEA-4 raised the possibility that this antigen, whose expression is characteristic of human ES cells as well as their malignant counterparts, embryonic carcinoma (EC) cells,21,24 might also mark the adult MSC population.
Characterization of MSC clone B9. (A) Growth curve of B9 cells obtained by single-cell sorting from the bulk population at 33 cell doublings. (B) Clonal cells have similar morphology to the parental population and stained positive for X-Gal. (C) Flow cytometric analysis of B9 cells at passage 4. Histogram profiles are gated on forward and side scatters, and PI negative (live) cells are shown in the first 2 panels. Plots show isotype control staining profile (thin line) versus specific antibody staining profile (thick line). Percentages represent the fraction of cells that express a given surface antigen. (D) Multilineage differentiation of clone B9 at passage 18 for fat (left) demonstrated by Oil Red O staining (top left) and expression of fat-specific markers in a time-course study (bottom left); for cartilage (middle) shown by Alcian Blue staining (top middle) and expression of cartilage markers (bottom middle); and for bone (right) demonstrated by Alizarin Red S staining (top right) and expression of bone-specific genes (bottom right). For panels B and D, original magnification was 10×/0.3 NA Ph1.
Characterization of MSC clone B9. (A) Growth curve of B9 cells obtained by single-cell sorting from the bulk population at 33 cell doublings. (B) Clonal cells have similar morphology to the parental population and stained positive for X-Gal. (C) Flow cytometric analysis of B9 cells at passage 4. Histogram profiles are gated on forward and side scatters, and PI negative (live) cells are shown in the first 2 panels. Plots show isotype control staining profile (thin line) versus specific antibody staining profile (thick line). Percentages represent the fraction of cells that express a given surface antigen. (D) Multilineage differentiation of clone B9 at passage 18 for fat (left) demonstrated by Oil Red O staining (top left) and expression of fat-specific markers in a time-course study (bottom left); for cartilage (middle) shown by Alcian Blue staining (top middle) and expression of cartilage markers (bottom middle); and for bone (right) demonstrated by Alizarin Red S staining (top right) and expression of bone-specific genes (bottom right). For panels B and D, original magnification was 10×/0.3 NA Ph1.
SSEA-4 expression on murine MSCs
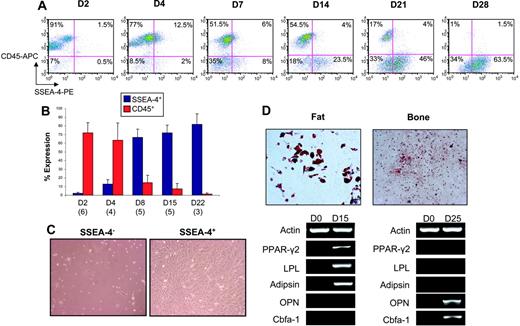
To determine whether SSEA-4 expression in these immortalized MSC lines relates to their transformation or reflects a true property of MSCs, we evaluated SSEA-4 expression in nontransformed, primary MSCs. We initially plated mouse bone marrow for mesenchymal stem cell outgrowth and monitored the expression of SSEA-4 as well as CD45 (a pan-leukocyte marker). As expected, 2 days after plating the majority of adherent cells were still of hematopoietic origin (CD45 ≅ 70%-90%) (Figure 3A-B); however, we could observe a small population that was positive for SSEA-4 (≅ 1%-2%) (Figure 3A-B). As hematopoietic cells were gradually eliminated from the adherent layer (reduction of CD45), cells expressing SSEA-4 took over the cultures (Figure 3A-B). Double-staining analyses revealed an increasing fraction of SSEA-4+CD45− cells (Figure 3A). A SSEA-4+CD45+ subpopulation also coexists in these cultures and, although it is increased in the first few days, it is depleted over time (Figure 3A). We then proceeded to sort and analyze SSEA-4+ and SSEA-4− cells from primary cultures 2 days after bone marrow was plated for mesenchymal stem cell growth. When purified cell populations were plated at the same cell density after sorting, SSEA-4− cells failed to grow, whereas SSEA-4+ cells rapidly adhered to the plastic and produced a homogeneous cell-monolayer characteristic of MSCs (Figure 3C). When compared with unpurified MSCs, SSEA-4+ cells exhibited superior growth (7.7-fold versus 2.11-fold) after a week in culture. In one double-staining experiment for SSEA-4 and CD45, we sorted the 4 cell fractions (SSEA-4+CD45−, SSEA-4+CD45+, SSEA-4−CD45+, and SSEA-4−CD45−). MSC growth was observed only in the SSEA-4+CD45− fraction (data not shown). Flow cytometry analyses revealed that SSEA-4+–sorted cells maintained high levels of SSEA-4 (approximately 90%, 3 days after sort), whereas their unpurified counterparts expressed only 30% (data not shown). When subjected to specific induction, sorted SSEA-4+ cells gave rise to cells of the adipocytic and osteoblastic lineages, as observed by appropriate staining and gene expression analyses (Figure 3D).
SSEA-4 marks murine MSCs. Time course for SSEA-4 and CD45 expression in BM adherent cells by flow cytometry. (A) A representative FACS profile of BM adherent cells analyzed for SSEA-4 and CD45 expression at 2, 4, 7, 14, 21, and 28 days in culture. Fluorescence intensity of SSEA-4 (PE-conjugated) is indicated on the x-axis, and CD45 (APC-conjugated) is shown on the y-axis. Percentages represent the fraction of cells that express a given surface antigen. (B) SSEA-4 and CD45 expression over time at days 2, 4, 8, 15, and 22. The numbers in parentheses below the bars represent the number of independent bone marrow cultures that were assayed at that time point. Error bars indicate standard error. (C) Two days after BM was harvested, adherent BM cells were sorted for SSEA4. Although SSEA4+ and SSEA-4− cells were plated at the same cell density, SSEA-4− cells failed to grow (left), whereas SSEA4+ cells demonstrated extensive growth (right). (D) Expanded SSEA4+ cells were capable of differentiating along fat and bone lineages when induced with appropriate media, as observed by Oil Red O (top left) and Alizarin Red S staining (top right), respectively, as well as by gene expression analyses (shown below). Original magnification for panels C and D, 10×/0.3 NA Ph1.
SSEA-4 marks murine MSCs. Time course for SSEA-4 and CD45 expression in BM adherent cells by flow cytometry. (A) A representative FACS profile of BM adherent cells analyzed for SSEA-4 and CD45 expression at 2, 4, 7, 14, 21, and 28 days in culture. Fluorescence intensity of SSEA-4 (PE-conjugated) is indicated on the x-axis, and CD45 (APC-conjugated) is shown on the y-axis. Percentages represent the fraction of cells that express a given surface antigen. (B) SSEA-4 and CD45 expression over time at days 2, 4, 8, 15, and 22. The numbers in parentheses below the bars represent the number of independent bone marrow cultures that were assayed at that time point. Error bars indicate standard error. (C) Two days after BM was harvested, adherent BM cells were sorted for SSEA4. Although SSEA4+ and SSEA-4− cells were plated at the same cell density, SSEA-4− cells failed to grow (left), whereas SSEA4+ cells demonstrated extensive growth (right). (D) Expanded SSEA4+ cells were capable of differentiating along fat and bone lineages when induced with appropriate media, as observed by Oil Red O (top left) and Alizarin Red S staining (top right), respectively, as well as by gene expression analyses (shown below). Original magnification for panels C and D, 10×/0.3 NA Ph1.
Expression of SSEA-4 on human MSCs and their prospective isolation
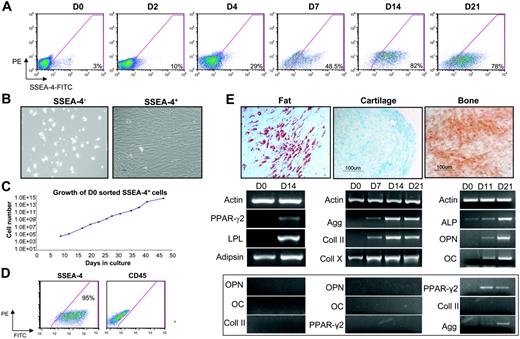
Next, we sought to investigate the expression of SSEA-4 in human bone marrow samples. Approximately 2% to 4% of human BM cells are positive for SSEA-4 on day 0 (Figure 4A). These results were observed in 5 independent BM samples. We then monitored the expression levels of SSEA-4 in human BM plated in standard mesenchymal growth conditions (Figure 4A; days 2-21; n = 3). As in the mouse, cells expressing SSEA-4 gradually took over the cultures (Figure 4A), whereas hematopoietic cells were eliminated from the adherent layer, as observed by the reduction in CD45+ cells (data not shown). High levels of SSEA-4 expression were also detected in standardized preparations of mesenchymal stem cells obtained from the Tulane Center for Gene Therapy. Although robust SSEA-4 expression was evident on human MSCs, levels of expression were significantly lower than human embryonal carcinoma (EC) cells (Figure 5). To test whether SSEA-4 expression could be used for prospective isolation of human mesenchymal stem cells, we isolated SSEA-4+ cells from fresh human bone marrow. SSEA-4+ and SSEA-4− cell populations were sorted from human bone marrow aspirates and plated at the same cell density after sorting (performed on 4 independent BM samples). SSEA-4− cells failed to grow, whereas SSEA-4+ cells adhered to the plastic and produced a homogeneous cell-monolayer characteristic of MSCs (Figure 4B). An equivalent number of unpurified BM cells plated at the same cell density did not successfully initiate MSC cultures. Such cultures could only be derived from higher densities of unpurified cells (data not shown). Growth kinetic analyses revealed that sorted SSEA-4+ BM cells could be grown for more than 7 weeks without senescence, and that their doubling time was 25.2 hours (Figure 4C). The growth rate remained stable over this time, as expected for normal primary untransformed cells. To confirm this, at passage 7 these cells were subjected to karyotype analysis, which revealed no abnormalities (data not shown).
SSEA-4 can be used to prospectively isolate human MSCs. (A) A representative FACS profile of time course analyses of SSEA-4 expression in fresh unsorted human BM (D0) and adherent cells at 2, 4, 7, 14, and 21 days in culture. Fluorescence intensity of FITC-labeled SSEA-4 is indicated on the x-axis. Percentages represent the fraction of cells that express SSEA-4 (background is subtracted). The y-axis represents PE, which provides a measure of autofluorescence. (B) Fresh BM was sorted based on SSEA-4 expression. SSEA4+ and SSEA-4− cells were plated at the same cell density (3 × 104 cells/cm2). SSEA-4− cells failed to grow (left), whereas SSEA4+ cells demonstrated extensive growth (right). (C) Growth curve of SSEA-4+ cells sorted on day 0 and plated at 6.3 × 104 cells. (D) SSEA-4 and CD45 expression on SSEA-4+–sorted cells at passage 2. (E) After appropriate induction, day-0 sorted SSEA4+cells were capable of differentiating along fat (left), cartilage (middle), and bone (right) lineages, as observed by Oil Red O, Alcian Blue, and Alizarin Red S staining, respectively (top row), as well as by relevant gene expression analyses (bottom row). Gene expression for unrelated lineages (controls) is shown below. Original magnification for panels B and E, 10×/0.3 NA Ph1.
SSEA-4 can be used to prospectively isolate human MSCs. (A) A representative FACS profile of time course analyses of SSEA-4 expression in fresh unsorted human BM (D0) and adherent cells at 2, 4, 7, 14, and 21 days in culture. Fluorescence intensity of FITC-labeled SSEA-4 is indicated on the x-axis. Percentages represent the fraction of cells that express SSEA-4 (background is subtracted). The y-axis represents PE, which provides a measure of autofluorescence. (B) Fresh BM was sorted based on SSEA-4 expression. SSEA4+ and SSEA-4− cells were plated at the same cell density (3 × 104 cells/cm2). SSEA-4− cells failed to grow (left), whereas SSEA4+ cells demonstrated extensive growth (right). (C) Growth curve of SSEA-4+ cells sorted on day 0 and plated at 6.3 × 104 cells. (D) SSEA-4 and CD45 expression on SSEA-4+–sorted cells at passage 2. (E) After appropriate induction, day-0 sorted SSEA4+cells were capable of differentiating along fat (left), cartilage (middle), and bone (right) lineages, as observed by Oil Red O, Alcian Blue, and Alizarin Red S staining, respectively (top row), as well as by relevant gene expression analyses (bottom row). Gene expression for unrelated lineages (controls) is shown below. Original magnification for panels B and E, 10×/0.3 NA Ph1.
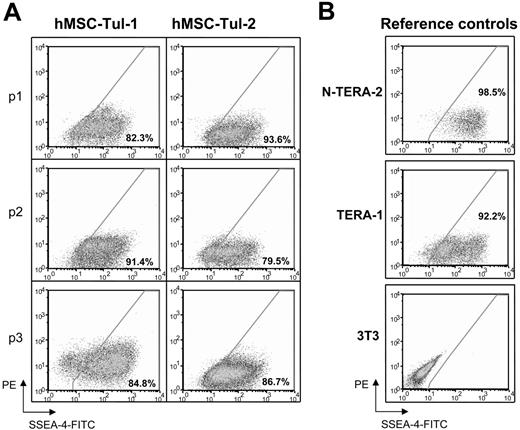
SSEA-4 levels in established human MSCs. Flow cytometry analyses of SSEA-4 expression in (A) standardized preparations of mesenchymal stem cells obtained from the Tulane Center for Gene Therapy at different passages and (B) reference controls for levels of expression; EC cell lines, N-TERA-2 and TERA-1 and 3T3 fibroblasts. Fluorescence intensity of SSEA-4 is observed in the x-axis. Percentages represent the fraction of cells that express SSEA-4. The y-axis represents PE, which provides a measure of autofluorescence.
SSEA-4 levels in established human MSCs. Flow cytometry analyses of SSEA-4 expression in (A) standardized preparations of mesenchymal stem cells obtained from the Tulane Center for Gene Therapy at different passages and (B) reference controls for levels of expression; EC cell lines, N-TERA-2 and TERA-1 and 3T3 fibroblasts. Fluorescence intensity of SSEA-4 is observed in the x-axis. Percentages represent the fraction of cells that express SSEA-4. The y-axis represents PE, which provides a measure of autofluorescence.
Flow cytometric analysis of SSEA-4+–sorted cells (Figure 4D) demonstrated the absence of hematopoietic cells in these cultures, as noted by the lack of CD45, whereas SSEA-4 was maintained at high levels. When the SSEA-4+ fraction from fresh aspirates was costained with CD45, only the SSEA-4+CD45− cell fraction (representing about 50% of total SSEA-4+ cells) grew out (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article).
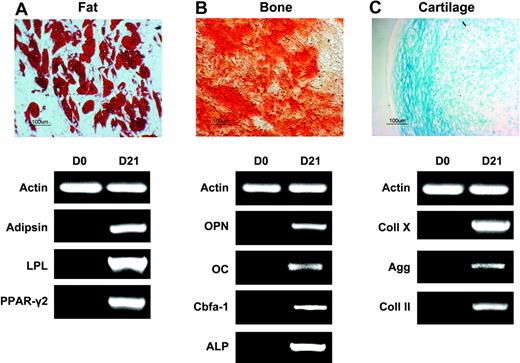
Multilineage differentiation potential of passage 7 SSEA-4+ BM cells was also investigated. When exposed to media containing hydrocortisone, expanded SSEA-4+ cells differentiated into adipocytes as demonstrated by Oil Red O staining and expression of fat-related genes, including PPAR-γ2 and LPL (Figure 4E, right). In the presence of media containing TGF-β, cells differentiated toward cartilage, as shown by Alcian Blue staining and expression of Aggregan and Collagen II, which first appeared at 7 days of induction (Figure 4E, middle). Differentiation into osteoblasts was confirmed by the presence of calcium deposition (Alizarin Red S staining) and expression of bone-associated genes, which were all present after 11 days in culture (Figure 4E, left).
Clonal analysis
To prove conclusively that cultures obtained from SSEA-4+–sorted cells contained true mesenchymal stem cells, as opposed to being mixed populations of lineage-specific progenitors, we performed clonal analysis on 2 independent samples. SSEA-4+ cells were sorted from BM and subjected to single-cell cloning by FACS after 2 or more passages (Table 3). We observed, not surprisingly, that cloning efficiency was superior at lower passages as well as in medium supplemented with growth factors (PDGF-BB and EGF) (Table 3). Individual clones were expanded, divided into 3 fractions each to be evaluated for bone, cartilage, and fat differentiation. The majority of these clones (76.5%, n = 17) differentiated along all 3 induced lineages (Figure 6). This percentage is superior to that previously described for unpurified MSCs (17% and 34%, with or without FGF-2, respectively).25
Cloning efficiency of SSEA-4+ MSC cultures
| Sample . | Growth factors, PDGF-BB and EGF (%) . | No growth factors (%) . | Total . |
|---|---|---|---|
| BM1 - p2 | 58/96 (60.4) | 33/96 (34.4) | 91/192 (47.4) |
| BM2 - p3 | 81/192 (42.2) | 64/288 (22.2) | 145/480 (30.2) |
| BM2 - p5 | 51/288 (17.7) | 36/384 (9.4) | 87/672 (13) |
| Sample . | Growth factors, PDGF-BB and EGF (%) . | No growth factors (%) . | Total . |
|---|---|---|---|
| BM1 - p2 | 58/96 (60.4) | 33/96 (34.4) | 91/192 (47.4) |
| BM2 - p3 | 81/192 (42.2) | 64/288 (22.2) | 145/480 (30.2) |
| BM2 - p5 | 51/288 (17.7) | 36/384 (9.4) | 87/672 (13) |
The number of wells with growth out of the total number of wells plated is indicated. Cloning efficiency, expressed as percentage, is shown in parentheses.
Clonal multipotentiality of SSEA-4+ cells. Multilineage differentiation of a representative mesenchymal stem cell clone obtained from SSEA4+ cells isolated from human BM aspirates. Fat (A), bone (B), and cartilage (C) differentiation were observed by Oil Red O, Alizarin Red S, and Alcian Blue staining (top), respectively, as well as by relevant gene expression analyses (bottom). Original magnification, 10×/0.3 NA Ph1.
Clonal multipotentiality of SSEA-4+ cells. Multilineage differentiation of a representative mesenchymal stem cell clone obtained from SSEA4+ cells isolated from human BM aspirates. Fat (A), bone (B), and cartilage (C) differentiation were observed by Oil Red O, Alizarin Red S, and Alcian Blue staining (top), respectively, as well as by relevant gene expression analyses (bottom). Original magnification, 10×/0.3 NA Ph1.
Transplantation of prospectively isolated human MSCs
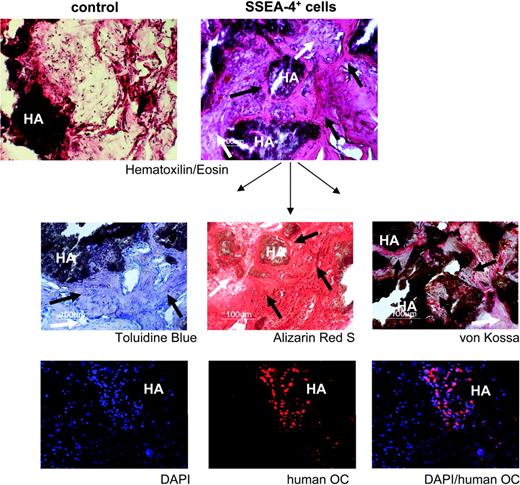
To demonstrate functional integrity of the MSC population obtained by prospective isolation with SSEA-4, expanded SSEA-4+ cells were evaluated for their capacity to produce an organized bone tissue in vivo (Figure 7). This assay was performed by ectopic transplantation of expanded SSEA-4+ cells (from 2 independent BM samples) into immunodeficient mice, using hydroxyapatite-collagen particles as a scaffold.22 Histologic analyses 8 weeks after transplantation revealed that all of the implants presented an organized bone tissue as well as contained a network of blood vessels and fibrous tissue (Figure 7, upper right). Control implants that received scaffold only (no cells) showed no bone formation, with only some sporadic cell infiltration (Figure 7, upper left). Further bone tissue characterization was performed by using Toluidine Blue (Figure 7, middle, left) for bone structure, and Alizarin Red S (Figure 7, middle, center), and von Kossa staining (Figure 7, middle, right), for mineral deposition. Immunofluorescence for human-specific osteocalcin (bottom) confirmed the presence of human cells undergoing bone differentiation. Osteocalcin-positive cells were always seen in the vicinity of the scaffold, the source of calcium and phosphorus for mineralization, as expected for physiologic bone formation.
In vivo bone formation by prospectively isolated human MSCs. Tissue sections from a representative transplant, stained as indicated, are shown. HA indicates hydroxyapatite scaffold. For each panel, black arrows indicate new bone formation; white arrows, fibrous tissue. The lower 3 panels show immunohistochemistry for human osteocalcin (OC) in red and nuclear staining with DAPI in blue. Original magnification: 10×/0.3 NA Ph1 (top 2 panels), 20×/0.4 NA Ph1 (lower panels).
In vivo bone formation by prospectively isolated human MSCs. Tissue sections from a representative transplant, stained as indicated, are shown. HA indicates hydroxyapatite scaffold. For each panel, black arrows indicate new bone formation; white arrows, fibrous tissue. The lower 3 panels show immunohistochemistry for human osteocalcin (OC) in red and nuclear staining with DAPI in blue. Original magnification: 10×/0.3 NA Ph1 (top 2 panels), 20×/0.4 NA Ph1 (lower panels).
Discussion
The results obtained in this study show that SSEA-4, a marker previously thought to be specific to very early embryonic development and to hES cells, also marks an adult mesenchymal stem cell population and can be used to prospectively isolate MSCs. Such prospective isolation is particularly attractive as current protocols used for isolating MSCs2,14,15 or for measuring their frequency (determined by the number of colony-forming unit fibroblasts; CFU-Fs)11,26,27 rely simply on their ability to adhere to tissue culture plastic. This invariably results in heterogeneous cultures. For instance, it has been shown that some CFU-Fs have a high ability for self-renewal and multipotentiality, whereas others have more limited potential,2,27,28 suggesting a high degree of heterogeneity within this clonogenic population.14,29 It is also known that hematopoietic progenitors can persist for a number of weeks in such cultures, with MSCs comprising a minority of the cell population.15 Furthermore, up to 30%1 or even more (C.A.F. and R.C.R.P., unpublished data, October 2004) of the cells that arise under CFU-F growth conditions are hematopoietic as evidenced by CD45 expression. Such heterogeneous cell preparations are far from ideal if one envisions the use of MSCs for regenerative medicine.
Attempts to obtain a pure population of MSCs have been made by several groups. For instance, a monoclonal antibody that binds to stromal cells, named Stro-1, has been produced.30 Although the surface antigen is still unknown, enrichment for clonogenic stromal cells (CFU-Fs) has been obtained with the use of this antibody (Stro-1+bright).27 Unfortunately, Stro-1 is also present, at low levels, on hematopoietic cells (Stro-1+dull).27 In the 14 years since Stro-1's discovery, no consensus has emerged regarding its use to purify MSCs. This is due particularly to the fact that other investigators have failed to detect this antigen on MSCs23 (and our observations). Another group has reported that human MSCs can be isolated based on the expression of CD49a, the α1-integrin subunit of the very late antigen (VLA-1), which is a receptor for collagen and laminin.26 According to the researchers, this population is CD49a+CD45med/low and contains all CFU-F progenitors present in the BM fraction. However, this population is heterogeneous and still contains many hematopoietic cells, not surprising because CFU-Fs represent a mixed-cell population with CD45 positivity. In contrast, SSEA-4 is expressed on both primary MSCs and MSC lines and, when used for prospective isolation, completely excludes the hematopoietic population.
The SSEA-3 and SSEA-4 antibodies were raised against preimplantation murine embryos and human teratocarcinoma cells, respectively.21,31 Their antigens have been characterized as overlapping carbohydrate epitopes on globo-series glycolipids.21 Their expression is restricted to preimplantation embryos because a switch from globo-series to lacto-series (identified by SSEA-1) synthesis occurs as the blastocyst develops.21,24 Human ES cell lines retain the earlier globo-series pathway and are SSEA-3/4+SSEA-1−,24 whereas mouse ES cell lines use the lacto-series pathway and are thus SSEA-3/4−SSEA-1+.24 With the exception of certain highly specialized cell types (erythrocytes, hence the 4% expression found in primary human BM, and certain neural ganglion cells),21,31,32 the globo-series ganglioside synthesis pathway has been thought restricted to the preimplantation embryo and not available to somatic cell types.21,31 Although the function of these carbohydrate antigens is still unknown, it is intriguing that in addition to pluripotent human ES and EC cells, they also present on the surface of multipotent MSCs. Although the evidence associating these markers with a “stemness” state is correlative, it suggests some overlap in specialized metabolic pathways between pluripotent human ES cells and multipotent MSCs. Regardless of its function, the fact that SSEA-4 is present on the surface of MSCs from human BM aspirates provides a scientific rationale for their purification. Homogeneous MSC preparations should result in less variability and superior biologic effects, benefiting future clinical applications.
Authorship
Contribution: E.J.G. performed the time course and prospective isolation experiments for SSEA-4 on human bone marrow; analyzed SSEA-4 expression in standardized preparations of human mesenchymal stem cells obtained from the Tulane Center for Gene Therapy; and performed the induction experiments of mouse and human MSCs into fat, bone, and cartilage. D.B. performed the induction experiments of mouse and human MSCs into bone, fat, and cartilage as well as the transplantation study. C.A.F. performed the time course and sorting experiments for SSEA-4 on mouse bone marrow. J.W.V. collaborated on the derivation of immortalized mesenchymal stem cell lines. R.C.R.P. derived the immortalized mesenchymal stem cell lines, designed and supervised all the experiments with SSEA-4 in mouse and human bone marrow, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
E.J.G. and D.B. contributed equally to this work.
Correspondence: Rita C.R. Perlingeiro, Center for Developmental Biology, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd, Dallas, TX 75390-9133; e-mail: rita.perlingeiro@utsouthwestern.edu.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank J. Graff and M. Kyba for critical reading of the manuscript, V. Aquino for providing human BM samples, and F. Elder for the karyotype analysis. The monoclonal antibodies to SSEA-1, SSEA-3, and SSEA-4 were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa. Some of the materials used in this work were provided by the Tulane Center for Gene Therapy through a grant from NCRR of the NIH (grant P40RR017447).
This work was supported by the Dr Bob and Jean Smith Foundation. C.A.F. was supported by a PhD fellowship from the State of Sao Paulo Research Foundation (FAPESP).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal