Abstract
Kaposi sarcoma–associated herpesvirus (KSHV) is etiologically linked to Kaposi sarcoma (KS), a tumor genetically akin to lymphatic endothelial cells (LECs). We obtained the immune transcriptional signature of KS and used KSHV-infected LECs (KLECs) as an in vitro model to determine the effects of KSHV on transcription and expression of genes involved in immunity. The antigen presentation, interferon (IFN) response, and cytokine transcriptomes of KLECs resemble those of KS. Transcription of genes involved in class I presentation is increased in KS and after infection of LECs, but MHC-I and ICAM-1 surface expression are down-regulated in KLECs. Inhibition of IFN induction of MHC-I transcription indicates that KSHV regulates MHC-I transcription. We show that MHC-I transcription is regulated by the KSHV-encoded viral FLICE inhibitory protein (vFLIP) and by viral IFN regulatory factor 1 (vIRF1). vFLIP up-regulates MHC-I and ICAM-1 through activation of NF-κB and stimulates T-cell proliferation, revealing a mechanism to prevent uncontrolled viral dissemination. In contrast, vIRF1 inhibits basal and IFN- and vFLIP-induced MHC-I transcription and surface expression through its interaction with the transcriptional coactivator p300, contributing to immune evasion. We propose that regulation of MHC-I by vFLIP and vIRF1 plays a crucial role in the host-pathogen equilibrium.
Introduction
Kaposi sarcoma (KS) lies at the interface of infection and malignancy.1-3 It is a neoplasm common in para-Mediterranean populations, endemic in parts of sub-Saharan Africa, and frequently seen in patients with AIDS.3 KS is a tumor of microvascular endothelium and gene-expression microarray (GEM) studies suggest that it belongs to the lymphatic endothelial lineage.4 KS-associated herpesvirus (KSHV) is linked to the etiopathogenesis of KS5,6 and certain lymphoproliferations, including primary effusion lymphoma (PEL)7 and a plasmablastic variant of multicentric Castleman disease (MCD).8 The risk of developing KS, PEL, and MCD is significantly higher during acquired or iatrogenic immunosuppression.3,9 Moreover, posttransplantation KS can resolve when immunosuppressive therapy is reduced,9 and the introduction of effective antiretroviral therapy for HIV infection has led to a decline in KS incidence.3 These observations indicate that disruption of host-pathogen equilibrium promotes the precipitation of these neoplasms.
Herpesviruses have evolved elaborate mechanisms to modulate host immune responses.10 EBV is the prototype of a cancer-inducing human herpesvirus.11-13 EBV modulates cellular antiviral responses in various ways, including down-regulation of major histocompatibility complex (MHC) proteins14 and blocking proteasomal degradation and antigen synthesis.15,16 However, EBV also enhances antiviral immune responses by way of its latent membrane protein 1 (LMP1), which up-regulates MHC-I.17,18 This leads to cytotoxic T-cell (CTL)–mediated elimination of EBV latency III cells, promoting the transition to latency I–infected B cells. As with KSHV, immunosuppression disturbs the host-virus equilibrium leading to an increased incidence of EBV-associated tumors.12
Several KSHV proteins regulate host innate or adaptive immune responses.19 Among these there are 5 viral proteins that block the innate antiviral interferon (IFN) response, including orf45,20 viral IL-6 (vIL6),21 viral interferon regulatory factors (vIRFs) 1 and 2,22-27 and the transactivator of the lytic cycle, RTA.28 Furthermore 2 viral modulators of immune response (vMIRs) act as E3 ubiquitin ligases and down-regulate MHC-I.29,30 vMIR2 also down-regulates ICAM-1 and CD86 by enhancing endocytosis, lysosomal targeting, and proteasome-mediated degradation31,32 and increases endocytosis of CD1d, leading to the escape of infected cells from NKT cells.33 The majority of these viral mechanisms are used during the lytic viral cycle, when most of these proteins are expressed and a vigorous host response occurs to curtail viral dissemination.
Immune regulation during KSHV latency remains insufficiently characterized. However, the KSHV lytic and latent gene profiles are not mutually exclusive. Some lytic proteins, such as RTA and vMIR2, are expressed during the initial stages of in vitro primary KSHV infection (up to 5 days after infection)34 and contribute to the establishment of latent infection. In PEL cells, it has been suggested that vIRF135,36 and vMIR137 are expressed during latency, although protein expression of these viral genes during latency has yet to be confirmed. Moreover, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis demonstrates that vIRF1 is also expressed in KS lesions38 and its expression clusters with 3 latent KSHV proteins, viral FLICE inhibitory protein (vFLIP), viral cyclin (vcyclin), and latent nuclear antigen (LANA), suggesting that vIRF1 is expressed during latency.
Here we show that KSHV has a profound effect on the immune transcriptome of lymphatic endothelial cells (LECs) and induces a “drift” of the LEC transcriptional profiles related to antigen presentation, IFN response, and cytokines toward those observed in KS. Although KSHV up-regulates transcription of genes involved in antigen presentation, their surface expression is in general unaffected or decreased after infection. We demonstrate that vFLIP up-regulates ICAM-1 and MHC-I by way of NF-κB activation and stimulates T-cell proliferation. In contrast, vIRF1 down-regulates MHC-I transcription and expression through its interaction with p300. Our findings reveal transcriptional mechanisms used by KSHV to achieve equilibrium between immune evasion and immune activation leading to optimal coexistence with its host.
Materials and methods
Cell culture
LECs were isolated from dermal microvascular endothelial cells as described39 or purchased from TCS Cellworks (Buckingham, United Kingdom). The lymphatic endothelial identity of the commercial cells was confirmed by staining with endothelial (CD31, von Willebrand factor, and VCAM) and lymphatic (PROX-1) markers and by GEM profiling (not shown). LECs were grown on fibronectin-coated plates in endothelial-cell growth medium MV (PromoCell, Heidelberg, Germany) supplemented with 10 ng/mL VEGF-C (R&D Systems, Abingdon, United Kingdom). Experiments were performed before passage 8. For the IFN induction of MHC-I expression, 105 cells were cultured in the presence of 150 U/mL IFN-α or 1000 U/mL IFN-γ (both from R&D Systems) for 24 hours. To generate KSHV we used a GFP-recombinant, BCBL-1 cell line40 grown in RPMI 1640 (Gibco, Paisley, United Kingdom) supplemented with 10% heat-inactivated fetal calf serum (FCS). To minimize production of wild-type virus, the BCBL-1 cell line was regularly purified by cell sorting (MoFlo; DakoCytomation, Ely, United Kingdom) to ensure that more than 99% of the BCBL-1 cells were infected with the GFP-expressing virus. 293T cells were grown in Dulbecco modified Eagle medium (Gibco), supplemented with 10% FCS.
KSHV production and infection of LECs
Virus production was induced by addition of tetradecanoyl phorbol acetate (Sigma, Dorset, United Kingdom) to the BCBL-1 cultures at 20 ng/mL final concentration. Four days after induction the supernatants were harvested, the virus was concentrated by ultracentrifugation as described,40 and resuspended in LEC media. We used 100-fold concentrated viral preparations to infect LECs at a multiplicity of infection of 250 to 500 viral copies/cell as described.4 This procedure reproducibly resulted in 30% to 50% LECs expressing GFP 3 to 4 days after infection.
GEM analysis
GEM profiles were obtained for 6 pairs of LECs and KLECs (3 or 4 days after infection and at least 50% positive for GFP expression), 5 nodular KS biopsies from skin, and total RNA derived from the normal skin of 5 healthy donors (AMS Biotechnology, Abingdon, United Kingdom). Total RNA was extracted from the KS biopsies with TRIzol solution (Gibco) followed by RNeasy mini kit purification (Qiagen, West Sussex, United Kingdom). Total RNA from cell cultures was isolated using the RNeasy mini kit. RNA quantity and integrity were assessed with RNA 6000 Nanochips (Agilent, West Lothian, United Kingdom). The cDNA synthesis and array procedure were performed as previously described.4 Sample RNA was hybridized to Affymetrix hg-u133+2 GeneChips. All sample arrays were background corrected, normalized, and summarized using default parameters of the RMA model.41 Array processing was performed using the affy package of the Bioconductor (http://www.bioconductor.org) suite of software for the R statistical programming language (http://www.r-project.org). The GEM data are available in the ArrayExpress database (accession number: E-MEXP-561).
Immune list compilation
Statistical analysis was performed on a subset of processed microarray data corresponding to 899 genes involved in immunity (1836 Affymetrix probe sets). The immune-related subset was compiled from publicly available databases (Gene Ontology http://www.geneontology.org/, IFN Stimulated Gene Database http://www.lerner.ccf.org/labs/williams/xchip-html.cgi), annotations made available by the microarray manufacturer (Affymetrix; http://www.affymetrix.com/), and the literature on endothelial-cell immunobiology. Compilation was performed prior to statistical analysis to avoid any selective bias resulting from the outcome of analysis. The immune-related genes were selected to create a robust system for the study of a pathogenic effect on the immune transcriptome of endothelial cells rather than to compile a comprehensive list of all genes involved in host-pathogen interactions. The class I antigen presentation pathway description (Figure 1D) is based on the KEGG pathway database (http://www.genome.jp/kegg/pathway.html) and the literature.42
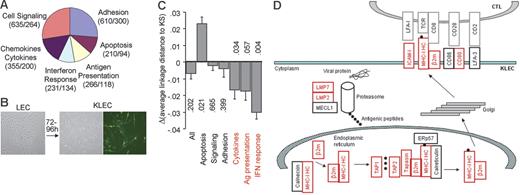
Analysis of the immune transcriptome of KLECs. (A) Schematic representation and functional grouping of the 899 genes used for immune transcriptome analysis. The numbers of Affymetrix hg-u133+2 GeneChip probe-sets and genes for each group are shown in parentheses. (B) Spindle-shaped KLECs were 40% to 50% GFP+ 3 to 4 days after infection. (C) Plot showing the difference between the average linkage distance of LEC and KLEC immune transcriptomes to that of KS, for the whole immune group, and for each functional group. Negative values indicate a drift of the transcriptome toward KS after infection. A drift toward KS is observed for the 3 immune-specific groups (in red). P values are shown to indicate significance (see “Materials and methods” for P value calculation). (D) Schematic representation of our GEM data showing that expression of genes involved in class I antigen presentation is higher in KLECs than LECs (q < 0.005). Up-regulated genes are shown in red, whereas unaffected genes are shown in black. Expression of β2-m, ICAM-1, LMP2, LMP7, TAP1, TAP2, and HLA-C is also significantly higher in KS compared to normal skin (q < 0.01). Black circles represent antigenic peptides. MHC-IHC indicates MHC-I heavy chain; LMP, large multifunctional peptidase; MECL1, multicatalytic endopeptidase complex subunit 1; TAP, transporter associated with antigen processing; ERp57, endoplasmic reticulum P58; β2m, β2-microglobulin; ICAM, intercellular adhesion molecule; LFA, lymphocyte function-associated antigen; TCR, T-cell receptor.
Analysis of the immune transcriptome of KLECs. (A) Schematic representation and functional grouping of the 899 genes used for immune transcriptome analysis. The numbers of Affymetrix hg-u133+2 GeneChip probe-sets and genes for each group are shown in parentheses. (B) Spindle-shaped KLECs were 40% to 50% GFP+ 3 to 4 days after infection. (C) Plot showing the difference between the average linkage distance of LEC and KLEC immune transcriptomes to that of KS, for the whole immune group, and for each functional group. Negative values indicate a drift of the transcriptome toward KS after infection. A drift toward KS is observed for the 3 immune-specific groups (in red). P values are shown to indicate significance (see “Materials and methods” for P value calculation). (D) Schematic representation of our GEM data showing that expression of genes involved in class I antigen presentation is higher in KLECs than LECs (q < 0.005). Up-regulated genes are shown in red, whereas unaffected genes are shown in black. Expression of β2-m, ICAM-1, LMP2, LMP7, TAP1, TAP2, and HLA-C is also significantly higher in KS compared to normal skin (q < 0.01). Black circles represent antigenic peptides. MHC-IHC indicates MHC-I heavy chain; LMP, large multifunctional peptidase; MECL1, multicatalytic endopeptidase complex subunit 1; TAP, transporter associated with antigen processing; ERp57, endoplasmic reticulum P58; β2m, β2-microglobulin; ICAM, intercellular adhesion molecule; LFA, lymphocyte function-associated antigen; TCR, T-cell receptor.
Statistical analysis
A moderated t-statistic43 was applied to processed gene-expression data to assess the significance of differential expression between sample groups for each gene probe-set present. The significance P values obtained were corrected for false discovery rate (FDR).44 The t-statistic and FDR correction were implemented in the limma and qvalue packages of Bioconductor and R, respectively. The average-linkage distance was used to assess similarity between 2 groups of gene-expression profiles, as described previously.4 The difference in distance between 2 groups of sample expression profiles to a third was assessed via the comparison of corresponding average linkage distances. For example, to identify a transcriptional drift of the LEC immune transcriptome toward KS on KSHV infection, the average-linkage distances between LEC and KS (dL) and KLEC and KS (dk) were compared to yield a measure of transcriptional difference: ΔKL = dK − dL. The error on such a comparison was estimated by combining the standard errors of the average-linkage distances involved (Figure 1C). To attach a measure of significance to any difference, ΔKL, we used a resampling method. For n ϵ N (where N is all Affymetrix hg-u133+2 probe-sets) microarray probe-sets corresponding to an immune functional group, a “null” distribution of distance differences was constructed by calculating Δ′KL on 10 000 randomly selected probe-set groups of the same size (n). The significance of the original observation, ΔKL, was assessed subsequently against the null distribution.
Antibodies and flow cytometry
Surface expression of proteins involved in antigen presentation was assessed by flow cytometry (FACSCalibur; Becton Dickinson, Oxford, United Kingdom). We used RPE-labeled antibodies against human HLA-A, -B, -C (MHC-I), CD40, CD1a, CD1d (all from BD PharMingen, Oxford, United Kingdom), CD80, CD86 (both from R&D Systems), HLA-DR (Sigma), and unconjugated ICAM-1 and mouse IgG (DakoCytomation) and HLA-E (provided by Dr Daniel Gerharty, Fred Hutchinson Cancer Research Center, Seattle, WA). Analyses were done after gating live cells.
Allogeneic T-cell proliferation assay
CD8+ T-cell proliferation was measured by labeling with carboxy-fluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Leiden, the Netherlands). Peripheral blood lymphocytes (PBLs) from healthy volunteers were labeled with 0.25 μM CFSE and mixed with LECs at a responder to stimulator ratio of 50:1. Cocultures were grown in 24-well plates in RPMI 1640 supplemented with 10% human AB serum, 2 mMl-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (all from Gibco). The cultures were incubated at 37°C, 5% CO2 for 7 days. IL-2 (R&D Systems) was added to the cells on day 3 at a final concentration of 12 U/mL. On day 7, cells were stained with an RPE-labeled anti-CD8 antibody (BD PharMingen) and the percent of proliferated CTLs was determined by flow cytometry as the CD8+/CFSElow cells.
RT-PCR and qRT-PCR
Expression of KSHV genes in KLECs and LECs infected with lentiviruses was determined by RT-PCR, using HotStart Taq DNA polymerase (Qiagen). Reactions included an enzyme activation step at 95°C for 15 minutes followed by 25 to 35 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds and a cycle of 72°C for 1 minute. The reaction products were separated on 2% agarose gels and visualized using the VersaDoc system. No products were detected using the KSHV gene-specific primers when using cDNA from LECs or LECs infected with pSIN lentiviral vector as template for PCR (40 cycles). (The KSHV gene primer sequences are available on request.) HLA-A expression was quantified by qRT-PCR using the SYBR Green Master Mix (ABI Prism 7700, Applied Biosystems, Warrington, United Kingdom). Optimized HLA-A forward and reverse primers were used at final concentration of 300 nM: forward 5′-TGTGGAGGAGGAAGAGCTCAGATA-3′ and reverse 5′-ACAAGGCAGCTGTCTCACACTTTA-3′. GAPDH was used as a reference gene as previously described.45 Reactions were performed in 25 μL with standard TaqMan cycling conditions. At least 2 dilutions of each cDNA preparation were assayed in duplicate and the relative expression of HLA-A was determined using the standard curve method. Similarly, we designed primers for qRT-PCR for KSHV genes (LANA, vFLIP, vMIR1, vMIR2, vIRF1, and ORF26) to determine their quantitative expression levels (sequences also available on request). Using these primers and cDNA from LECs, we did not detect amplification of any products before 40 cycles of qPCR (LEC cDNA produced similar GAPDH levels with KLEC samples).
Lentiviral expression of KSHV genes
KSHV genes and the vIRF1Δ1-82 mutant were cloned from the BC3 and BC1 PEL cell lines and were expressed using a modified, previously described lentiviral vector (pSIN),46 including a central polypurine tract, spleen focus forming virus promoter, KSHV gene, woodchuck hepatitis virus posttranscriptional regulatory element, and no selectable marker. Virions were produced by transient cotransfection of 293T cells for 5 hours with 2 μg pSIN, 1.5 μg p8.91, and 1.5 μg pMD.G plasmids using the standard FuGENE 6 (Roche, East Sussex, United Kingdom) protocol. Forty-eight hours later the virus containing supernatant was harvested, filtered (0.45 μm), and stored at −80°C. Typically, 1 mL virus preparation was used to infect 105 LECs for 5 hours at 37°C. For every KSHV gene, the same viral batch was used throughout the study unless otherwise stated. For the detection and quantification of lentiviral infection, we modified a previously described46 qPCR for the lentiviral packaging signal and GAPDH. The presence of an average of 2 to 5 copies/cell of every lentivirus was confirmed (not shown). A lentivirus containing GFP, instead of a KSHV gene, was used to assess infectivity of LECs, resulting in more than 90% infection as determined by flow cytometry. For coinfection experiments, LECs (105) were infected with 1 mL of each virus and control cells were mock-infected with the same total volume of pSIN lentivirus. MHC-I levels were assessed 3 days after infection. To block NF-κB transcription, LECs were incubated with 5 μM of BAY 11-7082 (Calbiochem, Beeston, United Kingdom) for 2 hours at 37°C and then infected with lentivirus as described. MHC-I levels were determined 24 hours after infection.
Results
KSHV infection modulates the LEC immune transcriptome
To determine the global effect of KSHV on the LEC immune transcriptome we used GEM analysis. We selected 899 immune-related genes (1836 probe-sets on the Affymetrix hg-u133+2 GeneChip) and classified them into 6 functional groups: antigen presentation, chemokines and cytokines, IFN response, adhesion, apoptosis, and cell signaling (Figure 1A; Table S1, available on the Blood website; see the Supplemental Tables link at the top of the online article). The first 3 groups were considered as immune-specific, whereas the last 3 functional groups include genes involved in host responses, but also other cellular processes. GEM analysis was performed for 6 pairs of primary LECs and KLECs (infected with a recombinant, GFP-expressing KSHV; Figure 1B), 5 nodular KS lesions, and 5 normal skin biopsies. Comparative bioinformatic analyses of the immune transcriptome were restricted to the 899 immune-related genes to exclude background noise generated by nonimmune genes.
Over 37% of immune-related gene probe-sets displayed significant differential regulation after KSHV infection of LECs (FDR threshold q < .005; Table S2). A particularly conservative threshold of significant regulation was used in this comparison to acknowledge minimal intragroup variation of the cell samples involved. The same analysis of all human genome probe-sets (54 675 in total) identified approximately 25% as significantly regulated after KSHV infection of LECs (not shown). Similarly, the immune profiles of KS biopsies and normal skin were compared to yield a KS immune signature. Approximately 13% of immune-related probe-sets were differentially regulated between KS and normal skin at q less than .01 (Table S3). In comparison, approximately 6% of all human genome probe-sets displayed significant regulation under the same criteria (not shown). Approximately 25% of immune-related probe-sets significantly regulated between KS and normal skin were similarly and significantly regulated after KSHV infection of LECs (Table S4). The differences between KS and normal skin biopsies describe the immune profile of KS tumor cells but are also influenced by the tumor microenvironment.
To investigate the contribution of KSHV infection to the immune transcriptome of KS we compared the group distances of the LEC and KLEC immune profiles to the immune profile of KS. The KLEC immune profile was more similar to that of KS than was the LEC immune profile (Figure 1C), but the effect was not statistically significant (P = .202). When the same analysis was repeated for the 6 functional subgroups of the immune-related genes, a “‘drift” toward KS after KSHV infection was observed for the antigen presentation (P = .057), IFN response (P = .004), and chemokine and cytokine (P = .034) functional groups (Figure 1C). The antigen presentation group describes the ability of infected cells to directly present antigen to T lymphocytes. KSHV infection leads to significant (q < .005) up-regulation of genes involved in class I antigen presentation (Figure 1D). Infection increases mRNA levels of MHC-I heavy chains (HLA-A, -B, -C) and β2-microglobulin (β2m), but also of other genes involved in antigen processing (LMP2 and LMP7 subunits of the immunoproteasome), antigen loading (TAP1, TAP2, tapasin), and costimulation of T lymphocytes (ICAM-1 and CD80). Expression of β2m, ICAM-1, LMP2, LMP7, TAP1, TAP2, and HLA-C is also significantly higher in KS compared to normal skin (q < .01), whereas HLA-A and HLA-B expression is higher in KS at significance levels of q < .05. Furthermore, the IFN response and cytokine groups include genes involved in primary antiviral responses, and the cytokine cellular microenvironment, which also affect antigen presentation. The antigen presentation, IFN response, and cytokine groups included over 80% of the genes that were significantly affected by KSHV infection and showed a similar pattern of differential expression between KS and normal skin (Table S4). These findings suggest that KSHV plays a crucial role in shaping the antigen presentation potential of infected cells during KS development.
The KLEC antigen presentation surface phenotype is significantly compromised
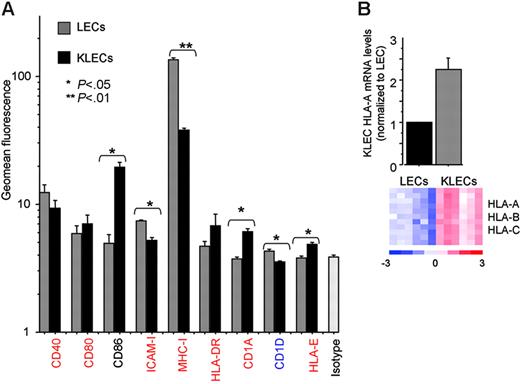
Next, we investigated the effect of KSHV on the surface antigen presentation phenotype of LECs. We determined the surface expression levels of 9 proteins involved in antigen presentation (Figure 2A). LECs express MHC-I, CD40, ICAM-1, and CD83 and low levels of CD80 and CD86, but do not express significant levels of HLA-DR or HLA-E (Figure 2A). Following infection (4 days after infection), MHC-I and ICAM-1 expression was significantly decreased, whereas the expression of CD86, CD1a, and HLA-E was significantly increased. The down-regulation of MHC-I expression contradicted the GEM data, and we determined LEC and KLEC HLA-A mRNA levels by qRT-PCR. Figure 1B shows that GFP-expressing KLECs isolated by cell sorting display higher HLA-A mRNA levels than noninfected controls. The agreement of GEM and qRT-PCR data for HLA-A is likely to also be the case for HLA-B and HLA-C, which share the same promoters with HLA-A47 and show similar GEM profile (Figure 2B). This shows that KSHV exhibits a broad compromising effect on the surface immunophenotype of LECs by using posttranscriptional mechanisms.
Surface immunophenotype of LECs and KLECs. (A) Surface expression levels of proteins involved in antigen presentation in LECs (⊡) and GFP-expressing KLECs (▪) were assessed by flow cytometry 4 days after infection. Error bars correspond to standard deviation, calculated after 3 independent infections with different batches of KSHV. Significant changes are indicated with asterisks (P values were calculated with a 2-sided t test). A representation of the corresponding GEM data are also shown (significantly up- and down-regulated gene names are shown in red and blue, respectively, whereas black corresponds to transcripts that were not significantly affected after infection). (B) HLA-A mRNA levels in LECs and GFP-expressing KLECs (purified by cell sorting, 4 days after infection) were determined by qRT-PCR. mRNA levels are normalized to LEC. A heat map of GEM data for HLA-A (2 probe-sets), HLA-B (1 probe-set), and HLA-C (7 probe-sets) in 6 pairs of LECs and KLECs is also shown. The heat map color scale indicates units of standard deviation from the mean expression of each row (red high and blue low expression).
Surface immunophenotype of LECs and KLECs. (A) Surface expression levels of proteins involved in antigen presentation in LECs (⊡) and GFP-expressing KLECs (▪) were assessed by flow cytometry 4 days after infection. Error bars correspond to standard deviation, calculated after 3 independent infections with different batches of KSHV. Significant changes are indicated with asterisks (P values were calculated with a 2-sided t test). A representation of the corresponding GEM data are also shown (significantly up- and down-regulated gene names are shown in red and blue, respectively, whereas black corresponds to transcripts that were not significantly affected after infection). (B) HLA-A mRNA levels in LECs and GFP-expressing KLECs (purified by cell sorting, 4 days after infection) were determined by qRT-PCR. mRNA levels are normalized to LEC. A heat map of GEM data for HLA-A (2 probe-sets), HLA-B (1 probe-set), and HLA-C (7 probe-sets) in 6 pairs of LECs and KLECs is also shown. The heat map color scale indicates units of standard deviation from the mean expression of each row (red high and blue low expression).
KSHV inhibits constitutive and IFN-induced expression of MHC-I
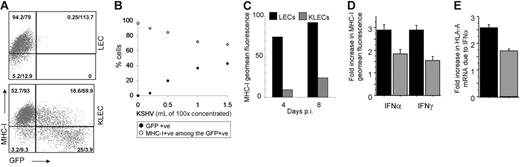
MHC-I down-regulation is a common immune evasion mechanism used by viruses.10 In KLECs, MHC-I down-regulation is proportional to GFP expression, which correlates with KSHV copy number per infected cell. The effect also correlates with the amount of virus used for infection (Figure 3B) and is maintained for up to 8 days after infection (Figure 3C).
Down-regulation of MHC-I expression by KSHV. Results represent at least 3 independent infections with different KSHV batches and error bars correspond to standard deviation from the mean. (A) Representative dot plot of MHC-I expression in LECs and in KLECs, showing that MHC-I down-regulation increases with GFP expression (axes are logarithmic). The percent of cells of the total and geometric mean (geomean) fluorescence are shown for each quadrant. (B) Increasing amounts of KSHV resulted in a proportionate increase of GFP-expressing cells and decrease of MHC-I+ cells among the infected cells; •, percentage of GFP-expressing cells after infection; ○, percentage of MHC-I–expressing cells among the GFP+ cells. (C) Representative experiment of levels of MHC-I fluorescence at 4 and 8 days after infection for LECs (▪) and GFP expressing KLECs (⊡). (D) Induction of MHC-I surface expression by IFN. LECs and KLECs (3 days after infection) were cultured for 24 hours in the presence of IFN-α (150 U/mL) or IFN-γ (1000 U/mL). The graph shows the fold increase in MHC-I expression in LECs and KLECs after IFN treatment compared to MHC-I expression of untreated LECs and KLECs, respectively. (E) Fold increase in HLA-A mRNA levels in LECs and KLECs after IFN-α treatment. Expression of HLA-A in noninfected and infected cells is normalized to HLA-A levels of untreated LECs and KLECs, respectively. In panels C-E, ▪, LEC levels; ⊡, KLEC levels.
Down-regulation of MHC-I expression by KSHV. Results represent at least 3 independent infections with different KSHV batches and error bars correspond to standard deviation from the mean. (A) Representative dot plot of MHC-I expression in LECs and in KLECs, showing that MHC-I down-regulation increases with GFP expression (axes are logarithmic). The percent of cells of the total and geometric mean (geomean) fluorescence are shown for each quadrant. (B) Increasing amounts of KSHV resulted in a proportionate increase of GFP-expressing cells and decrease of MHC-I+ cells among the infected cells; •, percentage of GFP-expressing cells after infection; ○, percentage of MHC-I–expressing cells among the GFP+ cells. (C) Representative experiment of levels of MHC-I fluorescence at 4 and 8 days after infection for LECs (▪) and GFP expressing KLECs (⊡). (D) Induction of MHC-I surface expression by IFN. LECs and KLECs (3 days after infection) were cultured for 24 hours in the presence of IFN-α (150 U/mL) or IFN-γ (1000 U/mL). The graph shows the fold increase in MHC-I expression in LECs and KLECs after IFN treatment compared to MHC-I expression of untreated LECs and KLECs, respectively. (E) Fold increase in HLA-A mRNA levels in LECs and KLECs after IFN-α treatment. Expression of HLA-A in noninfected and infected cells is normalized to HLA-A levels of untreated LECs and KLECs, respectively. In panels C-E, ▪, LEC levels; ⊡, KLEC levels.
In addition to the effect on basal MHC-I levels, we determined the effect of KSHV on IFN induction of MHC-I transcription and expression. LECs and KLECs (3 days after infection) were treated with IFN-α or IFN-γ for 24 hours prior to determining MHC-I levels. The IFNs caused a 3-fold up-regulation of MHC-I expression in LECs (Figure 3D). In contrast, IFN treatment of KLECs caused only a 1.5-fold increase in MHC-I surface expression (compared to basal MHC-I surface expression in KLECs; Figure 3D). The inhibitory effect of KSHV on the IFN-α–mediated HLA-A induction was reflected at the mRNA level (Figure 3E), indicating that KSHV is able to inhibit MHC-I transcription.
KSHV regulates MHC-I transcription by way of vIRF1 and vFLIP
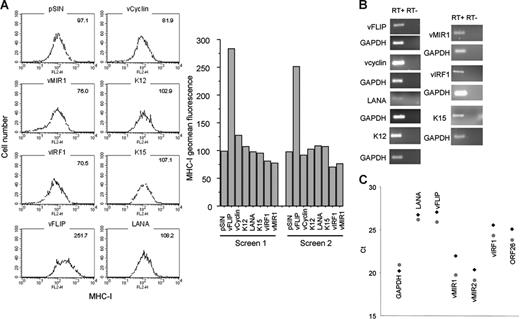
Next, we investigated which KSHV genes associated with viral latency affect MHC-I transcription and expression in LECs. The KSHV genes tested included vcyclin, vFLIP, K12, K15 (predominant), LANA, and vIRF1 (Figure 4A). vMIR1, a lytic gene,48 was used as a known negative regulator of MHC-I expression. All genes were expressed in LECs using a lentiviral system. For all the constructs, the presence of at least 2 copies of lentivirus per cell was confirmed by qPCR for the lentiviral vector (not shown) and gene expression was confirmed by RT-PCR (Figure 4B). Two independent screenings revealed that in addition to vMIR1, vIRF1 down-regulates MHC-I. The effect of vIRF1 is comparable to that of vMIR1 (corresponding to a reduction of expression by 20%-30%; Figure 4A), with both proteins displaying comparable expression levels (Figure 4B). In contrast, MHC-I is significantly up-regulated by vFLIP (Figure 4A). Notably, qRT-PCR analysis of KLECs demonstrates that vFLIP, vIRF1, and both vMIR1 and vMIR2 are expressed in this in vitro system (Figure 4C). As expected, expression levels of vFLIP cluster with LANA. The high expression of vMIRs could be due to the previously described “lytic burst” during primary KSHV infection34 or the presence of lytic cells among the infected cells confirmed by detection of expression of the late lytic KSHV ORF26. Overall, these data suggest that vIRF1 and the vMIRs contribute to the MHC-I regulation in this model.
Regulation of MHC-I by KSHV transcripts. (A) Histograms showing the levels of MHC-I expression in LECs expressing 6 KSHV genes. Geomean fluorescence is shown at the upper right corner of each histogram. The results from 2 independent screens are also shown (bar graphs). vMIR1 was used as a control for MHC-I down-regulation. MHC-I levels were determined 3 days after infection. (B) RT-PCR confirming expression of viral transcripts in LECs infected with the lentiviral constructs used for the screen. Second column shows the non-RT controls for each gene. GAPDH was used as a housekeeping control gene. (C) qRT-PCR analysis of KLECs (3 days after infection) for expression of GAPDH, LANA, vFLIP, vIRF1, vMIR1, vMIR2, and ORF26 in KLEC (♦) and purified by cell sorting (4 days after infection) GFP-expressing KLEC ( ). Ct values (y-axis) indicate the cycle number where PCR products become detectable. Lower Ct values correspond to higher average mRNA expression.
). Ct values (y-axis) indicate the cycle number where PCR products become detectable. Lower Ct values correspond to higher average mRNA expression.
Regulation of MHC-I by KSHV transcripts. (A) Histograms showing the levels of MHC-I expression in LECs expressing 6 KSHV genes. Geomean fluorescence is shown at the upper right corner of each histogram. The results from 2 independent screens are also shown (bar graphs). vMIR1 was used as a control for MHC-I down-regulation. MHC-I levels were determined 3 days after infection. (B) RT-PCR confirming expression of viral transcripts in LECs infected with the lentiviral constructs used for the screen. Second column shows the non-RT controls for each gene. GAPDH was used as a housekeeping control gene. (C) qRT-PCR analysis of KLECs (3 days after infection) for expression of GAPDH, LANA, vFLIP, vIRF1, vMIR1, vMIR2, and ORF26 in KLEC (♦) and purified by cell sorting (4 days after infection) GFP-expressing KLEC ( ). Ct values (y-axis) indicate the cycle number where PCR products become detectable. Lower Ct values correspond to higher average mRNA expression.
). Ct values (y-axis) indicate the cycle number where PCR products become detectable. Lower Ct values correspond to higher average mRNA expression.
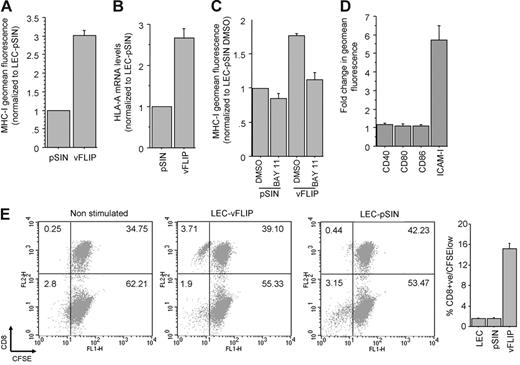
vFLIP expression up-regulates MHC-I through NF-κB activation and stimulates allogeneic T-cell proliferation
We confirmed the effect of vFLIP on MHC-I surface expression (Figure 5A) and showed that this occurs at the transcriptional level (Figure 5B). MHC-I up-regulation by vFLIP is through activation of the NF-κB pathway because the effect was significantly reduced when LECs were incubated with a chemical inhibitor of NF-κB prior to infection with the vFLIP lentivirus (Figure 5C). We also tested the effect of vFLIP on the expression of 4 other NF-κB responsive surface proteins involved in class I antigen presentation and observed a significant up-regulation of ICAM-1 (Figure 5D). Based on these findings we tested the ability of vFLIP to induce allogeneic CTL proliferation. LECs or LECs infected with empty lentiviral vector induce weak proliferation of allogeneic CTLs (1%-2% proliferated CD8+ cells after 7 days), whereas vFLIP expression significantly increases the allo-stimulatory activity of LECs (approximately 15% proliferated CD8+ T cells; Figure 5E).
vFLIP up-regulates MHC-I via activation of NF-κB. (A) MHC-I surface expression in LECs expressing vFLIP and the vector (pSIN). Mean and standard deviation were calculated after 3 independent infections (3 days after infection). (B) HLA-A mRNA levels of LECs expressing vFLIP and pSIN (3 days after infection). Expression is normalized to LECs infected with pSIN. (C) MHC-I expression in LECs treated with DMSO or BAY 11-7082 and then infected with vFLIP lentivirus or empty vector. MHC-I expression was determined 24 hours after infection and normalized to LECs treated with DMSO and infected with the empty lentiviral vector. (D) Surface expression of costimulatory class I proteins in vFLIP-expressing LECs. A 6-fold up-regulation of ICAM-1 is shown. (E) CD8+ T-cell proliferation after 7 days of coculture with LECs or LECs expressing vFLIP (see “Materials and methods” for details). Representative double staining (x-axis for CFSE and y-axis for CD8) dot plots are shown. The percent of cells in each quadrant is also shown. Average and standard deviation of CD8+ proliferated cells (CD8+/CFSElow) from triplicate experiments is shown on the adjacent bar graph. Error bars were calculated based on standard deviation.
vFLIP up-regulates MHC-I via activation of NF-κB. (A) MHC-I surface expression in LECs expressing vFLIP and the vector (pSIN). Mean and standard deviation were calculated after 3 independent infections (3 days after infection). (B) HLA-A mRNA levels of LECs expressing vFLIP and pSIN (3 days after infection). Expression is normalized to LECs infected with pSIN. (C) MHC-I expression in LECs treated with DMSO or BAY 11-7082 and then infected with vFLIP lentivirus or empty vector. MHC-I expression was determined 24 hours after infection and normalized to LECs treated with DMSO and infected with the empty lentiviral vector. (D) Surface expression of costimulatory class I proteins in vFLIP-expressing LECs. A 6-fold up-regulation of ICAM-1 is shown. (E) CD8+ T-cell proliferation after 7 days of coculture with LECs or LECs expressing vFLIP (see “Materials and methods” for details). Representative double staining (x-axis for CFSE and y-axis for CD8) dot plots are shown. The percent of cells in each quadrant is also shown. Average and standard deviation of CD8+ proliferated cells (CD8+/CFSElow) from triplicate experiments is shown on the adjacent bar graph. Error bars were calculated based on standard deviation.
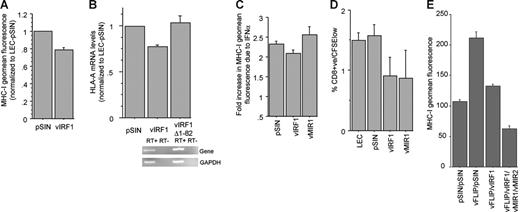
vIRF1 inhibits basal, IFN-induced, and vFLIP-induced MHC-I expression
In contrast to vFLIP, we confirmed that vIRF1 down-regulates MHC-I expression in LECs (Figure 6A). As in the case of vFLIP, MHC-I regulation is transcriptional (Figure 6B). vIRF1 blocks type I and II IFN-mediated transcriptional activation, by competing with the cellular IRF1 and IRF3 for binding the coactivators CBP and p300.23-25,27 The unique amino-terminus of vIRF1 is necessary for binding to p30023 and we show that a vIRF1 mutant lacking this domain (vIRF1Δ1-82) is unable to down-regulate MHC-I in LECs (Figure 6B). Furthermore, similarly to KLECs (although to a lesser extent), vIRF1-expressing LECs are less responsive to IFN-α–mediated induction of MHC-I expression, than LECs infected with the vector (Figure 6C). Notably, LECs expressing vMIR1 are more responsive to IFN-α treatment than the vector control, indicating that vMIR1 does not contribute to the observed effect in KLECs. vIRF1 and vMIR1 expression has a similar modest negative effect on the limited ability of LECs to stimulate allogeneic CTL proliferation (Figure 6D). Finally, coinfection of LECs with vIRF1 and vFLIP demonstrates that vIRF1 alone is able to significantly diminish the vFLIP-mediated up-regulation of MHC-I. Coexpression of vFLIP, vIRF1, vMIR1, and vMIR2 leads to overall MHC-I down-regulation compared to LECs infected with pSIN (Figure 6E).
vIRF1 down-regulates constitutive and IFN- and vFLIP-induced MHC-I expression. (A) MHC-I surface expression in LECs expressing vIRF1 and pSIN. Mean and standard deviation were calculated after 3 independent infections (3 days after infection). (B) HLA-A mRNA levels of LECs expressing vIRF1 and pSIN (3 days after infection). Expression is normalized to LECs infected with pSIN. LECs expressing a vIRF1 mutant lacking the p300-binding site (amino acids 1-82) show similar HLA-A mRNA levels to control cells (pSIN). Comparable levels of expression for vIRF1 and vIRF1Δ1-82 were confirmed by RT-PCR, using the same set of primers for both genes. (C) Plot showing the fold increase of MHC-I surface expression due to IFN-α in LECs expressing pSIN, vIRF1, and vMIR1. (D) Percent of CD8+/CFSElow cells among allogeneic PBLs cocultured with LECs infected with pSIN, vIRF1, or vMIR1 for 7 days (see “Materials and methods” for details). (E) vIRF1 and vMIRs reverse the effect of vFLIP on MHC-I. Bar graph shows MHC-I surface expression (geomean fluorescence and standard deviation) of LECs infected with pSIN (pSIN/pSIN), vFLIP and pSIN (vFLIP/pSIN), vFLIP and vIRF1 (vFLIP/vIRF1), and vFLIP, vIRF1, and the vMIRs (vFLIP/vIRF1/vMIR1/vMIR2). Error bars were calculated based on standard deviation.
vIRF1 down-regulates constitutive and IFN- and vFLIP-induced MHC-I expression. (A) MHC-I surface expression in LECs expressing vIRF1 and pSIN. Mean and standard deviation were calculated after 3 independent infections (3 days after infection). (B) HLA-A mRNA levels of LECs expressing vIRF1 and pSIN (3 days after infection). Expression is normalized to LECs infected with pSIN. LECs expressing a vIRF1 mutant lacking the p300-binding site (amino acids 1-82) show similar HLA-A mRNA levels to control cells (pSIN). Comparable levels of expression for vIRF1 and vIRF1Δ1-82 were confirmed by RT-PCR, using the same set of primers for both genes. (C) Plot showing the fold increase of MHC-I surface expression due to IFN-α in LECs expressing pSIN, vIRF1, and vMIR1. (D) Percent of CD8+/CFSElow cells among allogeneic PBLs cocultured with LECs infected with pSIN, vIRF1, or vMIR1 for 7 days (see “Materials and methods” for details). (E) vIRF1 and vMIRs reverse the effect of vFLIP on MHC-I. Bar graph shows MHC-I surface expression (geomean fluorescence and standard deviation) of LECs infected with pSIN (pSIN/pSIN), vFLIP and pSIN (vFLIP/pSIN), vFLIP and vIRF1 (vFLIP/vIRF1), and vFLIP, vIRF1, and the vMIRs (vFLIP/vIRF1/vMIR1/vMIR2). Error bars were calculated based on standard deviation.
Discussion
KS is a neoplasm characterized by vascular nodules composed of spindle-shaped tumor cells, a prominent vasculature and an inflammatory infiltrate. KSHV is latently expressed in most spindle cells, but a subpopulation also supports lytic infection.3 The surface markers and transcriptional profile of these spindle cells are closely related to LECs4,49 and we, therefore, used primary LECs as an in vitro primary infection model to obtain insight into the way KSHV modulates host immunity. We show that KSHV infection steers the cytokine, antigen presentation, and IFN signatures of LECs to those observed in KS lesions, suggesting that KSHV is important in shaping the transcriptional profiles of these immune-related genes in KS lesions. This analysis also validates the use of KLECs as a relevant in vitro model for studying the transcriptional regulation of antigen presentation by KSHV during KS development. In contrast, the apoptosis transcriptome of KLECs drifts away from the corresponding group in KS, reflecting the effect of other factors on the KS profile (such as the tumor microenvironment), the in vitro conditions used to culture infected LECs, and the differences in the viral gene-expression profile between KS and KLECs. Similar to other primary KSHV infection models,34 KLECs are characterized by prominent lytic viral gene expression (Figure 4C), which possibly also occurs during primary infection in vivo but is limited in KS lesions.
KSHV infection up-regulates the transcription of genes with promoters containing IFN and NF-κB responsive elements such as HLA-A, HLA-B, HLA-C, TAP1, TAP2, β2m, tapasin, and members of the immunoproteasome. A similar pattern is observed when KS biopsies are compared to normal skin. Despite this, surface expression of MHC-I and ICAM-1 is down-regulated in KLECs. In addition, KSHV also inhibits IFN-γ and IFN-α induction of MHC-I transcription and expression. These data indicate that KSHV uses both transcriptional and posttranscriptional mechanisms29 to modulate MHC-I. Although other human herpesviruses, such as EBV14,50 and human CMV51 use similar mechanisms during their lytic cycle, this is the first demonstration that KSHV regulates MHC-I transcription. Of note, surface expression of CD86, a protein that binds CD28 and is essential for T-lymphocyte proliferation and IL-2 production, is significantly induced by KSHV. This occurs despite the fact that vMIR2 down-regulates CD8631,32 and its expression is detectable in KLECs.
Mechanistic insights of KSHV modulation of MHC-I have concentrated on the posttranscriptional mechanisms used by 2 lytic viral proteins, vMIR1 and vMIR2.29,30 To identify latent KSHV genes, which contribute to MHC-I regulation by controlling its transcription, we tested the effect of 6 KSHV proteins associated with viral latency. We used a lentiviral vector system for viral gene expression. The advantages of this system are the efficient infection of slowly dividing LECs and the exclusion of artifacts due to protein overexpression. This could be particularly relevant for vIRF1 because its expression in latently infected tumor cells appears to be low.36 We confirm that vMIR1 down-regulates MHC-I expression in LECs. In addition, we identify that MHC-I transcription and surface expression are down-regulated and up-regulated by vIRF1 and vFLIP, respectively. These proteins were not identified as MHC-I regulators in the genetic screen that identified the vMIRs.29 This discrepancy could be due to the use of the HeLa cell line in that screen. HeLa cells are infected with human papillomavirus 18 and express the oncogenic protein E7, which down-regulates transcription of MHC-I52 and could potentially mask the effects of vIRF1 and vFLIP.
Like the EBV encoded LMP1,17 vFLIP up-regulates MHC-I transcription and expression. Both proteins activate NF-κB transcription53-56 and, as in the case of LMP1,18,57 we show that vFLIP up-regulates MHC-I by way of NF-κB activation. It also induces ICAM-1 and stimulates T-cell proliferation, inferring that transcriptional modulation of antigen presentation by this viral gene has functional implications. Expression of vFLIP is likely to contribute to the increased MHC-I transcription in KLECs. However, in this infection model (Figure 4C), possibly during early stages of primary infection in vivo, and in lytic cells in KS lesions, expression of vIRF1 with the vMIRs compensate for the induction of MHC-I by vFLIP (Figure 6E), leading to an overall down-regulation of MHC-I expression. Interestingly, these findings also provide insight into the interactions of LECs with immune cells and their function as antigen presenting cells. LECs express MHC-I and ICAM-1 and induce only weak CTL proliferation, which can be significantly enhanced by NF-κB activation on pathogenic stimulation (such as vFLIP).
The effect of vFLIP is partly inhibited by vIRF1. This viral gene is functionally related to other oncogenic viral proteins such as the adenovirus E1A,19 which also down-regulates MHC-I transcription.58 Our findings reveal vIRF1 as the first human herpesvirus protein capable of down-regulating basal and IFN-induced MHC-I transcription. The early lytic EBV protein BZLF1 exhibits similar activity,50 but has no direct effect on basal MHC-I expression.14 The unique effect of vIRF1 on constitutive MHC-I expression could be due to the use of endothelial cells because the endothelium of IRF1 knockout mice displays particularly reduced basal MHC-I expression compared to other tissues.59 Indeed, vIRF1 is likely to inhibit MHC-I transcription by blocking IRF1 because our data suggest that binding of vIRF1 to p300 is necessary for its function. Through the same mechanism, vIRF1 might also block transactivation of MHC-I transcription by NF-κB,47 explaining the inhibition of vFLIP-induced MHC-I up-regulation by vIRF1. Of note, the inhibition of IFN-mediated induction of MHC-I is more profound in KLECs than in vIRF1 expressing LECs. This could be due to lower levels of expression in the lentiviral-infected cells or to the fact that other KSHV proteins exhibit similar activity.
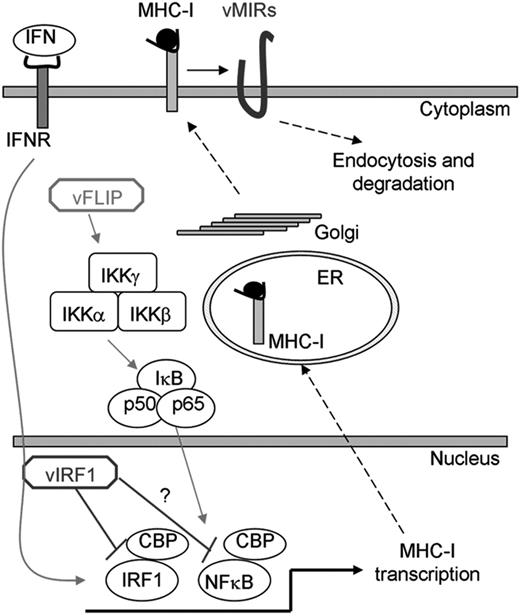
Although we cannot exclude the existence of other KSHV-encoded MHC-I regulators, these findings allow us to propose a model of MHC-I regulation by KSHV (Figure 7). During primary infection in immunocompetent individuals, when immune escape is vital for the virus, KSHV uses transcriptional and posttranscriptional mechanisms by way of vIRF1 and the vMIRs to evade antigen presentation. This results in establishment of latency and, thus, host-pathogen equilibrium. Although the precise cellular reservoirs and latency program of KSHV during persistent infection are still unknown, our findings suggest that the viral latency expression profile may be limited, possibly restricted to LANA60 and that vFLIP, by up-regulating MHC-I expression during limited phases of latency,61 stimulates CTL proliferation to curb viral dissemination. During states of immunodeficiency, the host-pathogen equilibrium is disturbed leading to a broader latency expression profile (sustained expression of vFLIP) and more pronounced and uncontrolled lytic replication (expression of vIRF1 and vMIRs). This results in proliferation of infected cells and KS development. Here, the majority of tumor cells are latently infected expressing vFLIP and MHC-I,62 rendering KS cells susceptible to CTL clearance, which contributes to KS regression during immune reconstitution.
Regulation of MHC-I expression by KSHV. MHC-I transcription is activated by IFN and NF-κB signaling. In addition to posttranscriptional MHC-I regulation by the vMIRs during lytic replication, KSHV regulates MHC-I transcription. Activation of NF-κB transcription by vFLIP leads to MHC-I up-regulation, ensuring controlled viral dissemination during latency. The vFLIP activity and IFN induction of MHC-I are regulated by vIRF1, which binds CBP and inhibits MHC-I expression by blocking IRF1 and possibly NF-κB transcription.
Regulation of MHC-I expression by KSHV. MHC-I transcription is activated by IFN and NF-κB signaling. In addition to posttranscriptional MHC-I regulation by the vMIRs during lytic replication, KSHV regulates MHC-I transcription. Activation of NF-κB transcription by vFLIP leads to MHC-I up-regulation, ensuring controlled viral dissemination during latency. The vFLIP activity and IFN induction of MHC-I are regulated by vIRF1, which binds CBP and inhibits MHC-I expression by blocking IRF1 and possibly NF-κB transcription.
Authorship
Contribution: D.L. designed and performed research, collected and analyzed data, and wrote the manuscript; M.W.B.T. analyzed data and cowrote the manuscript; R.J.V. constructed and contributed vital new reagents and collected data; H.-W.W. collected data and performed research; N.C.M. designed research; A.H. constructed and contributed vital new reagents; O.F. contributed vital reagents; F.G. designed research; and C.B. designed research and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Chris Boshoff, Cancer Research United Kingdom Viral Oncology Group, Wolfson Institute for Biomedical Research, University College London, London, Gower St, WC1E 6BT, London, United Kingdom; e-mail: c.boshoff@ucl.ac.uk.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was funded by the Medical Research Council, Cancer Research United Kingdom, and Wellcome Trust. R.J.V. is a PhD student supported by the BBSRC and an educational grant from Sanofi-Aventis.
We thank K. Alitalo for providing LECs, D. Gerharty for providing the anti-HLA-E antibody, J. Orenstein and the National Cancer Institute AIDS and Cancer Specimen Resource for providing the KS biopsies, J. Vieira for the GFP-KSHV virus, and D. Bourboulia, L. Martinez, and N. Presneau for technical help.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal