Abstract
The Children's Cancer Group (CCG) and the Pediatric Oncology Group (POG) joined to form the Children's Oncology Group (COG) in 2000. This merger allowed analysis of clinical, biologic, and early response data predictive of event-free survival (EFS) in acute lymphoblastic leukemia (ALL) to develop a new classification system and treatment algorithm. From 11 779 children (age, 1 to 21.99 years) with newly diagnosed B-precursor ALL consecutively enrolled by the CCG (December 1988 to August 1995, n = 4986) and POG (January 1986 to November 1999, n = 6793), we retrospectively analyzed 6238 patients (CCG, 1182; POG, 5056) with informative cytogenetic data. Four risk groups were defined as very high risk (VHR; 5-year EFS, 45% or below), lower risk (5-year EFS, at least 85%), and standard and high risk (those remaining in the respective National Cancer Institute [NCI] risk groups). VHR criteria included extreme hypodiploidy (fewer than 44 chromosomes), t(9;22) and/or BCR/ABL, and induction failure. Lower-risk patients were NCI standard risk with either t(12;21) (TEL/AML1) or simultaneous trisomies of chromosomes 4, 10, and 17. Even with treatment differences, there was high concordance between the CCG and POG analyses. The COG risk classification scheme is being used for division of B-precursor ALL into lower- (27%), standard- (32%), high- (37%), and very-high- (4%) risk groups based on age, white blood cell (WBC) count, cytogenetics, day-14 marrow response, and end induction minimal residual disease (MRD) by flow cytometry in COG trials.
Introduction
The treatment of childhood acute lymphoblastic leukemia (ALL) has advanced significantly over the past 3 decades, with overall survival rates progressing from 20% to 75%. This improvement can be attributed, in part, to intensification of therapy using agents previously shown to be effective in the treatment of ALL. While this approach has improved overall event-free survival (EFS), it is clear that a number of patients might have been cured with less aggressive therapy. However, patients identified at diagnosis as having better risk features still account for most relapses. In an effort to appropriately balance the risks and benefits of therapy, “risk-adapted therapy” has been adopted. EFS is predicted based on clinical and biologic variables, and treatment intensity is then modified according to expected EFS to maximize cure while minimizing toxicity.1-3
The Pediatric Oncology Group (POG) and Children's Cancer Group (CCG) adopted a common set of risk criteria in 1993 at an international conference supported by the National Cancer Institute (NCI).4 The NCI criteria were based on factors that had international acceptance and reproducibility: age, initial white blood cell (WBC) count, and the presence of extramedullary disease at diagnosis. To further refine therapy, both POG and CCG have also used additional risk factors that have been shown to have an impact on patient outcomes (eg, ploidy, blast karyotype, and early morphologic response). Recently, the POG and CCG merged to form the Children's Oncology Group (COG). This merger provided an opportunity to reassess individual approaches and develop a consensus classification strategy for treatment assignment. A COG ALL risk classification subcommittee developed a classification system that was (1) optimal for patient care, (2) amenable to asking biologic and therapeutic questions, and (3) functional so that risk-directed therapy could be offered to all eligible patients regardless of geographic location. This analysis included the clinical and biologic variables used in each legacy group, such as age, WBC count, sex, extramedullary disease, blast cytogenetics and ploidy, and early response to therapy. The resulting classification system incorporated the strongest prognostic indicators predictive of outcome in both groups, despite marked differences in treatment strategies, and delineated 4 risk groups with different outcomes. This paper presents the results of these analyses and the basis of the current classification system for COG ALL trials.
Patients, materials, and methods
Criteria for selection of risk factors
Variables were selected for inclusion in the final COG risk algorithm if both POG and CCG data revealed a strong correlation between the variable and outcome. A prognostic factor could also be selected for inclusion if there was a strong correlation within one of the 2 cooperative groups, with limited correlation shown by the other group, or through published literature. A second set of variables were excluded from the current COG ALL risk algorithm but were identified for further study (and validation) if there was a strong prognostic effect within one of the 2 groups with no published support or confirmation by the other group. Because data regarding the influence of minimal residual disease (MRD) on outcome were not available from the CCG or POG trials, we relied on published data to establish a level of MRD that would impact classification and protocol assignment.
Patients and treatment
The patients studied were children (age, 12 months to 21.99 years) with newly diagnosed B-precursor ALL consecutively enrolled on CCG (December 1988 to August 1995) and POG (January 1986 to November 1999) protocols. Eligibility for analysis in the current study for CCG patients included enrollment on CCG 1881, 1882, 1891, 1901, and 1922 for patients with B-lineage ALL or no assigned immunophenotype and with informative cytogenetics as defined below, under “Cytogenetic analysis.” POG patients included in these analyses were those enrolled on the ALinC 14 (POG 8602), ALinC 15 (POG 9005, 9006), and ALinC 16 (POG 9201, 9405, 9406, 9605) protocols for B-precursor ALL. Only patients with designated B-precursor immunophenotyping and with informative leukemic cell cytogenetic results were used in the analyses. Analyses for the presence of the TEL/AML1 translocation or trisomies of chromosomes 4, 10, and 17 and impact of combined day-7 and -14 marrow response and outcome were performed on a later data set from CCG protocols 1952 for NCI standard risk (1996 to 2000) and 1961 for NCI high risk (1996 to 2002), because these data were not available in the original data set. The details of each protocol can be obtained from published reports.5-17 In general, CCG studies used reinduction/reconsolidation pulses modified from the Berlin-Frankfurt-Munster (BFM) clinical trials consortium with further modifications in the augmented regimen,13 while POG protocols employed antimetabolite-based pulses emphasizing the use of “intermediate-dose” (1 g/m2 given intravenously over 24 hours) methotrexate with leucovorin rescue. All protocols were approved by the NCI and by the institutional review board (IRB) of each participating institution. Informed consent was obtained from the patients and/or families prior to enrollment.
Diagnosis
CCG.
The diagnosis of ALL was based on morphologic, cytochemical, and immunologic features of the cells, including lymphoblast morphology on Wright-Giemsa–stained bone marrow smears, positive nuclear staining for terminal deoxynucleotide transferase (TdT), negative staining for myeloperoxidase, and cell-surface expression of 2 or more B-cell–precursor lymphoid differentiation antigens. Immunophenotyping was performed centrally in the CCG ALL Biology Reference Laboratory by indirect immunofluorescence and flow cytometry. In some early trials no centrally performed immunophenotyping information was available for some patients. Patients were classified as B lineage if at least 30% of their leukemic cells were positive for CD19 or CD24 and if less than 30% of their leukemic cells were positive for one or more of the T-cell–associated antigens for CD2, CD3, CD5, or CD7.
POG.
Diagnostic criteria included cytochemical stains and morphology consistent with ALL and reference laboratory immunophenotyping consistent with B-precursor ALL as defined in the previous section for CCG patients.
Clinical and laboratory variables used to risk-stratify patients in legacy studies
Both CCG and POG assigned a preliminary risk group for induction therapy according to the NCI-Rome risk group definitions4 of standard risk (age less than 10 years and WBC count less than 50 × 109/L [50 000/μL]) and high risk (one or more of the following: age at least 10 years and WBC count at least 50 × 109/L [50 000/μL]). Infants less than 1 year of age were not included in the current analyses. Additional variables assessed include sex, hepatosplenomegaly, lymphadenopathy, and presence or absence of extramedullary disease at diagnosis. CCG measured early response by evaluating bone marrow morphology on day 7 and/or day 14 of induction therapy. Conventional morphologic criteria were used to assess blast content in these hypocellular marrows: M1, less than 5% blasts; M2, 5% to 25% blasts; and M3, more than 25% blasts. Central nervous system (CNS)–3 disease was defined as at least 5 nucleated cells per microliter with ALL blasts detected by cytospin. CNS-2 was defined as the presence of blasts by cytospin in cerebrospinal fluid (CSF) with fewer than 5 nucleated cells per microliter and CNS-1 as any number of nucleated cells per microliter with no ALL blasts detected by cytospin. Patients enrolled on the legacy POG ALinC 15 study were further risk classified at the end of induction based on DNA index and the presence of the t(9;22) or t(1;19). On the POG legacy study ALinC 16, modification of the initial NCI risk grouping was defined at end of induction as follows: Trisomies 4 and 10 (or DNA index above 1.16 if cytogenetic studies were not informative) promoted NCI standard risk to good risk and NCI high risk to standard risk. Any patient with CNS-3 or testicular disease or t(1;19), t(4;11), or t(9;22) was assigned to the poor-risk group.
Cytogenetic analysis
Analysis of bone marrow or unstimulated peripheral blood specimens obtained prior to initiation of remission induction therapy was evaluated according to standard protocols by CCG institutional and POG reference laboratories. Criteria for clonality were based on guidelines as defined by the International System for Cytogenetic Nomenclature (ie, 2 or more metaphase spreads with identical structural or additional chromosomes or 3 or more metaphases with identical chromosome loss).18 Members of the respective CCG and POG cytogenetics committees reviewed the cytogenetic reports, together with representational karyotypes, for each abnormal clone. Cytogenetic analyses were considered informative if, after review, an abnormal clone was identified or analysis of bone marrow karyotype revealed a minimum of 20 normal cells.
CCG and POG patient populations
From a total of 4986 eligible, noninfant patients with ALL treated on CCG protocols open from 1988 to 1995, 3056 (61.3%) had evaluable immunophenotyping data, and 1182 (38.7%) of those 3056 patients had evaluable cytogenetics (Figure 1). There are no significant differences in outcome between the total patient population of 4986 patients (5-year EFS, 76.2%), the subset of 3056 with evaluable immunophenotype data (5-year EFS, 75.9%), and the subgroup of 1182 with both evaluable immunophenotyping and cytogenetic data used for detailed analyses in this report (76.0%) (Table 1). Of a total of 6793 eligible patients (5-year EFS, 73.6%) enrolled on POG protocols from 1986 to 1999, 6780 (99.8%) had evaluable immunophenotype data (5-year EFS, 73.6%), and 5056 (74.6%) of these (5-year EFS, 73.0%) had evaluable cytogenetic data and were used for detailed analyses in this report. Eleven of the 5056 POG patients (3516 of whom were NCI standard risk; 1529 were NCI high risk) could not be classified into an NCI risk group due to missing WBC count at diagnosis and were not included in any analyses by NCI risk group, although they were included in the overall analyses. There appeared to be no selection bias with similar outcomes in the total groups and the analyzed subgroups.
ALL risk assignment algorithm. Algorithm is for analysis of POG and CCG data. The figure shows the number of evaluable patients in each of the analyses performed. Because the POG had a central immunophenotyping and molecular laboratory system and the CCG relied on peripheral laboratory results, the attrition of evaluable patients in the POG analysis was much lower.
ALL risk assignment algorithm. Algorithm is for analysis of POG and CCG data. The figure shows the number of evaluable patients in each of the analyses performed. Because the POG had a central immunophenotyping and molecular laboratory system and the CCG relied on peripheral laboratory results, the attrition of evaluable patients in the POG analysis was much lower.
Overall results (POG 1986 to 1999 and CCG 1988 to 1995)
| Group and NCI risk group . | No. patients . | 5-EFS (SE) . | 8-y EFS (SE) . | |||
|---|---|---|---|---|---|---|
| POG . | CCG* . | POG . | CCG . | POG . | CCG . | |
| All eligible† | ||||||
| Any | 6793§ | 3056 | 73.6 (0.7) | 75.9 (0.8) | 70.9 (0.9) | 72.7 (1.1) |
| Standard | 4740 | 2019 | 79.1 (0.7) | 80.2 (0.9) | 77.0 (1.0) | 78.3 (1.3) |
| High | 2040 | 1037 | 60.9 (1.4) | 67.5 (1.5) | 56.5 (1.9) | 64.9 (2.2) |
| All eligible with evaluable cytogenetics‡ | ||||||
| Any | 5056‖ | 1182 | 73.0 (0.8) | 76.0 (1.3) | 70.3 (1.1) | 74.2 (1.6) |
| Standard | 3516 | 776 | 78.7 (0.8) | 79.6 (1.5) | 76.6 (1.2) | 78.2 (2.1) |
| High | 1529 | 406 | 59.9 (1.6) | 69.1 (2.4) | 55.5 (2.3) | 66.5 (3.5) |
| Group and NCI risk group . | No. patients . | 5-EFS (SE) . | 8-y EFS (SE) . | |||
|---|---|---|---|---|---|---|
| POG . | CCG* . | POG . | CCG . | POG . | CCG . | |
| All eligible† | ||||||
| Any | 6793§ | 3056 | 73.6 (0.7) | 75.9 (0.8) | 70.9 (0.9) | 72.7 (1.1) |
| Standard | 4740 | 2019 | 79.1 (0.7) | 80.2 (0.9) | 77.0 (1.0) | 78.3 (1.3) |
| High | 2040 | 1037 | 60.9 (1.4) | 67.5 (1.5) | 56.5 (1.9) | 64.9 (2.2) |
| All eligible with evaluable cytogenetics‡ | ||||||
| Any | 5056‖ | 1182 | 73.0 (0.8) | 76.0 (1.3) | 70.3 (1.1) | 74.2 (1.6) |
| Standard | 3516 | 776 | 78.7 (0.8) | 79.6 (1.5) | 76.6 (1.2) | 78.2 (2.1) |
| High | 1529 | 406 | 59.9 (1.6) | 69.1 (2.4) | 55.5 (2.3) | 66.5 (3.5) |
The analyses of the CCG data do not include the patients from CCG 1952 and 1961, because they were included only in a limited analysis for TEL/AML1.
Includes all patients that had evaluable immunophenotyping and both those with and without evaluable cytogenetics.
Includes all patients who were eligible and had cytogenetic data.
Thirteen POG patients could not be classified into an NCI risk group due to missing WBC count at diagnosis and hence were only included in the overall (“Any”) analyses.
Eleven of the 5056 POG patients with evaluable cytogenetics could not be classified into an NCI risk group due to missing WBC count at diagnosis and hence were included only in the overall (“Any”) analyses.
Statistical methods
The primary outcome considered was event-free survival (EFS) calculated as the time from entry on a therapeutic trial to first event or date of last follow-up, where an event was defined as induction failure, relapse at any site, secondary malignancy, or death. Patients who did not fail were censored as of the date of last contact. Event-free survival curves were constructed using the Kaplan-Meier life table method. Standard deviations of the Kaplan-Meier estimates were based on the Peto variance estimates for the life table curves.19 The log-rank test was used for comparison of survival curves between groups.
Results
Identification of a very-high-risk group
To identify a very-high-risk subgroup for alternative therapies such as blood and marrow transplantation (BMT),20,21 we evaluated the entire cohort to identify factors associated with a projected 5-year EFS of 45% or below on previous POG and CCG protocols. Data on the occurrence of the BCR/ABL translocation (by karyotype, reverse transcriptase–polymerase chain reaction [RT-PCR], or fluorescence in situ hybridization [FISH]) are summarized in Table 2. Children with BCR/ABL or t(9;22) treated on POG or CCG protocols had overall 5-year EFS rates of 27.4% and 33.3%, respectively. Older age and higher WBC count (NCI high risk) have been associated with poorer prognosis as compared with NCI standard risk in Ph-positive ALL.22 This was confirmed in POG Ph-positive patients with 18.2% 5-year EFS for high-risk compared with 52.4% for standard-risk patients (Table 2).
Very-high-risk ALL criteria (evaluable cytogenetics; POG 1986 to 1999 and CCG 1988 to 1995)
| Group and NCI risk group . | No. patients . | 5-y EFS (SE) . | 8-y EFS (SE) . | |||
|---|---|---|---|---|---|---|
| POG . | CCG . | POG . | CCG . | POG . | CCG . | |
| All eligible* | 5056 | 1182 | 73.0 (0.8) | 76.0 (1.3) | 70.3 (1.1) | 74.2 (1.6) |
| Ph-positive | ||||||
| Any | 132‡ | 30 | 27.4 (4.4) | 33.3 (9.1) | 23.4 (5.5) | 28.6 (24.1) |
| Standard | 36 | 7 | 52.4 (8.8) | 28.6 (17.1) | 52.4 (11.4) | ND |
| High | 96 | 23 | 18.2 (5.0) | 34.8 (10.6) | 10.4 (4.9) | 29.0 (24.4) |
| Fewer than 45 chromosomes | ||||||
| Any | 58 | 14 | 46.5 (7.2) | 35.7 (12.8) | 46.5 (9.8) | 23.8 (14.7) |
| Standard | 27 | 6 | 44.4 (9.6) | 33.3 (19.3) | 44.4 (12.5) | 33.3 (27.2) |
| High | 31 | 8 | 48.2 (11.0) | 37.5 (17.1) | 48.2 (15.5) | 18.8 (16.9) |
| Other† | ||||||
| Any | 4866§ | 1138 | 74.6 (0.8) | 77.7 (1.3) | 71.9 (1.1) | 76.0 (1.8) |
| Standard | 3453 | 763 | 79.3 (0.8) | 80.4 (1.5) | 77.1 (1.2) | 79.0 (2.1) |
| High | 1402 | 375 | 63.1 (1.6) | 71.9 (2.4) | 58.8 (2.3) | 69.9 (3.5) |
| Group and NCI risk group . | No. patients . | 5-y EFS (SE) . | 8-y EFS (SE) . | |||
|---|---|---|---|---|---|---|
| POG . | CCG . | POG . | CCG . | POG . | CCG . | |
| All eligible* | 5056 | 1182 | 73.0 (0.8) | 76.0 (1.3) | 70.3 (1.1) | 74.2 (1.6) |
| Ph-positive | ||||||
| Any | 132‡ | 30 | 27.4 (4.4) | 33.3 (9.1) | 23.4 (5.5) | 28.6 (24.1) |
| Standard | 36 | 7 | 52.4 (8.8) | 28.6 (17.1) | 52.4 (11.4) | ND |
| High | 96 | 23 | 18.2 (5.0) | 34.8 (10.6) | 10.4 (4.9) | 29.0 (24.4) |
| Fewer than 45 chromosomes | ||||||
| Any | 58 | 14 | 46.5 (7.2) | 35.7 (12.8) | 46.5 (9.8) | 23.8 (14.7) |
| Standard | 27 | 6 | 44.4 (9.6) | 33.3 (19.3) | 44.4 (12.5) | 33.3 (27.2) |
| High | 31 | 8 | 48.2 (11.0) | 37.5 (17.1) | 48.2 (15.5) | 18.8 (16.9) |
| Other† | ||||||
| Any | 4866§ | 1138 | 74.6 (0.8) | 77.7 (1.3) | 71.9 (1.1) | 76.0 (1.8) |
| Standard | 3453 | 763 | 79.3 (0.8) | 80.4 (1.5) | 77.1 (1.2) | 79.0 (2.1) |
| High | 1402 | 375 | 63.1 (1.6) | 71.9 (2.4) | 58.8 (2.3) | 69.9 (3.5) |
ND indicates not done.
Includes all patients with evaluable immunophenotyping and evaluable cytogenetics.
Those with evaluable cytogenetics that do not have traits listed above it for those risk groups.
Six Ph-positive patients had fewer than 45 chromosomes and are excluded from the “fewer than 45 chromosomes” group.
Eleven of the 4866 POG patients could not be classified into an NCI risk group due to missing WBC count at diagnosis and hence were included only in the overall (“Any”) analyses.
Patients with hypodiploidy (fewer than 45 chromosomes) had a 5-year EFS of 46.5% for POG and 35.7% for CCG (Table 2). There was no significant effect of either WBC count or age in the marked hypodiploid patients. In our initial analysis, patients with hypodiploidy defined as fewer than 45 chromosomes were classified as very high risk; however, as described under “Discussion,” more recent data from a large international collaboration demonstrate that leukemias with 44 chromosomes are associated with an intermediate prognosis and, those with fewer than 44, a poor prognosis.23
A third variable associated with very high risk was failure to achieve remission at the end of induction therapy. Induction failure was defined as either an M3 (more than 25% blasts) response in the BM at the end of induction (CCG and POG) or an M2 (5% to 25% blasts) marrow at the end of induction followed by at least 5% blasts (M2/M3) at the end of extended induction (POG) or consolidation therapy (CCG). The relative frequencies of an M3 bone marrow at end of induction were 1% and 0.1% among high-risk and standard-risk patients, respectively, on CCG studies. Detailed analysis of this data set was limited because patients were removed from protocol therapy once they had failed induction, specific treatment data were not collected, and we were unable to make exclusions based on immunophenotyping and cytogenetics. There were 41 patients with an M3 marrow at the end of induction with 29 (70%) achieving remission after additional therapy. Of this group, 18 received a BMT (type indeterminate) and 11 received chemotherapy. The 5-year EFS for this CCG patient group was 44% overall with the BMT group performing better than the chemotherapy group (52.6% versus 33.3%, respectively). Of those failing to achieve a remission after additional therapy, all died within 9 months. On the POG studies, the overall induction failure rate was 1.3%. No data on subsequent therapy or relapses were captured on POG studies for induction failures, making analysis of later outcome impossible.
Analysis of other potential very-high-risk features
Other factors have been associated with a high risk of relapse in prior studies, including MLL rearrangements,24,25 monosomy 7,26 a balanced t(1;19) translocation,27 WBC count above 200 × 109/L (200 000/μL), and age above 15 years.28 POG and CCG molecular data documenting MLL rearrangements were limited. The incidence of t(4;11) among B-precursor ALL patients older than 12 months of age at diagnosis was about 1%. Patients with t(4;11) had a poor prognosis (Table 3). but did not meet the criteria for the very-high-risk group (5-year EFS, 45% or below). Evaluation of the impact of slow early response (SER) in patients treated on CCG 1961 (1996 to 2002) demonstrated that patients with t(4;11) and an M3 marrow at day 7 had an extremely poor prognosis, with one of 9 remaining in first remission. Patients with other 11q23 abnormalities did not have a poor prognosis.
Evaluation of 11q23 and t(1;19) as an ALL prognostic factor in children more than 12 months of age (evaluable cytogenetics; excludes those with fewer than 45 chromosomes and Ph-positive ALL)
| Group and NCI risk group . | No. patients . | 5-y EFS (SE) . | 8-y EFS (SE) . | |||
|---|---|---|---|---|---|---|
| POG . | CCG . | POG . | CCG . | POG . | CCG . | |
| t(4;11) | ||||||
| Any* | 44 | 13 | 49.9 (11.2) | 53.8 (14.9) | 49.9 (14.4) | 53.8 (21.1) |
| Standard | 6 | 4 | 50.0 (25.0) | 50.0 (35.4) | 50.0 (35.4) | ND |
| High | 38 | 9 | 49.9 (12.5) | 55.6 (16.6) | 49.9 (15.8) | 55.6 (21.4) |
| Other 11q23 | ||||||
| Any | 112 | 18 | 75.8 (4.9) | 71.8 (11.0) | 72.8 (7.4) | 71.8 (15.6) |
| Standard | 71 | 9 | 80.2 (5.6) | 66.7 (17.2) | 77.7 (8.9) | 66.7 (22.2) |
| High | 41 | 9 | 68.1 (9.3) | 77.8 (13.9) | 64.1 (12.8) | 51.9 (25.4) |
| Other† | ||||||
| Any | 4710 | 1107 | 74.8 (0.8) | 78.0 (1.3) | 72.1 (1.1) | 76.4 (1.8) |
| Standard | 3376 | 750 | 79.3 (0.8) | 80.8 (1.5) | 77.2 (1.2) | 79.3 (2.1) |
| High | 1323 | 357 | 63.3 (1.7) | 72.2 (2.5) | 58.9 (2.4) | 70.0 (3.6) |
| Balanced t(1;19) | ||||||
| Any‡ | 69 | 18 | 68.2 (7.3) | 50.0 (12.5) | 68.2 (10.3) | 50.0 (17.7) |
| Standard | 38 | 9 | 70.1 (9.3) | 44.4 (16.6) | 70.1 (12.1) | 44.4 (23.4) |
| High | 31 | 9 | 65.8 (11.6) | 55.6 (18.5) | 65.8 (19.2) | 55.6 (26.2) |
| Unbalanced t(1;19) | ||||||
| Any | 194 | 33 | 74.1 (4.1) | 81.8 (6.7) | 68.5 (6.1) | 81.8 (7.8) |
| Standard | 94 | 12 | 80.1 (5.1) | 83.3 (10.8) | 75.7 (7.3) | 83.3 (12.0) |
| High | 100 | 21 | 68.4 (6.6) | 81.0 (8.6) | 61.4 (10.2) | 81.0 (10.2) |
| Other† | ||||||
| Any | 4603§ | 1087 | 74.7 (0.8) | 78.0 (1.3) | 72.0 (1.1) | 76.3 (1.9) |
| Standard | 3321 | 742 | 79.3 (0.9) | 80.8 (1.5) | 77.2 (1.2) | 79.4 (2.1) |
| High | 1271 | 345 | 62.6 (1.7) | 71.8 (2.6) | 58.4 (2.4) | 69.5 (3.8) |
| Group and NCI risk group . | No. patients . | 5-y EFS (SE) . | 8-y EFS (SE) . | |||
|---|---|---|---|---|---|---|
| POG . | CCG . | POG . | CCG . | POG . | CCG . | |
| t(4;11) | ||||||
| Any* | 44 | 13 | 49.9 (11.2) | 53.8 (14.9) | 49.9 (14.4) | 53.8 (21.1) |
| Standard | 6 | 4 | 50.0 (25.0) | 50.0 (35.4) | 50.0 (35.4) | ND |
| High | 38 | 9 | 49.9 (12.5) | 55.6 (16.6) | 49.9 (15.8) | 55.6 (21.4) |
| Other 11q23 | ||||||
| Any | 112 | 18 | 75.8 (4.9) | 71.8 (11.0) | 72.8 (7.4) | 71.8 (15.6) |
| Standard | 71 | 9 | 80.2 (5.6) | 66.7 (17.2) | 77.7 (8.9) | 66.7 (22.2) |
| High | 41 | 9 | 68.1 (9.3) | 77.8 (13.9) | 64.1 (12.8) | 51.9 (25.4) |
| Other† | ||||||
| Any | 4710 | 1107 | 74.8 (0.8) | 78.0 (1.3) | 72.1 (1.1) | 76.4 (1.8) |
| Standard | 3376 | 750 | 79.3 (0.8) | 80.8 (1.5) | 77.2 (1.2) | 79.3 (2.1) |
| High | 1323 | 357 | 63.3 (1.7) | 72.2 (2.5) | 58.9 (2.4) | 70.0 (3.6) |
| Balanced t(1;19) | ||||||
| Any‡ | 69 | 18 | 68.2 (7.3) | 50.0 (12.5) | 68.2 (10.3) | 50.0 (17.7) |
| Standard | 38 | 9 | 70.1 (9.3) | 44.4 (16.6) | 70.1 (12.1) | 44.4 (23.4) |
| High | 31 | 9 | 65.8 (11.6) | 55.6 (18.5) | 65.8 (19.2) | 55.6 (26.2) |
| Unbalanced t(1;19) | ||||||
| Any | 194 | 33 | 74.1 (4.1) | 81.8 (6.7) | 68.5 (6.1) | 81.8 (7.8) |
| Standard | 94 | 12 | 80.1 (5.1) | 83.3 (10.8) | 75.7 (7.3) | 83.3 (12.0) |
| High | 100 | 21 | 68.4 (6.6) | 81.0 (8.6) | 61.4 (10.2) | 81.0 (10.2) |
| Other† | ||||||
| Any | 4603§ | 1087 | 74.7 (0.8) | 78.0 (1.3) | 72.0 (1.1) | 76.3 (1.9) |
| Standard | 3321 | 742 | 79.3 (0.9) | 80.8 (1.5) | 77.2 (1.2) | 79.4 (2.1) |
| High | 1271 | 345 | 62.6 (1.7) | 71.8 (2.6) | 58.4 (2.4) | 69.5 (3.8) |
ND indicates not done.
Data are from POG 1986 to 1999 and CCG 1988 to 1995.
t(4;11) versus 11q23 versus other; any (POG, P < .001; CCG, P = .004); standard risk (POG, P = .039; CCG, P = .002); high risk (POG, P = .009; CCG, P = .91).
Those with evaluable cytogenetics that do not have traits listed above it for those risk groups.
Balanced t(1;19) versus unbalanced t(1;19) versus other: any (POG, P = .235; CCG, P = .02); standard risk (POG, P = .294; CCG, P = .05); high risk (POG, P = .677; CCG, P = .41).
Eleven of the POG patients could not be classified into an NCI risk group due to missing WBC count at diagnosis and hence were included only in the overall (“Any”) analyses.
Previously, the t(1;19)(q23;p13) had been reported to be an adverse prognostic factor,29 and the CCG has reported that a balanced t(1;19) translocation was associated with significantly worse prognosis than the unbalanced variant.27 In our current analysis of outcome on CCG trials, patients with a balanced t(1;19) had a 5-year EFS of 50% versus 81.8% for patients with an unbalanced t(1;19) (Table 3), but it was based on only 51 t(1;19) patients and 18 with the balanced t(1;19). Analysis of 273 patients with a t(1;19) treated on POG trials showed no significant outcome difference between those with the balanced (68.2%) or unbalanced (74.1%) variant. Thus, t(1;19) was not included in the risk stratification algorithm.
Other variables, including race, sex, age above 15 years, and WBC count above 200 × 109/L (200 000/μL) (Table 4), were evaluated to determine whether these features defined a very-high-risk group. These comparisons included only NCI high-risk patients. When each factor was considered individually, POG patients with a WBC count above 200 × 109/L (200 000/μL) had a 5-year EFS of 38.6%, but this was not confirmed by CCG data.
Evaluation of clinical factors as potential ALL high-risk prognostic factors (all patients with evaluable cytogenetics; POG 1986 to 1999 and CCG 1988 to 1995)
| Group . | NCI risk group . | No. patients . | 5-y EFS (SE) . | 8-y EFS (SE) . | |||
|---|---|---|---|---|---|---|---|
| POG . | CCG . | POG . | CCG . | POG . | CCG . | ||
| Sex | |||||||
| Male | High | 849 | 208 | 54.1 (2.2) | 68.5 (3.4) | 49.3 (3.0) | 65.7 (5.1) |
| Female | High | 680 | 198 | 67.1 (2.3) | 69.8 (3.4) | 63.2 (3.3) | 67.4 (4.9) |
| Race | |||||||
| African American | High | 165 | 36 | 53.1 (5.0) | 56.9 (8.6) | 51.5 (6.8) | 53.4 (18.2) |
| Hispanic | High | 225 | 29 | 54.3 (4.8) | 67.7 (9.3) | 52.2 (9.3) | 67.7 (17.2) |
| Other | High | 1139 | 340 | 61.9 (1.8) | 70.7 (2.8) | 56.8 (2.5) | 68.0 (3.7) |
| Age | |||||||
| 15.00 y or less | High | 1360 | 343 | 60.9 (1.7) | 71.0 (2.6) | 56.7 (2.3) | 68.3 (3.7) |
| More than 15.00 y | High | 169 | 63 | 51.1 (5.4) | 59.0 (6.6) | 44.1 (8.2) | 57.0 (10.0) |
| WBC count | |||||||
| 200 × 109/L | High | 1421 | 367 | 61.6 (1.6) | 70.2 (2.5) | 56.8 (2.3) | 67.3 (3.7) |
| More than 200 × 109/L | High | 108 | 39 | 38.6 (6.0) | 58.5 (8.2) | 38.6 (8.4) | 58.5 (11.9) |
| CNS status at diagnosis* | |||||||
| CNS-1 | Any | 4382‡ | 1037 | 75.6 (7.9) | 78.4 (1.3) | 72.9 (1.1) | 76.6 (1.9) |
| CNS-2 | Any | 326 | 70 | 65.0 (3.4) | 67.0 (6.1) | 62.6 (5.3) | 67.0 (7.4) |
| CNS-3 | Any | 105‡ | 21 | 64.6 (5.4) | 76.2 (9.3) | 62.6 (7.0) | 76.8 (11.2) |
| CNS-1 | Standard | 3193 | 712 | 79.9 (0.9) | 81.2 (1.5) | 77.7 (1.2) | 79.6 (2.1) |
| CNS-2 | Standard | 176 | 38 | 70.1 (4.5) | 68.2 (8.0) | 69.1 (6.7) | 68.2 (10.3) |
| CNS-3 | Standard | 48 | 8 | 71.8 (7.6) | 75.0 (15.3) | 71.8 (9.5) | 75.0 (16.8) |
| CNS-1 | High | 1181 | 325 | 64.0 (1.8) | 72.2 (2.6) | 59.6 (2.5) | 69.8 (3.9) |
| CNS-2 | High | 150 | 32 | 59.0 (5.3) | 65.6 (9.3) | 54.8 (8.4) | 65.6 (10.7) |
| CNS-3 | High | 55 | 13 | 58.7 (7.5) | 76.9 (11.7) | 55.0 (9.9) | 76.9 (15.1) |
| Testicular status at diagnosis* | |||||||
| No disease | Any | 2667 | 618 | 71.4 (1.1) | 76.5 (1.8) | 68.4 (1.5) | 74.9 (2.4) |
| Disease† | Any | 10 | 8 | 90.0 (12.7) | 62.5 (17.1) | 90.0 (16.4) | 62.5 (17.1) |
| Group . | NCI risk group . | No. patients . | 5-y EFS (SE) . | 8-y EFS (SE) . | |||
|---|---|---|---|---|---|---|---|
| POG . | CCG . | POG . | CCG . | POG . | CCG . | ||
| Sex | |||||||
| Male | High | 849 | 208 | 54.1 (2.2) | 68.5 (3.4) | 49.3 (3.0) | 65.7 (5.1) |
| Female | High | 680 | 198 | 67.1 (2.3) | 69.8 (3.4) | 63.2 (3.3) | 67.4 (4.9) |
| Race | |||||||
| African American | High | 165 | 36 | 53.1 (5.0) | 56.9 (8.6) | 51.5 (6.8) | 53.4 (18.2) |
| Hispanic | High | 225 | 29 | 54.3 (4.8) | 67.7 (9.3) | 52.2 (9.3) | 67.7 (17.2) |
| Other | High | 1139 | 340 | 61.9 (1.8) | 70.7 (2.8) | 56.8 (2.5) | 68.0 (3.7) |
| Age | |||||||
| 15.00 y or less | High | 1360 | 343 | 60.9 (1.7) | 71.0 (2.6) | 56.7 (2.3) | 68.3 (3.7) |
| More than 15.00 y | High | 169 | 63 | 51.1 (5.4) | 59.0 (6.6) | 44.1 (8.2) | 57.0 (10.0) |
| WBC count | |||||||
| 200 × 109/L | High | 1421 | 367 | 61.6 (1.6) | 70.2 (2.5) | 56.8 (2.3) | 67.3 (3.7) |
| More than 200 × 109/L | High | 108 | 39 | 38.6 (6.0) | 58.5 (8.2) | 38.6 (8.4) | 58.5 (11.9) |
| CNS status at diagnosis* | |||||||
| CNS-1 | Any | 4382‡ | 1037 | 75.6 (7.9) | 78.4 (1.3) | 72.9 (1.1) | 76.6 (1.9) |
| CNS-2 | Any | 326 | 70 | 65.0 (3.4) | 67.0 (6.1) | 62.6 (5.3) | 67.0 (7.4) |
| CNS-3 | Any | 105‡ | 21 | 64.6 (5.4) | 76.2 (9.3) | 62.6 (7.0) | 76.8 (11.2) |
| CNS-1 | Standard | 3193 | 712 | 79.9 (0.9) | 81.2 (1.5) | 77.7 (1.2) | 79.6 (2.1) |
| CNS-2 | Standard | 176 | 38 | 70.1 (4.5) | 68.2 (8.0) | 69.1 (6.7) | 68.2 (10.3) |
| CNS-3 | Standard | 48 | 8 | 71.8 (7.6) | 75.0 (15.3) | 71.8 (9.5) | 75.0 (16.8) |
| CNS-1 | High | 1181 | 325 | 64.0 (1.8) | 72.2 (2.6) | 59.6 (2.5) | 69.8 (3.9) |
| CNS-2 | High | 150 | 32 | 59.0 (5.3) | 65.6 (9.3) | 54.8 (8.4) | 65.6 (10.7) |
| CNS-3 | High | 55 | 13 | 58.7 (7.5) | 76.9 (11.7) | 55.0 (9.9) | 76.9 (15.1) |
| Testicular status at diagnosis* | |||||||
| No disease | Any | 2667 | 618 | 71.4 (1.1) | 76.5 (1.8) | 68.4 (1.5) | 74.9 (2.4) |
| Disease† | Any | 10 | 8 | 90.0 (12.7) | 62.5 (17.1) | 90.0 (16.4) | 62.5 (17.1) |
Excludes those with fewer than 45 chromosomes and Ph-positive ALL.
Six boys with testicular disease were known to be NCI standard risk, and 7 were NCI high risk. Two failed, one per risk group, both in year 1.
A total of 4813 POG patients (excluding those with fewer than 45 chromosomes and Ph-positive ALL) had a known CNS status at diagnosis. Of these, 10 patients (8 CNS-1 and 2 CNS-3) could not be classified into an NCI risk group due to missing WBC count at diagnosis and hence were included only in the overall (“Any”) analyses.
The impact of central nervous system (CNS) disease at the time of diagnosis was analyzed. CNS-3 and CNS-2, NCI standard-risk POG patients, had a 5-year EFS of 71.8% and 70.1%, respectively, compared with 79.9% for CNS-1 (Table 4). NCI high-risk POG patients had a similar trend with a 5-year EFS of 58.7% and 59.0% among CNS-3 and CNS-2 patients, respectively, compared with 64.0% for CNS-1 (Table 4). In CCG studies, only CNS-2 patients had a lower 5-year EFS (67.0%), with EFS in CNS-1 and CNS-3 patients of 78.4% and 76.2%, respectively. Only 18 patients were diagnosed as having testicular disease at diagnosis, precluding meaningful analysis.
Conventional measures of early marrow response to therapy
Rapidity of response was used to identify patients for treatment intensification on CCG trials based on the presence of a slow early response (SER), defined as an M3 bone marrow on day 7 (high risk) or an M2/M3 marrow on day 14 (standard risk) of induction. The CCG trials included in the primary analyses in this report determined either day-7 or day-14 marrow response—not both. We analyzed data from B-precursor ALL patients enrolled in CCG 1952 (standard risk) and CCG 1961 (high risk) in which a day-7 marrow response was determined and had a day-14 marrow response assessed if the day-7 marrow was not M1 (approximately half of patients). Evaluation of NCI high-risk patients treated on CCG 1961 showed that both day-7 SER (M3) and day-14 M2/M3 patients had an inferior outcome, even though patients with a day-7 M3 marrow were nonrandomly assigned to augmented therapy (Table 5). Similarly, early response was a strong predictor of outcome among standard-risk ALL patients treated on CCG 1952 with SER patients, defined as a day-14 M3 marrow, (who were nonrandomly assigned to augmented therapy) having an inferior outcome to those with a rapid early response (RER) (5-year EFS, 66.5% versus 84.4%). Rapid response to induction therapy was also an important predictor of outcome in patients with the presence of favorable features, including TEL/AML1 fusion or trisomies of chromosomes 4, 10, and 17 (data not shown).
Evaluation of rapidity of response as an ALL prognostic factor (evaluable cytogenetics; excludes those with fewer than 45 chromosomes and Ph-positive ALL; CCG 1952 [1996 to 2000] and CCG 1961 [1996 to 2002])
| Group . | NCI risk group . | No patients. . | 5-y EFS (SE) . | 8-y EFS (SE) . |
|---|---|---|---|---|
| Day-7 marrow | ||||
| RER, M1/M2 | Standard | 713 | 84.3 (1.6) | 82.0 (8.0) |
| SER, M3 | Standard | 233 | 75.7 (3.1) | 74.6 (12.5) |
| RER, M1/M2 | High | 704 | 76.4 (2.5) | 74.6 (21.7) |
| SER, M3 | High | 268 | 69.4 (4.8) | 60.7 (38.0) |
| Day-14* marrow | ||||
| RER, M1* | Standard | 835 | 84.4 (1.4) | 82.4 (6.7) |
| SER, M2/M3 | Standard | 107 | 66.6 (5.0) | 65.4 (38.5) |
| RER, M1* | High | 819 | 77.0 (2.4) | 73.4 (21.9) |
| SER, M2/M3 | High | 113 | 59.0 (7.4) | 59.0 (37.8) |
| Group . | NCI risk group . | No patients. . | 5-y EFS (SE) . | 8-y EFS (SE) . |
|---|---|---|---|---|
| Day-7 marrow | ||||
| RER, M1/M2 | Standard | 713 | 84.3 (1.6) | 82.0 (8.0) |
| SER, M3 | Standard | 233 | 75.7 (3.1) | 74.6 (12.5) |
| RER, M1/M2 | High | 704 | 76.4 (2.5) | 74.6 (21.7) |
| SER, M3 | High | 268 | 69.4 (4.8) | 60.7 (38.0) |
| Day-14* marrow | ||||
| RER, M1* | Standard | 835 | 84.4 (1.4) | 82.4 (6.7) |
| SER, M2/M3 | Standard | 107 | 66.6 (5.0) | 65.4 (38.5) |
| RER, M1* | High | 819 | 77.0 (2.4) | 73.4 (21.9) |
| SER, M2/M3 | High | 113 | 59.0 (7.4) | 59.0 (37.8) |
These data were obtained from patients treated on CCG 1961 high-risk (n = 946) and CCG 1952 standard-risk (n = 1874) protocols only because day-7 and -14 marrow response data were not available on earlier studies. These data are not available for POG studies.
Day-14 bone marrows were not performed if the day 7 bone marrow was M1. Day-7 M1 marrows were included in the M1 marrows at day 14.
Identification of a lower-risk group
Patients with a lower risk of relapse were defined as those having a 5-year EFS of at least 85%. Hyperdiploidy is associated with an improved survival although, more recently, specific chromosome trisomies are more important than ploidy (as determined by chromosome number or DNA index).30,31 POG previously reported that the combination of trisomies 4 and 10 was associated with a good prognosis.5 CCG had identified trisomies 10, 17, and 18 as each independently prognostic with the best prognosis associated with both trisomies 10 and 17.30 Recently, evaluation of CCG and POG patients using identical methodology showed that simultaneous trisomy of chromosomes 4, 10, and 17 (“triple trisomies”) was associated with an excellent prognosis.32 Patients in the NCI standard-risk group with triple trisomies had a 5-year EFS of 89.3% in POG and 91.5% in CCG (Table 6).
Evaluation for good-risk ALL prognostic factors in NCI standard risk (evaluable cytogenetics; excludes those with fewer than 45 chromosomes and Ph-positive ALL; POG 1988 to 1999 and CCG 1996 to 2000)
| Group . | No. patients . | 5-Y EFS (SE) . | 8-y EFS (SE) . | |||
|---|---|---|---|---|---|---|
| POG . | CCG . | POG . | CCG . | POG . | CCG . | |
| Trisomies 4, 10, and 17 | 747 | 108 | 89.3 (1.4) | 91.5 (2.8) | 88.3 (2.1) | 91.5 (3.7) |
| Other* | 2706 | 655 | 76.6 (1.0) | 78.6 (1.7) | 74.1 (1.3) | 77.0 (2.3) |
| TEL/AML1† | 115 | 81 | 85.1 (10.4) | 86.2 (4.0) | NA | 86.2 (16.0) |
| Other* | 345 | 359 | 77.7 (6.8) | 80.5 (2.3) | NA | 78.4 (12.2) |
| Group . | No. patients . | 5-Y EFS (SE) . | 8-y EFS (SE) . | |||
|---|---|---|---|---|---|---|
| POG . | CCG . | POG . | CCG . | POG . | CCG . | |
| Trisomies 4, 10, and 17 | 747 | 108 | 89.3 (1.4) | 91.5 (2.8) | 88.3 (2.1) | 91.5 (3.7) |
| Other* | 2706 | 655 | 76.6 (1.0) | 78.6 (1.7) | 74.1 (1.3) | 77.0 (2.3) |
| TEL/AML1† | 115 | 81 | 85.1 (10.4) | 86.2 (4.0) | NA | 86.2 (16.0) |
| Other* | 345 | 359 | 77.7 (6.8) | 80.5 (2.3) | NA | 78.4 (12.2) |
All patients were in the standard risk group.
NA indicates not applicable.
Those with evaluable cytogenetics that do not have traits listed above it for those risk groups (hypodiploid and t(9;22) excluded). Twelve patients with trisomies 4, 10, and 17 had extramedulary disease (EMD) (CNS or testicular), and all but one are in continuous complete remission (CCR).
TEL/AML1 status was determined retrospectively in a subset of the patients on POG studies 9201, 9405, and 9605 (using Southern blot) and the CCG 1952 study.
Previously, the cryptic t(12;21) TEL/AML1 fusion, first reported in 1995,33,34 was associated with a very good outcome. TEL/AML1 fusion was assessed in only a subset of patients treated on more recent trials, reflecting the smaller numbers and limited duration of follow-up compared with the other analyses performed for this paper. The 5-year EFS of NCI standard-risk patients with TEL/AML1 fusion was 85.1% on POG protocols 9201, 9405, and 9605 and 86.2% on CCG protocol 1952 (Table 6).
A risk assignment algorithm for B-precursor ALL
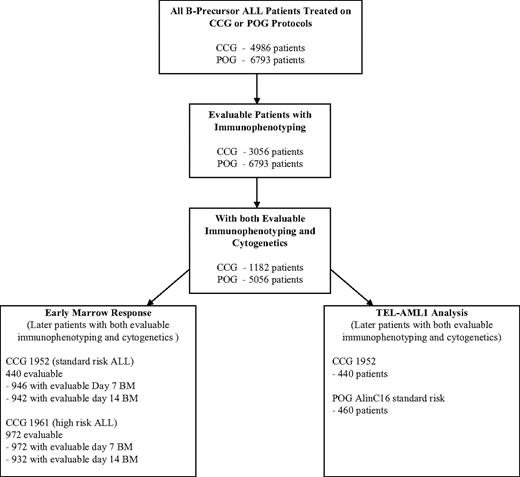
Using the previous analyses and emerging data from other studies, a classification system was developed and implemented as COG AALL03B1 (Classification of Acute Lymphoblastic Leukemia).35 The algorithm uses a number of biologic markers, some of which are determined at each COG center and others at central COG laboratories. The algorithm initially classifies patients and assigns induction chemotherapy as either NCI standard (3-drug induction) or high risk (4-drug induction) based on age and presenting WBC count. At the end of induction therapy, risk stratification is refined based on biologic features of the leukemia (the presence of extramedullary disease, ploidy, triple trisomies, TEL/AML1 or BCR/ABL fusion) and early treatment response (day-15 and -29 morphology determined at local centers and day-29 MRD determined at COG reference laboratories by flow cytometry) to assign patients into lower- (Figure 2A), high- (Figure 2B), standard-, and very-high- (Figure 2C) risk subgroups. The predictive ability of the COG ALL risk classification algorithm is illustrated with the later CCG (CCG 1952, standard risk; 1961, high risk) trials in Figure 3A and POG (POG 9201, lower risk; 9405, standard risk; 9406, high risk; and 9605, standard risk) trials in Figure 3B.
Risk assignment algorithms. Each of the algorithms shows the criteria for placement of patients in (A) standard-risk, (B) high-risk, and (C) very-high-risk assignment. Very-high-risk assignment occurs independently of initial NCI risk assignment (standard or high risk). Patients who have received less than 48 hours of oral or intravenous steroids during the week immediately prior to diagnosis are eligible for classification if the results of a complete blood count (CBC), obtained prior to the initiation of steroid therapy (less than 72 hours prior to steroids), are available and the necessary FISH, cytogenetic, and molecular data are interpretable. The “presteroid” CBC and age of the patient are used to determine NCI-Rome risk classification (standard risk versus high risk [SR versus HR]). If patients have received more than 48 hours of oral or intravenous steroids (and a presteroid CBC is available to assign NCI risk group), they are treated as an SER and nonrandomly assigned to the augmented regimen. In the absence of a presteroid CBC, patients who have received less than 48 hours of steroids are assigned to the HR protocol. These patients are eligible for randomization on the HR protocol. As expected, patients with a slow early response will be assigned to the full augmented BFM treatment arm. In the absence of a presteroid CBC, patients who have received more than 48 hours of steroids are treated as an SER on the HR study and assigned to the full augmented arm. Inhalational steroids are not considered as pretreatment. Both SR and HR patients with identified MLL translocations, CNS-3, or testicular disease, receive augmented SR therapy.
Risk assignment algorithms. Each of the algorithms shows the criteria for placement of patients in (A) standard-risk, (B) high-risk, and (C) very-high-risk assignment. Very-high-risk assignment occurs independently of initial NCI risk assignment (standard or high risk). Patients who have received less than 48 hours of oral or intravenous steroids during the week immediately prior to diagnosis are eligible for classification if the results of a complete blood count (CBC), obtained prior to the initiation of steroid therapy (less than 72 hours prior to steroids), are available and the necessary FISH, cytogenetic, and molecular data are interpretable. The “presteroid” CBC and age of the patient are used to determine NCI-Rome risk classification (standard risk versus high risk [SR versus HR]). If patients have received more than 48 hours of oral or intravenous steroids (and a presteroid CBC is available to assign NCI risk group), they are treated as an SER and nonrandomly assigned to the augmented regimen. In the absence of a presteroid CBC, patients who have received less than 48 hours of steroids are assigned to the HR protocol. These patients are eligible for randomization on the HR protocol. As expected, patients with a slow early response will be assigned to the full augmented BFM treatment arm. In the absence of a presteroid CBC, patients who have received more than 48 hours of steroids are treated as an SER on the HR study and assigned to the full augmented arm. Inhalational steroids are not considered as pretreatment. Both SR and HR patients with identified MLL translocations, CNS-3, or testicular disease, receive augmented SR therapy.
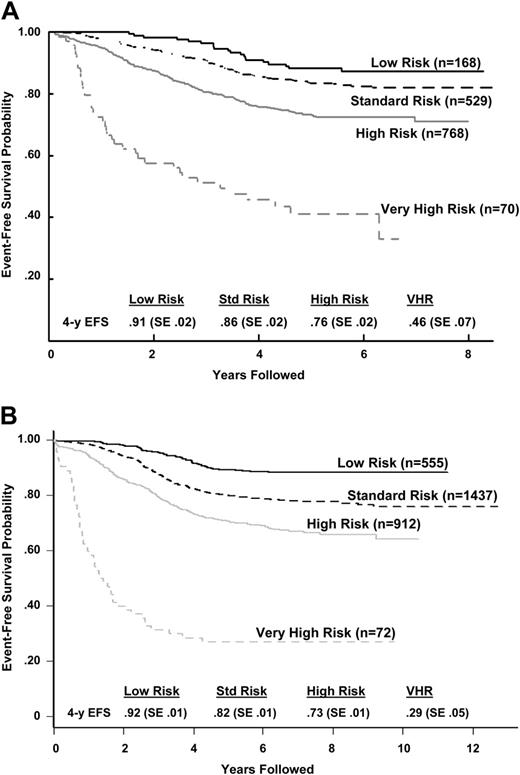
Outcome after classification by the COG risk classification algorithm. (A) CCG 1950s/1960s B-precursor ALL event-free survival outcome by COG risk classification algorithm. The P value for the log-rank test was less than .001. Hazard ratios (with low risk being the baseline) were 1.53 for standard risk, 2.73 for high risk, and 8.82 for very high risk. (B) POG ALinC 16 B-precursor ALL event-free survival outcome by COG risk classification algorithm (does not include rapidity of response because those data were not collected for these studies). The P value for the log-rank test was less than .001. Hazard ratios (with the low-risk group being the baseline) were 2.05 for standard risk, 3.34 for high risk, and 15.02 for very high risk.
Outcome after classification by the COG risk classification algorithm. (A) CCG 1950s/1960s B-precursor ALL event-free survival outcome by COG risk classification algorithm. The P value for the log-rank test was less than .001. Hazard ratios (with low risk being the baseline) were 1.53 for standard risk, 2.73 for high risk, and 8.82 for very high risk. (B) POG ALinC 16 B-precursor ALL event-free survival outcome by COG risk classification algorithm (does not include rapidity of response because those data were not collected for these studies). The P value for the log-rank test was less than .001. Hazard ratios (with the low-risk group being the baseline) were 2.05 for standard risk, 3.34 for high risk, and 15.02 for very high risk.
Discussion
This study, designed to identify prognostic factors from the most recent era of completed CCG/POG clinical trials, includes more than 6000 children with newly diagnosed B-precursor ALL. This represents the largest reported ALL prognostic factor analysis in children to date allowing for the validation of a number of markers previously identified in smaller analyses. This large number of patients has allowed for the limited analyses to identify smaller risk groups and is unique in that it was performed on patients receiving different approaches to ALL therapy. Treatment on CCG studies was based on modifications of the BFM treatment regimen with reinduction/reconsolidation courses (“delayed intensification”), with further intensification in patient subsets (“augmented” therapy).13 The POG protocols were predominantly antimetabolite based, with intensification using intermediate-dose methotrexate and no delayed intensification phases. One major difference between the 2 groups was the extensive use of central reference laboratories by the POG, resulting in a higher percentage of available cytogenetic and/or molecular analyses and providing larger patient numbers for the current analyses. Another major difference between the 2 groups was the use of distinct protocols for T-cell and B-precursor ALL by the POG compared with common protocols by the CCG. The latter difference was not a factor, because we limited analyses to B-precursor ALL. In our analyses, age and WBC count remained important variables in trials conducted by both groups. In contrast, other variables previously predictive of outcome, including sex, race, hepatosplenomegaly, mediastinal mass, and French-American-British (FAB) morphology, have been replaced by genetic analyses of the leukemic blast and evaluations of early treatment response.
Despite different approaches by POG and CCG, there were striking consistencies among certain prognostic factors suggesting underlying biologic features that mediate treatment outcome. Both groups demonstrated that Ph-positive ALL and extreme hypodiploidy were very poor risk factors and that TEL/AML1 fusion and triple trisomies conveyed a very good prognosis. Originally, patients with fewer than 45 chromosomes or a DNA index below 0.81 were classified in the very-high-risk group, but an analysis of a large international cohort revealed that only hypodiploid patients with fewer than 44 chromosomes had a very poor outcome. Subsequently, additional international analyses of larger groups of hypodiploid patients, including these COG ALL patients, found that patients with 43 or fewer chromosomes had a very poor prognosis, while those with 44 or 45 chromosomes did not (EFS, more than 45%).23 Based on this, we modified our definition of extreme hypodiploidy for inclusion in the very-high-risk group to fewer than 44 chromosomes or a DNA index below 0.81. In our initial analysis, 11q23 translocations alone or the t(4;11) in particular did not meet criteria for very high risk. However, on the basis of outcome of t(4;11) patients with an SER in CCG 1961, we expanded our definition of very-high-risk patients to include those with an 11q23 (MLL) translocation and a poor response to induction chemotherapy. More effective therapy appears to have negated the prognostic importance of t(1;19)(q23;p13)29 or a balanced t(1;19).27
There were some outcome differences between the 2 cooperative groups. For example, NCI high-risk patients with CNS-3 disease at diagnosis treated on CCG trials had a 10% to 15% better 5-year EFS than on POG trials. However, with a relatively small number of patients, definitive conclusions could not be drawn. CNS-2 status at diagnosis in both groups was consistently worse than that of CNS-1 patients and no better than that of CNS-3 patients. Because CNS-3 patients received cranial irradiation and CNS-2 patients did not, CNS-directed therapy should be considered for CNS-2 patients in future trials.
Early response based on bone marrow morphology has been consistently reliable in predicting outcome,36,37 and augmentation of therapy for poor early response has had a major impact on outcome. CCG 1882 randomized SER patients (M3 day 7) to receive standard treatment or an augmented regimen resulting in a significantly better outcome (5-year EFS, 74% ± 3.8% versus 55% ± 4.5%, respectively).13 Similar findings, based on the results of a day-14 bone marrow, were seen in standard-risk patients treated on CCG 1952. In CCG 1952, patients with an M3 marrow at day 14 (M3/M3) received augmented therapy, while patients with M2 at day 14 (M3/M2) received standard therapy. The CCG 1952 data demonstrate that the outcome for the M3/M3 patients (augmented therapy) was significantly better than that of the M3/M2 patients (standard therapy).38 Thus, morphologic assessment of tumor burden continues to play an important role in the current classification system. In current COG trials, patients with NCI high- or standard-risk ALL are considered to be an SER if the day-14 bone marrow is M2 or M3. SER patients who do not meet criteria for the very-high-risk group will receive the augmented BFM regimen regardless of their initial NCI risk classification. Uniform use of the day-15 marrow was selected because it separates patients with greater specificity into groups with a 5-year EFS that is 15% to 20% lower than with an RER (Table 5).
A major recent advance in childhood ALL therapy has been the development of tools to measure subclinical levels of MRD. The clinical trials used for the analyses in this report did not include MRD determinations. While data were not mature enough for outcome analysis at the time we designed the current generation of COG trials, the ongoing POG 9900 generation of trials had established the feasibility of real-time MRD determination via flow cytometry in a central reference laboratory. In the more than 3000 patients enrolled on POG 9900, informative MRD results were available for more than 95% of patients within 24 hours of sample receipt. With no mature POG or CCG MRD data available to construct the ALL classification system, we relied on published data that demonstrated a strong correlation between end induction MRD and outcome using either flow cytometric or molecular techniques. Patients with low-end induction MRD burden (10−3 or less to 10−4 blasts per cells counted) have an excellent outcome, compared with a high-end induction MRD burden (more than 10−2) having a poor outcome.39 In the COG classification scheme (AALL03B1), we included both early morphologic response and end induction MRD levels. Patients are defined as an SER if the day-15 marrow contains at least 5% blasts by morphology (M2 or M3) or if the day-29 MRD burden is at least 0.1%. Such patients are nonrandomly assigned to receive the augmented BFM (A-BFM) regimen that has been shown to improve the outcome of patients with a poor early marrow response in CCG 1882.13 Patients with either an M2 marrow or at least 1% MRD by flow cytometry at day 29 receive 2 additional weeks of extended induction chemotherapy followed by response assessment. The continued use of an extended induction for M2- or MRD-positive patients has been maintained because both POG and CCG used additional postinduction therapy in previous protocols.
The current analysis of almost 6000 children and adolescents with ALL has provided an unprecedented opportunity to create an algorithm for identifying prognostic patient groups. The application of this algorithm (Figure 2) for risk classification in current cooperative group trials will divide patients with B-precursor ALL into low-risk (about 27% of the total), standard-risk (32%), high-risk (37%), and very-high-risk (4%) groups. This classification scheme balances the risks and benefits of intensified therapy by selecting patients who have a higher risk and will benefit most from treatment intensification, and it facilitates identification of very-high-risk patients who are candidates for experimental therapy. Because our analyses did not identify a group with more than 95% expected EFS, the current generation of COG ALL trials asks randomized questions of treatment intensification in low-, standard-, and high-risk groups on therapy backbones of different intensity. With the exclusion of patients with CNS-2 disease and the addition of MRD criteria in the classification algorithm, we expect to identify low-risk patients with a more than 95% chance of cure who might be candidates for a reduction of therapy question in future COG ALL trials.
After initial NCI risk classification, the COG end induction risk stratification algorithm requires immunophenotypic and molecular cytogenetic data as well as early response data, which must be available by day 35 of induction. In the current risk stratification schema, COG B-precursor ALL patient groups with varying prognoses are assigned to induction therapies according to data identified at the local center: age, initial WBC count, and immunophenotype. After blast genotype and early response criteria become available, in conjunction with data on the presence of extramedullary disease, patients are risk assigned to different postinduction therapies and randomized questions. Patients with NCI standard-risk ALL are subdivided into standard risk-low, standard risk-average, and standard risk-high groups (Figure 2A). Standard risk-low and -average patients are eligible for randomization whereas standard risk-high patients are nonrandomly assigned to the most augmented regimen in that study. NCI high-risk patients with an SER, CNS, or testicular disease are nonrandomly assigned to the augmented arm of that study, with all others randomized (Figure 2B). Very-high-risk patients, identified at the end of induction, include those with Ph-positive ALL, severe hypodiploidy, MLL translocation with an SER, and induction failures from all risk groups (Figure 2C). All are eligible for the COG VHR ALL trial, starting at the end of induction, which includes aggressive chemotherapy, HLA-matched sibling donor BMT and, for Ph-positive ALL, the BCR/ABL kinase inhibitor, imatinib.
The COG risk stratification algorithm requires an intensive evaluation for clinical, immunophenotypic, cytogenetic, and molecular markers with a rapid turnaround for risk classification and protocol assignment. COG expanded the POG reference laboratory classification system to include 2 additional central reference laboratories for cytogenetic, molecular, immunophenotyping, and MRD analyses with sample analysis of 2000 ALL patients per year from more than 230 COG centers in the United States, Canada, Australia, New Zealand, and Western Europe. These population-based data sets, inclusive of patients regardless as to geographic location or socioeconomic status, allow for ongoing analysis of the factors described in this paper and other potential markers. For example, monosomy 726 may confer a poor prognosis. The current generation of COG ALL clinical trials will assess the relative importance of prognostic variables with shared treatment arms on the low-risk, standard-risk, and high-risk studies to allow for the determination of end induction MRD in the context of variables like blast genotype (eg, TEL/AML1). The application of this strategy may be of limited use to either single centers or smaller cooperative groups that do not have adequate resources.
Many of the current risk assessment variables are imprecise surrogate markers for underlying biologic factors of the host and blasts. The COG is using newer technologies to examine blast gene expression profiles and host gene polymorphisms that may provide better predictive tools and lead to a mechanistic understanding of treatment success or failure.40-44 Because individual biologic and clinical factors are dependent on the therapy delivered, further therapeutic optimization may result in factors losing their prognostic impact, underscoring the importance of ongoing assessment.
Authorship
Contribution: K.R.S. (the primary author), D.J.P., M.J.B., A.J.C., N.A.H., J.E.R., M.L.L., E.A.R., N.J.W., S.P.H., W.L.C., P.S.G., and B.M.C. designed and performed research and wrote the paper; and H.N.S. (a CCG statistician), J.J.S., and M.D. (POG statisticians) analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kirk R. Schultz, Department of Pediatrics, Division of Hematology/Oncology/Bone Marrow Transplantation, University of British Columbia, BC Children's Hospital, 4480 Oak St, Vancouver, BC, V6H 3V4, Canada; e-mail: kschultz@interchange.ubc.ca.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by POG grant CA 30969, CCG grant CA 13539, and COG grant CA 98543. A complete listing of grant support for research conducted by CCG and POG before initiation of the COG grant in 2003 is available online at http://www.childrensoncologygroup.org/admin/grantinfo.htm.

![Figure 2. Risk assignment algorithms. Each of the algorithms shows the criteria for placement of patients in (A) standard-risk, (B) high-risk, and (C) very-high-risk assignment. Very-high-risk assignment occurs independently of initial NCI risk assignment (standard or high risk). Patients who have received less than 48 hours of oral or intravenous steroids during the week immediately prior to diagnosis are eligible for classification if the results of a complete blood count (CBC), obtained prior to the initiation of steroid therapy (less than 72 hours prior to steroids), are available and the necessary FISH, cytogenetic, and molecular data are interpretable. The “presteroid” CBC and age of the patient are used to determine NCI-Rome risk classification (standard risk versus high risk [SR versus HR]). If patients have received more than 48 hours of oral or intravenous steroids (and a presteroid CBC is available to assign NCI risk group), they are treated as an SER and nonrandomly assigned to the augmented regimen. In the absence of a presteroid CBC, patients who have received less than 48 hours of steroids are assigned to the HR protocol. These patients are eligible for randomization on the HR protocol. As expected, patients with a slow early response will be assigned to the full augmented BFM treatment arm. In the absence of a presteroid CBC, patients who have received more than 48 hours of steroids are treated as an SER on the HR study and assigned to the full augmented arm. Inhalational steroids are not considered as pretreatment. Both SR and HR patients with identified MLL translocations, CNS-3, or testicular disease, receive augmented SR therapy.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/3/10.1182_blood-2006-01-024729/5/m_zh80030707220002.jpeg?Expires=1770346300&Signature=2262k8WYL4KUJ9h83V0usGA2Md8GUzyPCN6g4sOATKOGaX9-5roNT-XE9-Bbt6i5nbWz1RHVintIIfT9hB5nYqhbOYBM09XYMYeBmGIT5iJlcFsR1oSisPftMJBPTTLb3XNNZ-DeW8qKgwfcJw~cSV9RKD27UoDy~hbY6Rb6TCr9UdyK6gNtdgp~o2Pzz3Jujh7o~iXtotPNSrSn8KMwHlferZkzWnj5Xfg8bCu-dobcMDae4Oj~eRv-etAQuR59RKflhumyP9-sk8cJKkW8EbbsbUqStGdgypx79fwCelDwC1sqXi-l2ZNxHwLFSgnzo9rMJd0B~qr1kgGPzQ2OaQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal