Abstract
Expression of a bcr-3 isoform of retinoic acid receptor α–promyelocytic leukemia (RARα-PML) in mice expressing a bcr-1 isoform of PML-RARα is associated with increased penetrance of murine acute promyelocytic leukemia (APL) and the frequent acquisition of an interstitial deletion of one copy of mouse chromosome 2 (del(2)). To determine whether the isoform of RARα-PML is important for these effects, we created mice that expressed a bcr-1 isoform of RARα-PML. Coexpression with the bcr-1 isoform of PML-RARα did not increase the penetrance of APL (7 of 45 animals developed APL with PML-RARα alone vs 12 of 44 with both transgenes; P = .19). Furthermore, the frequency of del(2) in APL cells from doubly transgenic mice was not different from that of mice expressing PML-RARα alone (3 of 6 vs 6 of 12, respectively—P = 1.38—compared with 11 of 11 for mice coexpressing PML-RARα and bcr-3 RARα-PML). The bcr-1 and bcr-3 isoforms of RARα-PML, therefore, have different biological activities that may be relevant for the pathogenesis of murine APL.
Introduction
We previously created a model of acute promyelocytic leukemia (APL) in transgenic animals by targeting the expression of a human bcr-1 isoform of promyelocytic leukemia–retinoic acid receptor α (PML-RARα) cDNA to early myeloid cells with the use of regulatory sequences from the human cathepsin G (hCG) gene.1 Myeloproliferative disease develops in all mice expressing this transgene, but APL develops in only approximately 15% to 30% of mice (after a latency of 6-13 months).1 RARα-PML reciprocal translocation products are expressed in up to 80% of patients with APL.2,3 The bcr-1 isoform of RARα-PML results from the fusion of RARα at intron 2 with PML at intron 6, whereas the bcr-3 isoform of RARα-PML fuses RARα at intron 2 with PML at intron 3 (Figure 1A). Mice that coexpress the reciprocal human bcr-3 isoform RARα-PML cDNA in the same early myeloid compartment as the bcr-1 isoform of PML-RARα display an APL penetrance of approximately 60% (with the same latency).4 Most APL tumors that develop in mice expressing both transgenes acquire an interstitial deletion on one copy of chromosome 2 (del(2)).5 The importance of del(2) as a progression factor in murine APL has been further supported by its association with radiation-induced AML and the increased penetrance of APL observed after irradiation of young PML-RARα expressing mice (which also induces del(2)).6,7 These data suggest that the expression of a bcr-3 RARα-PML fusion gene, or exposure to irradiation, facilitates the development of del(2) in PML-RARα–expressing mice. The minimal del(2) interval defined to date contains the PU.1 gene, which is directly relevant for AML pathogenesis.6,8-13
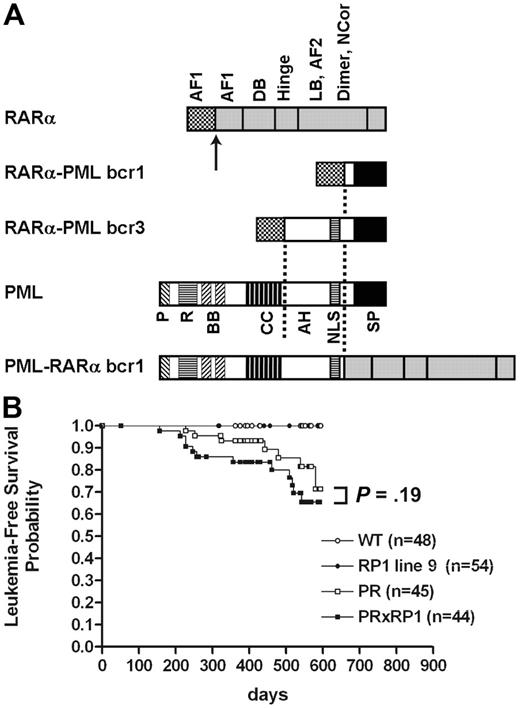
RARα-PML cDNA isoforms and Kaplan-Meier analysis of APL disease in transgenic animals. (A) Diagram of the structural features of bcr-1 and bcr-3 isoforms. The identical portion of RARα is expressed in both isoforms of RP (breakpoint in RARα is indicated by the arrow), whereas the expressed PML domains are different because of the locations of the breakpoints in PML (indicated by the dashed lines). DB indicates DNA binding; LB, ligand binding; Dimer, dimerization; P, proline rich; R, ring finger; BB, B-box; CC, coiled-coil; AH, alpha helix; NLS, nuclear localization signal; SP, serine-proline rich. (B) Data represent the complete study of the first intercross (PR×RP1, line 9). The number of animals per group is indicated. The cumulative probability of death resulting from APL is not significantly different in PR animals than it is in PR×RP1 animals (P = .19).
RARα-PML cDNA isoforms and Kaplan-Meier analysis of APL disease in transgenic animals. (A) Diagram of the structural features of bcr-1 and bcr-3 isoforms. The identical portion of RARα is expressed in both isoforms of RP (breakpoint in RARα is indicated by the arrow), whereas the expressed PML domains are different because of the locations of the breakpoints in PML (indicated by the dashed lines). DB indicates DNA binding; LB, ligand binding; Dimer, dimerization; P, proline rich; R, ring finger; BB, B-box; CC, coiled-coil; AH, alpha helix; NLS, nuclear localization signal; SP, serine-proline rich. (B) Data represent the complete study of the first intercross (PR×RP1, line 9). The number of animals per group is indicated. The cumulative probability of death resulting from APL is not significantly different in PR animals than it is in PR×RP1 animals (P = .19).
In our original study, a bcr-3 isoform of RARα-PML was chosen because of its ready availability and because it contains a large amount of sequence from PML (including its nuclear localization signal and an Rb-interacting domain14 ). However, because it is not the reciprocal translocation partner of bcr-1 PML-RARα, we have now expressed a reciprocal bcr-1 in mice with the same human cathepsin G–targeting cassette. The lack of a phenotype indicates that different reciprocal isoforms may have different biological functions relevant for the pathogenesis of APL in mice.
Materials and methods
We have previously described the generation of bcr-1 hCG-PML-RARα and bcr-3 hCG-RARα-PML transgenic mice in the C3H × C57Bl/6 background.1,4 A bcr-1 isoform of an RARα-PML cDNA (derived from the PML3 isoform of PML15 ), containing RARα exon 2 fused in-frame to PML exons 7a-8, was ligated into the hCG cassette for this study. Sample size calculations were based on detecting a 30% change in survival proportion at a significance level of α = 0.05 (2-tailed) and a power of 80%. The generation of 2 independently derived bcr-1 RARα-PML founder mice, the generation of 2 intercrossed family cohorts for study, the maintenance of founder lines in the C3H × C57Bl/6 background, the detection of transgene mRNA expression from whole bone marrow cells, the cryopreservation of APL cells, and an analysis of transgenic animals were performed as previously described.1,4,7,8 Array-based comparative genomic hybridization (aCGH) was performed using a custom BAC array produced by the Genome Sequencing Center at Washington University. The array encompassed the complete tiling path for mouse chromosome 2 (1996 BACs spotted in duplicate). Sample labeling, hybridization, washing, imaging, and analysis were performed as previously described.16
Results and discussion
We have reported previously the phenotype of transgenic animals coexpressing a bcr-3 isoform of RARα-PML with a bcr-1 isoform of PML-RARα.4 To define the biological role of a bcr-1 isoform of RARα-PML, we inserted this cDNA into our standard hCG regulatory cassette at the ClaI-NotI site in the 5′UT. We identified 2 founder mice that transmitted the transgene through the germline (lines 9 and 11). The level of bcr-1 RARα-PML mRNA expression in whole bone marrow samples from transgenic mice was similar to that of PML-RARα expression (relative expression compared with PML-RARα: line 9, 0.78; line 11, 2.72), which was similar to the level of bcr-3 RARα-PML expression previously reported.4 Expression level of the hCG portion of the transgene (common to both constructs) in the bone marrow of doubly transgenic animals was appropriately increased compared with PML-RARα or bcr-1 RARα-PML–expressing mice alone.
Bcr-1 RARα-PML (RP1) mice have normal peripheral blood counts compared with age-matched littermate wild-type (WT) mice, nonleukemic PML-RARα (PR), and nonleukemic PML-RARα × RARα-PML (PR×RP1) mice. Expression of bcr-1 RARα-PML does not increase the penetrance of APL in bcr-1 PML-RARα–expressing mice, nor does it increase the latency of APL development (Figure 1B). In the first completed intercross (line 9), APL occurred in 0 of 48 WT animals, 0 of 54 RP1 animals, 7 of 45 (15.5%) PR animals, and 12 of 44 (27%) PR×RP1 animals after 18 months of observation (PR vs PR×RP1; P = .19). The second intercross (line 11) was less mature than the first intercross, but the results were similar. No significant difference was observed in APL development in PR animals (8 of 22) compared with PR×RP1 animals (14 of 33) after 12 months of observation (P = .5). Peripheral blood counts and splenic morphology of APL cells were similar for PR and PR×RP1 animals (data not shown).
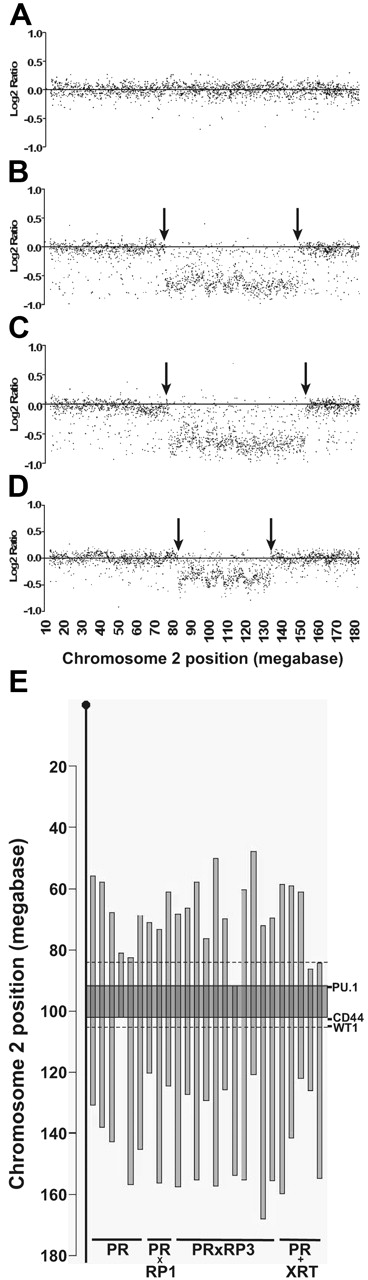
We next analyzed cells derived from the spleens of leukemic animals for the presence of del(2) with the use of array-based comparative genomic hybridization (aCGH). The frequency of del(2) could not be assessed in RP1 mice because none of these animals developed leukemia. DNA was obtained from cryopreserved splenic APL cells from animals in this study and previously banked APL specimens from PR, PR×RP3, or irradiated PR animals (PR+XRT).4,7 Six of 12 PR samples, 11 of 11 PR×RP3 samples, 3 of 6 PR×RP1 samples, and 5 of 6 PR+XRT samples contained del(2) by aCGH (PR×RP3 compared with PR×RP1 (P = .03) (Figure 2A-E). The minimally deleted region on chromosome 2 mapped between megabase 90 and 101 (encompassing 82 genes/ESTs), which significantly narrowed the previously defined deleted region, which spanned 21 Mb (including 474 genes/ESTs).6 Importantly, PU.1 is on the minimally deleted region in all deletions identified by aCGH; the CD44 and WT1 genes fall outside this minimally deleted region, corroborating the observation that haploinsufficiency for PU.1 is relevant for APL penetrance, whereas haploinsufficiency for CD44 or WT1 is not (Figure 2E).8
Chromosome 2 complete tiling path aCGH plots and mapping of the minimally deleted region. Genomic DNA was fragmented and labeled by random priming in a reaction containing a Cy3-dCTP (green test samples, APL) or a Cy5-dCTP (red reference samples, control DNA) fluorophore. Cohybridization with the test and reference DNA was performed overnight, followed by washing and imaging. The resultant green-to-red log2 ratios were calculated and plotted by chromosomal location. Fifteen of the 35 samples analyzed with aCGH were previously analyzed with spectral karyotyping (SKY). We detected del(2) in 3 of 5 tumors that did not contain del(2) with SKY. Six of the 35 samples were previously analyzed with fluorescence in situ hybridization (FISH), and 1 PR+XRT sample that contained del(2) by FISH was not detected using aCGH. This sample contained del(2) in only 20% of cells, which is below the limit of detection using aCGH (M.J.W., R.E.R., T.J.L., unpublished observations, November 2006). (A) Log2 plot from an APL sample with a normal chromosome 2. (B-D) APL samples with interstitial deletions on chromosome 2 produce a negative log2 ratio for BAC probes located within the deletion. Arrows indicate deletion boundaries. (E) Twenty-five APL samples from various genotypes of mice contained del(2) by aCGH. Vertical bars indicate the boundaries of the interstitial deletions. Dashed lines indicate the boundaries of del(2) based on a radiation-induced AML model (84-105 megabases) that used simple sequence length polymorphisms to map deletion end points.6 Gray shaded rectangle (90-101 megabases) identifies the minimally deleted region (MDR) defined by aCGH. PU.1 is located within the aCGH defined MDR, whereas CD44 and WT1 genes are located distal to the telomeric border.
Chromosome 2 complete tiling path aCGH plots and mapping of the minimally deleted region. Genomic DNA was fragmented and labeled by random priming in a reaction containing a Cy3-dCTP (green test samples, APL) or a Cy5-dCTP (red reference samples, control DNA) fluorophore. Cohybridization with the test and reference DNA was performed overnight, followed by washing and imaging. The resultant green-to-red log2 ratios were calculated and plotted by chromosomal location. Fifteen of the 35 samples analyzed with aCGH were previously analyzed with spectral karyotyping (SKY). We detected del(2) in 3 of 5 tumors that did not contain del(2) with SKY. Six of the 35 samples were previously analyzed with fluorescence in situ hybridization (FISH), and 1 PR+XRT sample that contained del(2) by FISH was not detected using aCGH. This sample contained del(2) in only 20% of cells, which is below the limit of detection using aCGH (M.J.W., R.E.R., T.J.L., unpublished observations, November 2006). (A) Log2 plot from an APL sample with a normal chromosome 2. (B-D) APL samples with interstitial deletions on chromosome 2 produce a negative log2 ratio for BAC probes located within the deletion. Arrows indicate deletion boundaries. (E) Twenty-five APL samples from various genotypes of mice contained del(2) by aCGH. Vertical bars indicate the boundaries of the interstitial deletions. Dashed lines indicate the boundaries of del(2) based on a radiation-induced AML model (84-105 megabases) that used simple sequence length polymorphisms to map deletion end points.6 Gray shaded rectangle (90-101 megabases) identifies the minimally deleted region (MDR) defined by aCGH. PU.1 is located within the aCGH defined MDR, whereas CD44 and WT1 genes are located distal to the telomeric border.
In addition, del(2) has been described in AML syndromes induced by irradiation,6 benzene,17 and overexpression of Hox genes in bone marrow progenitors.18 Although del(2) appears to facilitate APL/AML progression, it is not required; indeed, many PML-RARα–expressing mice do not have it.4,7,19 The genetic differences between bcr-1 and bcr-3 isoforms of RARα-PML in doubly transgenic mice may contribute to their different abilities to facilitate del(2). The α helical domain (AH) and the nuclear localization signal (NLS) of PML are present in the bcr-3 isoform but not in the bcr-1 isoform of RARα-PML used in these studies (Figure 1A). These domains are expressed from both the PR1 and the RP3 transgenes in PR×RP3 mice, but they are present only in the PR1 transgene in PR1×RP1 mice (Figure 1A).
Although the relative levels of mRNA expression were similar between the RP3 and RP1 transgenes, small differences in gene dosage could also potentially contribute to differences in disease penetrance between the models. We think this is unlikely because 2 different bcr-3 RARα-PML transgene insertions (with different expression levels) increased disease penetrance.4 Although the mechanism is not yet clear, our data show that the additional genetic information contained in the RP3 cDNA may be relevant for APL pathogenesis and the acquisition of del(2). However, it remains possible that the ability of RP3 to increase leukemia penetrance in the PR1×RP3 mice could reflect a nonphysiologic interaction between PR1 and RP3.
APL patients expressing a bcr-3 isoform of PML-RARα appear to undergo additional cytogenetic changes compared with patients expressing the bcr-1 isoform,20 but this finding is controversial.21-23 Our results also suggest that the expression of a bcr-3 isoform of RARα-PML (RP3) may facilitate the acquisition of cytogenetic changes, predominantly manifested by the frequent acquisition of del(2) in mice. The mechanism by which RP3 facilitates the interstitial deletion of chromosome 2 is unknown, but the study of this mechanism may provide valuable insight into the ways by which cytogenetic changes involving chromosomal copy number may occur in some patients with AML.
Authorship
Contribution: M.J.W., R.E.R., J.R.A., E.R.M., and T.J.L. designed the research. M.J.W., R.E.R., J.R.A., and J.S.P. performed the research. J.R.A. and E.R.M. contributed vital new reagents or analytical tools. M.J.W., R.E.R., J.R.A., and J.S.P. collected data. M.J.W., R.E.R., J.R.A., J.S.P., E.R.M., and T.J.L. analyzed the data. M.J.W. and T.J.L. wrote the paper.
Conflict-of-interest disclosure: the authors declare no competing financial interests.
Correspondence: Timothy J. Ley, Division of Oncology, Section of Stem Cell Biology, Washington University School of Medicine, Campus Box 8007, 660 S Euclid Ave, St Louis, MO 63110-1093; e-mail: tley@im.wustl.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants CA83962 and CA101937 and grant 1K08HL083012 (M.J.W.), the Bakewell Cancer Research Fund, the Buder Charitable Foundation (T.J.L.), a Fellowship grant from the Leukemia Research Foundation, and an American Society of Hematology Fellow Scholar Award.
We thank Francesco Contegno and Pier Giuseppe Pelicci for providing the bcr-1 RARα-PML cDNA, Mieke Hoock for excellent colony management, Patrick Cahan for assistance in preparing Figure 2E, and Nancy Reidelberger for expert editorial assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal