Abstract
Functional studies show that programming of CD8+ T cells occurs early after initial antigen encounter within as little as 2 hours. To define the molecular basis of these events, we transferred TCR transgenic T cells from F5 Rag−/− mice into naive recipients and stimulated them with recombinant vaccinia expressing the immunodominant influenza epitope NP366-374. Transcription in epitope-specific cytotoxic T lymphocytes (CTLs) was analyzed using Affymetrix 430 2.0 GeneChips and quantitative polymerase chain reaction (PCR). We demonstrated an early transcriptional burst with the greatest number of genes reaching peak expression 12 hours after stimulation. Using in vivo cytotoxicity assays we demonstrated that early up-regulation of cytolytic genes was accompanied by acquisition of killing capacity within 24 hours of stimulation. However, T-cell proliferation was not observed until 48 hours. We therefore conclude that clonal expansion rather than acquisition of effector function is the rate-limiting step in the development of a primary CTL response.
Introduction
Following primary exposure to viruses, antigen-specific CD8+ T cells undergo clonal proliferation and differentiation to form an expanded effector population with the capacity to control infection. This initial phase of T-cell expansion is followed by a period of contraction after clearance of antigen and by the generation of memory.1-4 Although attention has focused on these later events because of their importance in vaccination, the early period following antigen exposure is increasingly recognized as being instrumental in determining the course of this response.
Several studies have shown that a short period of antigenic stimulation is sufficient to initiate a sustained program of proliferation, differentiation, contraction, and memory formation in CD8+ T cells. These have defined the concept of T-cell programming.5-8 In particular, it has been demonstrated that only 2 hours of antigen exposure is sufficient to promote proliferation and acquisition of effector function.5 Neither longer stimulation nor more abundant antigen increases the proliferative capacity of the cell,6,8 although it is also likely that factors such as signal strength modulate this capacity.9 Once initiated, this program of differentiation follows a relatively constant pattern, the kinetics of which are unperturbed by alterations in antigen load.8
Defining the transcriptional changes that occur early after activation and underpin T-cell programming will be of fundamental importance in understanding the development of the CD8+ T-cell response. Previous attempts have been made to correlate the transcriptional activity of T cells with their phenotype, but these have been limited to a few genes.10,11 More recently, the use of gene expression microarrays has allowed the study of global changes in transcription by the comparison of gene expression profiles between naive, effector, and memory cell populations in adoptively transferred transgenic murine cytotoxic T lymphocytes (CTLs).12 These studies compared the relative gene expression of naive, effector, and memory CTLs following stimulation with LCMV and demonstrated that changes in expression of genes associated with cellular functions including signaling, cytotoxicity, and migration had occurred within the CD8+ T-cell population by the time of the primary effector response, 8 days after antigenic stimulation. However, little is yet understood about the transcriptional events that occur within that period. Whether this is characterized by gradual changes in gene expression that peak at the time of the primary effector response or whether early bursts of transcription program cellular differentiation remains unknown.
Likewise, relatively little is known about the timing of acquisition of CD8+ T-cell effector functions. The ability to rapidly kill infected cells is one of the prime functions gained during the course of the primary response13-15 and maximal killing has been seen to coincide with the peak proliferative response.12,16 However, it is still unclear how early cytotoxic capacity develops and whether it is acquired independently of proliferation.
In view of the evidence suggesting that the T-cell response is programmed early after antigen encounter, we studied the molecular basis of these events by analyzing the transcriptional and functional changes that occur shortly after TCR engagement. Using an adoptive transfer model we analyzed gene expression in epitope-specific cells in vivo at multiple time points up to 96 hours after stimulation with Affymetrix GeneChips and quantitative polymerase chain reaction (PCR). In parallel, we examined the acquisition of antigen-specific cytotoxic capacity using in vivo cytotoxicity assays. Using this approach we demonstrate successive waves of gene expression occurring early during the primary CD8+ T-cell response and correlate these changes with the unexpected acquisition of CTL function prior to cell division.
Materials and methods
Mice and viruses
F5 RAG−/− and C57BL/6 mice were bred and maintained at the Biomedical Services facility, John Radcliffe Hospital (Oxford, United Kingdom). Males between 8 and 12 weeks of age were used. F5 RAG−/− splenocytes (107) were adoptively transferred to C57BL/6 mice by intravenous injection. Twenty-four hours following adoptive transfer, mice were stimulated using 107 PFU recombinant vaccinia virus by intraperitoneal injection. A recombinant WR-strain vaccinia expressing the NP(met)366-374 epitope (rVV-NP366) was used for immunization. Recombinant vaccinia expressing the OVA peptide (rVV-OVA) was kindly provided by Keith Gould (Imperial College, London, United Kingdom). All animal work was carried out under license in accordance with the Animals (Scientific Procedures) Act 1986.
CFSE and cell surface staining
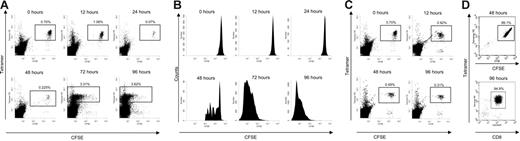
Prior to adoptive transfer, F5 RAG−/− splenocytes were stained with CFSE as previously described.6 For FACS analysis, splenocytes were stained using Db-NP366-374 tetramer and anti-CD8 PerCP (BD PharMingen, San Diego, CA). Cell sorting was carried out using a FACSDiVa high-speed cell sorter (Becton Dickinson, San Jose, CA). Sorted cells were gated on the double-positive CFSE+ tetramer-positive population, or CD8+ tetramer-positive population. Around 95% of recovered cells were epitope-specific CTLs (Figure 1D).
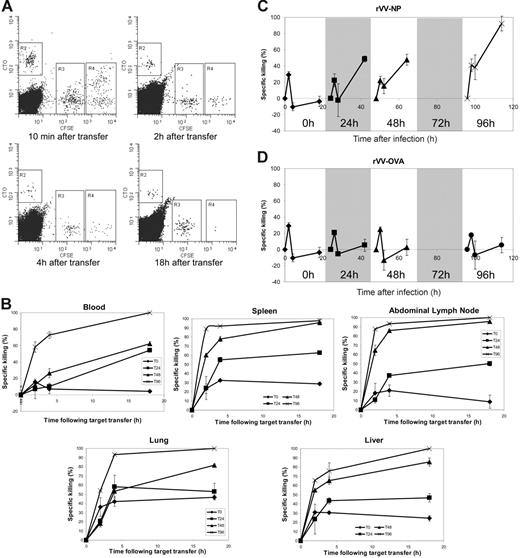
Isolation of NP366-374 specific CD8+ T cells following stimulation with recombinant vaccinia virus. CFSE-labeled splenocytes (107) were transferred from F5 Rag−/− to C57BL/6 mice and the recipients infected 24 hours later with 107 PFU recombinant vaccinia virus expressing NP366-374 by intraperitoneal injection. (A) Epitope-specific cells were tracked using CFSE and tetramer-PE. The size of the transferred epitope-specific population is shown as percentage of lymphocytes. (B) CFSE profiles of the transferred CTL population are shown at each time point. (C) Epitope-specific cells were analyzed following infection with recombinant vaccinia expressing the OVA peptide (0-, 24-, 48-, and 96-hour time points are shown). Identical results were seen with sham infection using PBS alone. (D) FACS gave up to approximately 99% purity. All plots show data representative of at least 6 individuals in the “lymphocyte” gate.
Isolation of NP366-374 specific CD8+ T cells following stimulation with recombinant vaccinia virus. CFSE-labeled splenocytes (107) were transferred from F5 Rag−/− to C57BL/6 mice and the recipients infected 24 hours later with 107 PFU recombinant vaccinia virus expressing NP366-374 by intraperitoneal injection. (A) Epitope-specific cells were tracked using CFSE and tetramer-PE. The size of the transferred epitope-specific population is shown as percentage of lymphocytes. (B) CFSE profiles of the transferred CTL population are shown at each time point. (C) Epitope-specific cells were analyzed following infection with recombinant vaccinia expressing the OVA peptide (0-, 24-, 48-, and 96-hour time points are shown). Identical results were seen with sham infection using PBS alone. (D) FACS gave up to approximately 99% purity. All plots show data representative of at least 6 individuals in the “lymphocyte” gate.
Total RNA extraction and labeling
Total RNA was extracted using the Qiagen (Valencia, CA) RNeasy micro kit according to the manufacturer's instructions. Up to 23 animals were pooled for each sample, with equal numbers of cells derived from each mouse. RNA was extracted from a total of 106 epitope-specific cells for each sample. Amplification and labeling were carried out using the small-sample protocol by Affymetrix (Santa Clara, CA). Labeled cRNA was hybridized to Affymetrix Mouse Genome 430 2.0 GeneChips (a global array with 45 101 probe sets). Eleven pooled samples were hybridized: 3 biologic replicates from F5 Rag−/− spleens before adoptive transfer, one from adoptively transferred recipients at each time point 0, 12, 24, 48, and 96 hours after infection, and 3 biologic replicates from recipients at 72 hours.
Affymetrix analysis
Fluorescence data were imported into 3 analysis packages: GeneSpring (Agilent Technologies, Santa Clara, CA), R Bioconductor with the Robust Multichip Average (RMA) module,17 and DNA-Chip analyzer.18 Gene expression levels were calculated using the Affymetrix MicroArray Suite (MAS) algorithm or within RMA or dChip. For each experiment, normalizations were carried out between the relevant chips. This was achieved using 3 techniques: GeneSpring's per-chip and per-gene normalization, the quantile normalization method in RMA, and normalization to an invariant gene set in dChip. Further analysis of the resulting normalized gene expression values was undertaken using GeneSpring. Genes that had been flagged as absent or marginal by MAS were discarded. Quality assessment of the chips demonstrated a background fluorescence level of around 150, and any genes with a raw expression level below this level were also discarded. Genes of interest were then selected using fold-changes in expression. Each normalization method led to a different set of such genes and only those common to all 3 sets were analyzed further. Cluster analysis in the time-course experiment was performed using K-means clustering, the optimal number of clusters having been determined by the “K-means best” algorithm in GeneSpring. Pathway analysis was performed using ArrayXPath (http://www.snubi.org/software/ArrayXPath/) using gene lists derived from cluster analysis and genes with more than 2-fold differential expression. Microarray data are MIAME compliant and available in Array Express (ID: E-MIMR-38).
Quantitative PCR
Epitope-specific cells were purified as before from individual animals with no pooling. RNA was extracted using the Qiagen RNeasy system and reverse transcribed using Superscript II (Invitrogen, Paisley, United Kingdom) according to the manufacturer's protocols. Quantitative PCR was performed using the Lightcycler Faststart DNA Master Plus kit (Roche, Indianapolis, IN). Primers for granzyme A, granzyme B, perforin, IL-1β, IL-2, IFN-γ, TNF-α, CCL3, CXCR3, CCR6, IL-1R2, CDK4, cyclin A2, and cyclin G2 were obtained from previously published sequences or the PrimerBank database (http://pga.mgh.harvard.edu/primerbank/). The sequences and Lightcycler program are available on request. A standard curve of relative concentration was calculated for each reaction run using serial dilutions of cDNA derived from unsorted mouse splenocytes. The resulting calculated “concentrations” were normalized against the housekeeping genes aminolevulinic acid synthase 1 (ALAS1) and glucose-6-phosphate 1-dehydrogenase X (G6PDX).
In vivo cytotoxicity assay
Splenocytes from naive C57BL/6 mice were stained with CFSE 1 μM or 1 nM (Molecular Probes, Eugene, OR) or CellTracker Orange 8 μM (CTO, Molecular Probes). Each population was incubated with peptide NP366 or PA224 at 1 μM, or medium alone, respectively, for 2 hours at 37°C. Labeled cells (5 × 106 cells of each population) were transferred by intravenous injection into infected mice. Individual animals were followed longitudinally by blood sampling immediately and at 2, 4, and 18 hours after transfer. Lymphocytes were isolated by density gradient centrifugation using Lympholyte-Mammal (Cedarlane Laboratories, Hornby, ON, Canada) and target populations distinguished by CFSE and CTO staining. The observed loss of NP366-pulsed cells was attributed to antigen-specific CTL lysis and the rate of killing calculated as follows: 1 − (% NP366 pulsed at assay time point/% PA224 pulsed at assay time point)/(% NP366 pulsed at previous assay time point/% PA224 pulsed at previous assay time point) × 100. Similarly, spleen, abdominal lymph node, lung, and liver tissues were also analyzed for transferred target cells. In these tissues, antigen-specific cell loss at each time point was quantified as follows: 1 − (% NP366 pulsed/% PA224 pulsed) × 100.
Results
Generation of the primary CD8+ T-cell response
The study of the early primary immune response is constrained principally by the low frequency of antigen-specific precursors. For this reason, we used an adoptive cell transfer model based on the F5 Rag−/− mouse. This strain expresses a TCR specific for the immunodominant influenza virus peptide NP366-374 presented in the context of H-2Db; more than 75% of its lymphocytes are epitope-specific. CFSE-labeled splenocytes from F5 Rag−/− mice were adoptively transferred into C57BL/6 mice and these were infected with a recombinant vaccinia virus expressing the NP366-374 peptide (rVV-NP366) by intraperitoneal injection 24 hours after transfer.
Quantitative analysis of splenocytes after infection revealed a characteristic response. Animals were culled at 6 time points up to 96 hours after infection (0, 12, 24, 48, 72, and 96 hours) and epitope-specific cells were identified by CFSE and tetramer staining (Figure 1A). Between 0 and 12 hours following infection, the size of the transferred CD8+ population remained stable, followed by an apparent decrease in the number of these cells at 24 hours. By 48 hours the CD8+ cells had begun to divide until at 96 hours the CFSE concentration had fallen below the limits of detection and tetramer-positive cells made up more than 3% of total splenic lymphocytes. Analysis of CFSE dilution indicated that the vast majority of transferred epitope-specific CTLs underwent concurrent cell division and very few remained unstimulated (Figure 1B). This response was not seen in either unstimulated controls or controls infected with recombinant vaccinia virus expressing the irrelevant OVA peptide (rVV-OVA); neither the apparent initial fall in number of transferred CTLs nor the subsequent proliferation of the transferred CTL population was observed (Figure 1C), thereby confirming the specificity of the response for the NP366-74 peptide.
Experiments were undertaken to investigate the cause of the apparent cell loss. Quantitative analysis of other tissue compartments failed to demonstrate a reciprocal increase in CTL numbers in blood, lymph nodes, liver, or lung at the 24-hour time point (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Although we were able to show that a proportion of transferred CTLs underwent apoptotic cell death, this could not account for the majority of the apparent loss of CTLs (data not shown). However, analysis of ungated fluorescence-activated cell sorting (FACS) plots revealed that the majority of the CTLs were still present within the “monocyte” gate at the 24 hours time point (Figure S2). Only epitope-specific CD8+ T cells found in the lymphocyte gate could be used for transcriptional analysis, and epitope-specific CD8+ T cells at each time point were sorted by flow cytometry with a purity of more than 94% in all cases (Figure 1D).
Effects of adoptive transfer alone on gene expression
Previous studies analyzing CD8+ T cells with microarrays have used naive transgenic cells prior to adoptive transfer as their baseline controls. We were concerned that the possible effect on transcription of adoptive transfer alone had not been taken into account. We therefore compared the expression profiles of epitope-specific cells after adoptive transfer but before antigen challenge with those from F5 Rag−/− splenocytes before adoptive transfer. Cells from individual F5 Rag−/− spleens were used to generate cRNA for hybridization to 3 Affymetrix 430.2 GeneChips. Hierarchical cluster analysis demonstrated good concordance between the 3 biologic replicates (data not shown). For the posttransfer sample, cells from 18 mice were pooled to obtain sufficient total RNA and this was hybridized to one chip. Gene expression levels were calculated using 3 different software packages (MAS, RMA, and dChip) followed by normalization within each package and filtering in GeneSpring (see “Materials and methods”). Only those genes with more than 2-fold differential expression between the 2 conditions irrespective of normalization method were analyzed.
A total of 153 genes were shown in this way to be differentially expressed between the epitope-specific CD8+ T cells before and after transfer (Table S1). The up-regulated genes (eg, Jun and Fos) suggested a picture of cellular activation, which we attribute to the stress of ex vivo manipulation. For all subsequent analyses, both to ensure comparability with previous studies and to control for the effect of transfer, we rested adoptively transferred cells for 24 hours before stimulation and used transferred prestimulation samples as baseline controls. Quantitative PCR was used to analyze expression of selected genes in a longitudinal fashion in unstimulated controls to address concerns regarding persistent changes in gene expression (see “Validation using quantitative PCR”).
Microarray analysis of effector CD8+ T cells 96 hours after infection
In an initial analysis we compared the gene expression profile of effector cells (96 hours after infection) with unstimulated control (0 hours). Data were processed as described and genes with a fold-change of more than 2 were selected. A total of 498 genes fulfilled these criteria, with 371 genes up-regulated and 127 genes down-regulated (Table S2). Several groups of functionally related genes were identified; in particular, clusters of genes relating to signaling, metabolism, transcription, cell cycle, and immune function.
Gene expression profiling of a 96-hour time course
We next sought to perform a detailed analysis of the early transcriptional events that characterize priming. Animals were established as before, culled at 6 time points up to 96 hours after infection, and the lymphocyte subpopulation purified. Up to 23 individual mice were pooled for each sample to obtain sufficient RNA.
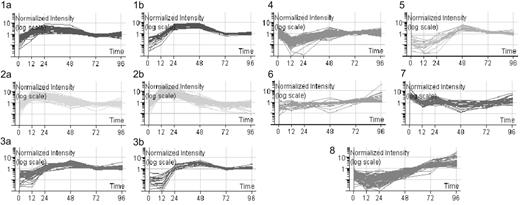
Gene expression levels were processed as described. Selection of genes with differential expression more than 2-fold at any time point (12-96 hours after infection) compared with baseline (0 hours) produced a list of 3409 genes, almost 7 times the number identified when we directly compared naive and effector cells. For further stringency, we only analyzed genes with more than 4-fold differential expression. These 819 genes were grouped using K-means clustering, resulting in 8 clusters in total (Figure 2). The profiles of the clusters suggested that peak expression of many genes was reached early during the primary immune response rather than at the final 96-hour time point.
K-means clustering of 819 differentially expressed genes. Gene expression levels were calculated using MAS, RMA, and dChip, then normalized using the associated algorithms. Genes with a raw expression level below 150 were excluded. Genes with more than 4-fold change in expression compared to baseline (0 hours) were selected. Only those genes selected irrespective of normalization method were further analyzed. Eleven clusters were identified by the “K-means best” algorithm and 3 pairs of clusters (1a and b, 2a and b, and 3a and b) were merged due to their similar patterns.
K-means clustering of 819 differentially expressed genes. Gene expression levels were calculated using MAS, RMA, and dChip, then normalized using the associated algorithms. Genes with a raw expression level below 150 were excluded. Genes with more than 4-fold change in expression compared to baseline (0 hours) were selected. Only those genes selected irrespective of normalization method were further analyzed. Eleven clusters were identified by the “K-means best” algorithm and 3 pairs of clusters (1a and b, 2a and b, and 3a and b) were merged due to their similar patterns.
NCBI and GO databases were probed for functional information. On the basis of these data, genes were assigned to functional categories within each cluster (Table S3). Furthermore, pathways analysis was performed using ArrayXPath, which assigned genes from each cluster to known signaling pathways and determined the significance of these by Fisher exact test (Table S4). The resulting analyses indicated that several clusters were strongly dominated by genes belonging to a few functionally related groups and signaling pathways. For example, in cluster 1, where gene expression peaked at 24 hours and remained at a relatively high level, 37% of genes were involved in DNA replication and repair. Cluster 2, in which gene expression rose rapidly to peak at 12 to 24 hours and then fell, was dominated by genes involved in protein synthesis pathways. Cluster 3, which included genes peaking later, at 48 hours, was dominated by genes that regulate cell cycling.
Temporal changes in gene expression of functional categories
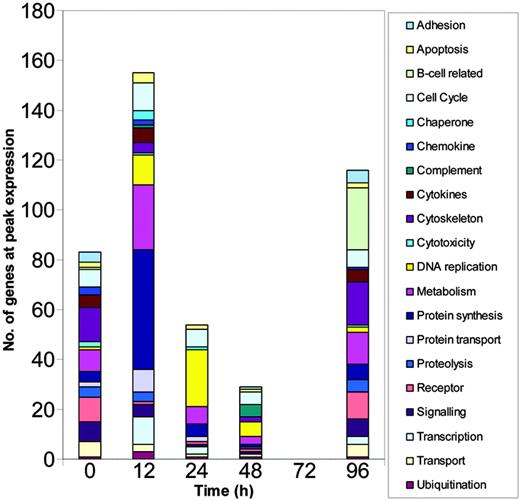
The cluster analysis showed that cellular functions followed distinct kinetic profiles during the early response and that expression of many genes peaked early. To examine the kinetics in more detail, we grouped genes according to their putative functions and calculated the number of genes in each functional category that reached peak expression at each time point (Figure 3). Although many genes did peak in their expression profile at 96 hours, most genes reached peak levels of expression much earlier than this with the greatest number reaching peak levels of expression at 12 hours.
Numbers of functionally related genes undergo changing profiles over the course of the primary response. Gene expression levels were calculated using MAS, RMA, and dChip, normalized using the associated algorithms and filtered using GeneSpring. Genes with more than 4-fold change in expression compared to baseline (0 hours) were selected. Only those genes selected irrespective of normalization method were further analyzed and these genes were grouped according to their known or putative functions. The analysis represents the cumulative numbers of genes at peak expression in each functional group at each time point.
Numbers of functionally related genes undergo changing profiles over the course of the primary response. Gene expression levels were calculated using MAS, RMA, and dChip, normalized using the associated algorithms and filtered using GeneSpring. Genes with more than 4-fold change in expression compared to baseline (0 hours) were selected. Only those genes selected irrespective of normalization method were further analyzed and these genes were grouped according to their known or putative functions. The analysis represents the cumulative numbers of genes at peak expression in each functional group at each time point.
Multiple patterns of gene expression within functional groups
In an alternative analysis method, we constructed gene lists of known importance in the CD8+ T-cell response grouped according to function. These groups encoded transcription factors, cell cycle genes, cytokines, chemokines and their receptors, and cytotoxic effectors. The lists were then applied to the microarray data and genes that demonstrated more than 2-fold change in expression compared with baseline at any point were selected (Table S5) and analyzed by K-means clustering (Figure 4).
Gene expression patterns of selected functional groups. Gene expression levels were calculated using MAS, RMA, and dChip, normalized using the associated algorithms, and filtered using GeneSpring. Genes with more than 2-fold change in expression compared to baseline (0 hours) were selected. Only those genes selected irrespective of normalization method were further analyzed. Genes were analyzed by functional group and cluster, which identified a number of differing patterns of gene expression in each group. The resulting groups of genes were therefore further refined into those related by function ([A] transcription factors, [B] cell cycle genes, [C] cytokines, [D] cytokine receptors, [E] chemokine receptors, and [F] cytotoxic effectors) and by change in expression during the primary response. Graphs represent the mean expression level of genes within each group as generated using RMA. Histograms indicate the number of genes contributing to each group.
Gene expression patterns of selected functional groups. Gene expression levels were calculated using MAS, RMA, and dChip, normalized using the associated algorithms, and filtered using GeneSpring. Genes with more than 2-fold change in expression compared to baseline (0 hours) were selected. Only those genes selected irrespective of normalization method were further analyzed. Genes were analyzed by functional group and cluster, which identified a number of differing patterns of gene expression in each group. The resulting groups of genes were therefore further refined into those related by function ([A] transcription factors, [B] cell cycle genes, [C] cytokines, [D] cytokine receptors, [E] chemokine receptors, and [F] cytotoxic effectors) and by change in expression during the primary response. Graphs represent the mean expression level of genes within each group as generated using RMA. Histograms indicate the number of genes contributing to each group.
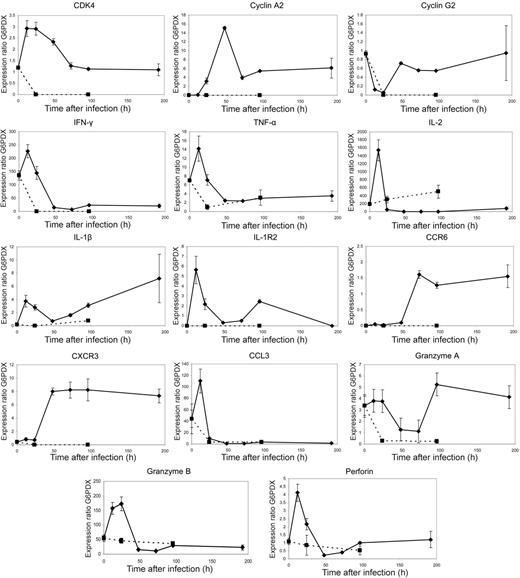
The majority of transcription factors were highly up-regulated early in the course of the primary response at 12 and 24 hours after stimulation (Figure 4A). These included the Myc gene, which is crucial in the control of cellular proliferation. The increased expression level of most transcription factors was transient and returned to baseline long before the proliferation peaked at 96 hours after stimulation.
Similar patterns of early transient expression were observed in genes regulating the cell cycle (Figure 4B). These included cyclins and cyclin-dependent kinases involved in the control of checkpoints during mitosis. Although there was little or no cell division up to 24 hours after stimulation (Figure 1A), our data indicated substantial early activity in these genes. From 48 hours, rapid cell division occurred, suggesting that genes expressed during this period, such as cyclin A2, were driving proliferation. A few genes, including cyclin G2, were highly expressed both in naive cells and at 96 hours. It has been suggested that these genes are involved in halting the cell cycle19-21 consistent with the observed pattern of expression.
Cytokines and chemokines were expressed primarily in one of 2 patterns (Figure 4C): either with an early peak or in a biphasic manner, with an initial fall and then high expression at 96 hours. Genes expressed early included IFN-γ, TNF-α, IL-2, CCL3, and CCL4, which are essential for the generation of a fully functional cytotoxic response. In all cases, peak expression occurred very early and gene expression was not sustained. Conversely, cell surface molecules, such as the cytokine and chemokine receptors, gradually increased in expression up to 96 hours (Figure 4D-E).
Of the cytotoxic effector molecules, granzymes A and B and perforin were seen in this analysis to be differentially expressed on primary stimulation (Figure 4F). Both granzyme B and perforin were most highly expressed early at 12 to 24 hours, although the level of expression of perforin was substantially less than that of the granzymes. Granzyme A was relatively highly expressed at baseline, an effect attributable to adoptive transfer, but showed further peaks at 24 and 96 hours.
Validation using quantitative PCR
To validate the microarray data and assay the expression of selected genes further, a number of representative genes were chosen for analysis using quantitative PCR. Fresh samples were taken from individuals at time points up to 96 hours as well as at 192 hours. Epitope-specific CD8+ T cells were purified as before. ALAS1 and G6PDX demonstrated no substantial change in expression over the duration of the microarray experiment and were therefore used as housekeepers.
The gene expression profiles of all selected genes closely matched the microarray data (Figure 5 and S3). Early expression of genes encoding cell cycle proteins, cytokines, chemokines, and cytotoxic effector molecules was confirmed. In addition, results from the 192-hour time point indicated that changes in gene expression after the 96-hour time point occurred more slowly. These data further supported the picture of a highly dynamic early transcriptional program.
PCR data. Quantitative PCR data correlate closely with microarray expression profiles emphasizing large scale changes in expression prior to the peak proliferative response at 96 hours after stimulation, whereas unstimulated controls do not show this pattern. F5 Rag−/− splenocytes were transferred by intravenous injection into C57BL/6 mice that were infected 24 hours later with recombinant vaccinia virus expressing NP366-374. Spleens were collected from these mice at time points up to 192 hours after infection and epitope-specific cells sorted by flow cytometry. Quantitative PCR of 14 selected genes was performed using the Roche Lightcycler system. Gene expression ratios were calculated using G6PDX as an housekeeping gene. Dotted lines represent quantitative PCR of the same genes in unstimulated controls. In the cases of CDK4, cyclin A2, cyclin G2, IL-1β IL-1R2, CXCR3, and CXCR6, gene expression ratios were very low at time point 0 and were not detectable in unstimulated control mice at any further time points. Graphs represent data from 3 mice per time point, with error bars showing the standard error.
PCR data. Quantitative PCR data correlate closely with microarray expression profiles emphasizing large scale changes in expression prior to the peak proliferative response at 96 hours after stimulation, whereas unstimulated controls do not show this pattern. F5 Rag−/− splenocytes were transferred by intravenous injection into C57BL/6 mice that were infected 24 hours later with recombinant vaccinia virus expressing NP366-374. Spleens were collected from these mice at time points up to 192 hours after infection and epitope-specific cells sorted by flow cytometry. Quantitative PCR of 14 selected genes was performed using the Roche Lightcycler system. Gene expression ratios were calculated using G6PDX as an housekeeping gene. Dotted lines represent quantitative PCR of the same genes in unstimulated controls. In the cases of CDK4, cyclin A2, cyclin G2, IL-1β IL-1R2, CXCR3, and CXCR6, gene expression ratios were very low at time point 0 and were not detectable in unstimulated control mice at any further time points. Graphs represent data from 3 mice per time point, with error bars showing the standard error.
Our data demonstrated changes in 153 genes in adoptively transferred cells prior to stimulation (see “Effects of adoptive transfer alone on gene expression”). Although this was a small proportion of the overall number of genes that were subsequently shown to be differentially expressed, concerns remained regarding any persistent changes and their effect on subsequent alterations caused by antigenic stimulation. For this reason, we also analyzed the expression of these genes in adoptively transferred cells from uninfected controls. These cells had not undergone any expansion (Figure 1C) and quantitative PCR failed to demonstrate any significant up-regulation of these genes over time (Figure 5 dotted line). Indeed, in contrast to the observations in infected mice, levels of expression of these genes tended to decrease gradually over time.
Acquisition of killing capacity
Our transcriptional data indicated substantial changes in gene expression occurring early in the course of the primary response. These included a number of genes encoding important effector molecules including perforin and granzyme B, both of which were up-regulated within 12 to 24 hours after stimulation (Figure 5). To correlate these transcriptional profiles with function, in vivo cytotoxicity assays were performed. Adoptive transfer and infection of recipient mice were carried out as previously and in vivo cytotoxicity assays were carried out at 0, 24, 48, and 96 hours after infection. Animals were monitored by blood and tissue sampling immediately, and at 2, 4, and 18 hours after transfer of labeled target cells (Figure 6A). Antigen-specific cell loss in blood, spleen, abdominal lymph nodes, lung, and liver were quantified by comparing the disappearance of target cells pulsed with the cognate peptide (NP366) or control peptide (PA224) (Figure 6B). Whereas only a small number of NP366-pulsed targets were lost over the course of the 18 hours in the unstimulated animals, a significantly higher number were destroyed even in animals infected just 24 hours previously. With the onset of cell division, the amount of specific killing increased accordingly and the most rapid loss of NP366-pulsed cells was seen in mice with the largest effector population (that is, the mice infected 96 hours previously).
CTL killing capacity is acquired between 24 and 48 hours after stimulation. F5 Rag−/− splenocytes were transferred by intravenous injection into C57BL/6 mice that were infected 24 hours later with recombinant vaccinia virus expressing NP366-374. In vivo cytotoxicity assays were performed using C57BL/6 splenocytes labeled with CFSE or CTO and coated with NP366-374 (R4: high concentration CFSE), PA224-233 (R3: low concentration CFSE), or medium alone (R2: CTO). Blood samples were taken 10 minutes, and 2, 4, and 18 hours following transfer of targets and the resulting lymphocytes analyzed using FACS. Spleen, abdominal lymph node, liver, and lung samples were taken at 2, 4, and 19 hours after transfer. Loss of NP366-pulsed CFSEhi cells was attributed to antigen-specific CTL lysis. Killing capacity was assessed in mice at 0, 24, 48, and 96 hours after infection. (A) Representative FACS plots of cytotoxicity assays at 96 hours after infection are shown. (B) Specific loss of NP366-pulsed target cells at each sampling time following injection of target cells in each compartment was calculated using the formula: 1 − (% NP366 pulsed/% PA224 pulsed) × 100. Graphs represent data from at least 3 mice with standard error shown. (C) Rate of specific killing over the period between each sampling point was calculated using the formula: 1− (% NP366 pulsed at assay time point/% PA224 pulsed at assay time point)/(% NP366 pulsed at previous assay time point/% PA224 pulsed at previous assay time point) × 100. The results are plotted on a timeline from the time of infection, with the results of each assay in their relative position. Graphs represent data from at least 3 mice used in each cytotoxicity assay with standard error shown. (D) Rates of specific killing were calculated as described for control animals infected with recombinant vaccinia expressing the OVA peptide.
CTL killing capacity is acquired between 24 and 48 hours after stimulation. F5 Rag−/− splenocytes were transferred by intravenous injection into C57BL/6 mice that were infected 24 hours later with recombinant vaccinia virus expressing NP366-374. In vivo cytotoxicity assays were performed using C57BL/6 splenocytes labeled with CFSE or CTO and coated with NP366-374 (R4: high concentration CFSE), PA224-233 (R3: low concentration CFSE), or medium alone (R2: CTO). Blood samples were taken 10 minutes, and 2, 4, and 18 hours following transfer of targets and the resulting lymphocytes analyzed using FACS. Spleen, abdominal lymph node, liver, and lung samples were taken at 2, 4, and 19 hours after transfer. Loss of NP366-pulsed CFSEhi cells was attributed to antigen-specific CTL lysis. Killing capacity was assessed in mice at 0, 24, 48, and 96 hours after infection. (A) Representative FACS plots of cytotoxicity assays at 96 hours after infection are shown. (B) Specific loss of NP366-pulsed target cells at each sampling time following injection of target cells in each compartment was calculated using the formula: 1 − (% NP366 pulsed/% PA224 pulsed) × 100. Graphs represent data from at least 3 mice with standard error shown. (C) Rate of specific killing over the period between each sampling point was calculated using the formula: 1− (% NP366 pulsed at assay time point/% PA224 pulsed at assay time point)/(% NP366 pulsed at previous assay time point/% PA224 pulsed at previous assay time point) × 100. The results are plotted on a timeline from the time of infection, with the results of each assay in their relative position. Graphs represent data from at least 3 mice used in each cytotoxicity assay with standard error shown. (D) Rates of specific killing were calculated as described for control animals infected with recombinant vaccinia expressing the OVA peptide.
The rate of loss of labeled cells that occurred in blood between sample points was calculated by comparing the frequency of surviving labeled cells at one sampling time with the frequency at the previous sampling time. The resulting profiles expressed the amount of antigen-specific cell loss that occurred over the course of each assay and between sampling intervals (Figure 6C).
All 4 profiles showed between 20% and 40% specific loss of NP366 peptide-treated cells within 2 hours of target cell transfer. In unstimulated animals, no further cell loss could be demonstrated, but in animals that had been stimulated with antigen 96 hours earlier around 40% of the remaining targets were lost within the next 2 hours and almost 100% had been destroyed by the end of the assay (18 hours). The profiles at 24 and 48 hours after infection were similar to each other and demonstrated significantly increased killing. In both cases, around 50% of remaining targets were lost in the last 14 hours of the assay. In vivo killing assays performed in control animals infected with rVV-OVA showed no accentuation of antigen-specific cell loss, indicating that the cell loss induced by priming was indeed antigen-specific and was not due to bystander activation or other nonspecific effects of the rVV (Figure 6D).
We attribute the excess loss of NP366-pulsed cells to CD8+ T cell-mediated lysis. If this interpretation is correct, this implies that acquisition of CTL killing capacity occurred between 24 and 48 hours after infection. During this period, there was no demonstrable increase in the overall number of epitope-specific CD8+ effector cells (Figure 1), indicating that any changes in cytotoxicity could be attributable only to changes in transcription rather than an increase in effector frequency. These functional data correlated closely with the rapid up-regulation of cytolytic genes by 48 hours after infection (Figure 5).
Discussion
Programming by antigen encounter early in the primary CD8+ T-cell response has been demonstrated in several systems and suggests that important changes in T cells occur within 24 hours of stimulation.5-8,22-24 Here we have shown that the development of a primary CD8+ T-cell response is not characterized by gradual or persistent increases in gene expression but more by transient bursts of transcription that occur soon after antigen contact. In the case of the cytotoxicity molecules, we demonstrated that early gene expression correlated with the acquisition of killing capacity 24 hours after stimulation. This preceded the onset of clonal expansion, which occurred at approximately 48 hours after stimulation.
Microarrays have made it possible to quantify transcriptional changes in multiple cellular pathways simultaneously. A recent study focused on the differential expression of genes between established populations of naive, effector, and memory CD8+ T cells.12 However, by comparing naive cells derived directly from transgenic mice and day 8 effectors, the dynamics of expression of those genes identified remained unclear.
Our preliminary experiments showed that adoptive transfer alone could lead to altered transcription in transferred cells. The impact on gene expression of adoptive transfer has not previously been demonstrated although many studies assume that any changes induced will have subsided by the time of stimulation 24 hours after the transfer of cells. We were concerned that these changes might persist and affect our interpretation of further transcriptional data. However, quantitative PCR of 14 genes, including several that were up-regulated following adoptive transfer alone, confirmed that these changes were transient and that no further changes occurred in the absence of epitope-specific stimulation.
To maintain the stringency and comparability of our results, we chose to use mRNA from adoptively transferred nonstimulated cells as our baseline control. A direct comparison between these and the effector populations revealed differentially expressed genes in numbers that were similar to previously published data, with 498 genes differentially expressed more than 2-fold. However, the more detailed time course showed that actually over 7 times this number of genes were differentially expressed over the course of the primary response. This result indicated that a vast number of genes underwent transient changes in expression within the first 96 hours that were undetectable at the end of the period.
The differential expression of genes occurred in 2 main phases: an early phase of transient gene expression that peaked at 12 to 24 hours after stimulation, followed by a second wave of increasing expression that persisted beyond 96 hours. Quantitative analysis of the epitope-specific response had revealed an apparent decrease in number of transferred CTLs detected in the spleen by 24 hours following transfer, which might suggest that the biphasic pattern was due to the analysis of 2 separate subpopulations generated during differential migration or cell death. However, analysis of the ungated splenocyte population revealed that these cells had not, in fact, disappeared but had moved to the “monocyte” gate. This is consistent with recent work suggesting that T-cell priming occurs in 3 distinct phases: an initial phase involving short encounters with dendritic cells (DCs), a second phase involving formation of stable conjugates, and a third phase characterized by less stable contacts and the onset of proliferation.25 Our gene expression analysis was performed on antigen-specific CD8+ T cells isolated from the lymphocyte gate (to prevent contamination with other cell types) and so did not include CTLs forming stable conjugates with DCs. We suggest therefore that the gene expression profiles at the 12-hour time point result from the first phase, that the apparent low frequency of CTLs within the lymphocyte gate at the 24-hour time point reflects their stable engagement with DCs, and that the gene expression profiles at later time points occur following the end of this second phase. Our results support the idea that the early short-lived CTL contacts occurring during phase 1 induce early and important cellular events.
The detection of unexpected changes in expression of genes, such as CD19 and immunoglobulins, not previously reported in T cells is a commonly reported issue in microarray studies. Although it has been suggested that transcription of some genes may occur without translation to the protein level, the most likely possibility in this case is minor contamination (< 5%) of samples during sorting. Plasma cells express extremely high levels of mRNA for immunoglobulins and hence changes in expression of immunoglobulin genes within microarray studies must be treated with extreme skepticism because they likely reflect only tiny numbers of contaminating cells. Fluctuations in levels of immunoglobulins have been seen in previous T-cell microarray studies and, in those cases, were also not felt to have been relevant.12 Overall comparison of our expression data on immunoglobulin gene expression with results obtained from other groups studying activated B cells indicates that the absolute mRNA expression level for immunoglobulins was very low in our sample, consistent with low level (< 5%) contamination. Conversely, genes up-regulated in our data such as granzymes A and B and perforin are not up-regulated in other cell types supporting the validity of our data.26
Our results strongly implied that changes in T-cell function occurred earlier than previously anticipated. Major changes in expression of mRNA for proteins involved in regulating the cell cycle were seen within 48 hours of antigen encounter and correlated with the onset of cell division from 48 hours after stimulation, the time lag to proliferation being consistent with that seen in other systems.27-30 Genes encoding cytotoxic proteins were also shown to be up-regulated at an early stage. These were well validated using quantitative PCR and we therefore aimed to determine whether these transcriptional changes correlated with acquisition of cytolytic function.
Using in vivo cytotoxicity assays we observed that transferred but unstimulated CD8+ T cells had low killing capacity, which was not sustained. However, between 24 and 48 hours after infection these cells acquired significant specific killing before any measurable expansion of the effector population. Although the rate of acquisition of cytolytic capacity and the killing rates themselves were substantially slower than in a recall response,31 these experiments showed that during the primary response CTLs develop killing capacity within 24 hours of antigen contact, substantially earlier than has previously been suggested.32
The rapidity with which naive CD8+ T cells acquire cytolytic capacity in our system is novel and surprising. There has traditionally been a dichotomy between cells of the innate immune system, which act early, and those of the acquired immune system, which act later. However, our data suggest that the cause of this distinction is not delayed differentiation of CD8+ T cells, because priming of naive CTLs induces rapid up-regulation of cytolytic genes and those cells that do exist are able to kill. Rather it is the relatively small number of effector cells present during the early period that prevents larger-scale activity by CTLs. The rate-limiting step in the development of an active CD8+ T-cell response is therefore proliferation and not acquisition of function.
Authorship
Contribution: C.C. performed research, analyzed data, and wrote the paper; A.G.H. analyzed data; V.C. contributed vital reagents and advice; A.J.M. contributed vital reagents and advice; C.R.B. contributed to the study design; and M.F.C.C. designed the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christopher Chiu, Department of Immunology, Wright-Fleming Institute, Imperial College, Norfolk Pl, London W2 1PG, United Kingdom; e-mail: c.chiu@imperial.ac.uk.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by the Wellcome Trust, United Kingdom. Our thanks to Eric O'Connor for assistance with FACS sorting and Helen Fletcher, Laurence Game, and members of the MRC Microarray Centre, Hammersmith Hospital, London for their advice and assistance.

![Figure 4. Gene expression patterns of selected functional groups. Gene expression levels were calculated using MAS, RMA, and dChip, normalized using the associated algorithms, and filtered using GeneSpring. Genes with more than 2-fold change in expression compared to baseline (0 hours) were selected. Only those genes selected irrespective of normalization method were further analyzed. Genes were analyzed by functional group and cluster, which identified a number of differing patterns of gene expression in each group. The resulting groups of genes were therefore further refined into those related by function ([A] transcription factors, [B] cell cycle genes, [C] cytokines, [D] cytokine receptors, [E] chemokine receptors, and [F] cytotoxic effectors) and by change in expression during the primary response. Graphs represent the mean expression level of genes within each group as generated using RMA. Histograms indicate the number of genes contributing to each group.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/3/10.1182_blood-2006-03-011643/5/m_zh80030707530004.jpeg?Expires=1767846641&Signature=nhAnCMwEEkF802ZVLcXLlogBcit9zCiooDp374gNh88UlPguIyM~ecfBiZHlzDKuxzDw7hY0GiDiPbRO90yX3fzddZJuhBjfIXp7M6P3ZGVrwe1qtuNCQEPaU9N~6-Hue7U379GQAqqxzdr2wWs5aeVKDycUVdsQPvTk2nAKgGP9-gsCBZj7jadOpol3f1dMRW4aU4rMhYY0fE6vLYgcEmg29L2FPfgWkf2AFHVETsmuAQbRZM14p1jhp4tYrYv0rWQn47qvG9cnXDRJfYNWVc~0f53vPeSRLKcbpfMlHGixw5O30evUY02Y2NXmKcQ~Ru0AWT6lyokRHOXX0EUnWw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal