Abstract
The RAS proteins undergo farnesylation of a carboxyl-terminal cysteine (the “C” of the carboxyl-terminal CaaX motif). After farnesylation, the 3 amino acids downstream from the farnesyl cysteine (the -aaX of the CaaX motif) are released by RAS-converting enzyme 1 (RCE1). We previously showed that inactivation of Rce1 in mouse fibroblasts mislocalizes RAS proteins away from the plasma membrane and inhibits RAS transformation. Therefore, we hypothesized that the inactivation of Rce1 might inhibit RAS transformation in vivo. To test this hypothesis, we used Cre/loxP recombination techniques to simultaneously inactivate Rce1 and activate a latent oncogenic K-RAS allele in hematopoietic cells in mice. Normally, activation of the oncogenic K-RAS allele in hematopoietic cells leads to rapidly progressing and lethal myeloproliferative disease. Contrary to our hypothesis, the inactivation of Rce1 actually increased peripheral leukocytosis, increased the release of immature hematopoietic cells into the circulation and the infiltration of cells into liver and spleen, and caused mice to die more rapidly. Moreover, in the absence of Rce1, splenocytes and bone marrow cells expressing oncogenic K-RAS yielded more and larger colonies when grown in methylcellulose. We conclude that the inactivation of Rce1 worsens the myeloproliferative disease caused by oncogenic K-RAS.
Introduction
Activating mutations in RAS genes result in constitutive signaling of the RAS proteins and are implicated in the pathogenesis of many human cancers, including several hematologic malignancies.1,2 For example, activating mutations in RAS are present in as many as 44% of patients with acute myeloid leukemia.3 Hematologic malignancies also occur when RAS signaling is increased, as observed in diseases such as neurofibromatosis (NF1) in which a genetic abnormality develops in a RAS-interacting protein.4
The RAS proteins undergo several posttranslational processing steps, beginning with the farnesylation of the cysteine residue (the “C” of the CaaX motif) by protein farnesyltransferase (FTase). After this “lipidation” step, the last 3 amino acids of the protein (the -aaX of the CaaX motif) are released by RAS-converting enzyme 1 (RCE1), and the newly exposed farnesyl cysteine is methylated by isoprenyl cysteine carboxyl methyltransferase (ICMT). Several RAS isoforms (but not K-RAS4B) are also palmitoylated at nearby cysteine residues.5
One strategy to block RAS-induced oncogenic transformation is to mistarget the RAS proteins within cells by inhibiting the enzymes that carry out the posttranslational modifications of the RAS proteins. Early preclinical trials of FTase inhibitors (FTIs) against RAS-induced tumors demonstrated significant efficacy with low toxicity.6 However, in clinical trials of human solid tumors, FTIs have not met the high expectations, in part because RAS proteins can be isoprenylated by a related isoprenyl transferase, geranylgeranyl transferase type I (GGTase-I), in the presence of an FTI.7
We have evaluated the possibility of inhibiting RCE1 as a strategy to prevent RAS-induced oncogenic transformation. One potential advantage of this strategy is that RCE1 inhibition would interfere with the processing of the RAS proteins regardless of whether they are farnesylated or geranylgeranylated. In addition, this strategy would not be expected to cause significant toxicity in vivo. Indeed, we have inactivated Rce1 in the liver, spleen, and bone marrow of mice and not observed significant adverse effects.8 Ayiagari et al9 showed that Rce1-deficient fetal liver cells are capable of rescuing hematopoiesis in lethally irradiated mice.
Several lines of investigation suggest that the inhibition of RCE1 would inhibit RAS-induced oncogenic transformation. First, the inactivation of Rce1 mislocalized the RAS proteins and reduced cell proliferation and the anchorage-independent growth of RAS-transformed fibroblasts in soft agar and nude mice.8,10 Second, in the absence of Rce1, skin carcinoma cells grew slowly and were highly sensitive to the effects of an FTI.8 Third, several potent RCE1 inhibitors have been developed,11,12 and 2 of them reduced the anchorage-independent growth of K-RAS–transformed cells in vitro.13,14 However, nothing is known about the effects of inhibiting RCE1 on the growth of RAS-induced malignancies in vivo.
In this study, we determined if the inactivation of Rce1 in mice would inhibit the development of K-RAS–induced myeloproliferative disease (MPD) in vivo. To accomplish this, we used Cre recombinase to simultaneously inactivate the expression of Rce1 and activate the expression of oncogenic K-RASG12D in hematopoietic cells.
Materials and methods
Breeding mice for in vivo experiments
Mice with a conditional Rce1 knockout allele (Rce1fl) have been described.8 Mice with a Kras2LSL allele15 have an activating mutation (G12D) in the Kras2 gene and a “floxed” transcriptional terminator sequence upstream in the promoter (LSL; loxP-STOP-loxP). Cre expression results in the removal of the STOP cassette, which turns on the expression of K-RASG12D. For our experiments, Rce1fl/fl mice with a Kras2LSL allele were bred with Rce1fl/+ mice harboring an interferon-inducible Mx1-Cre transgene16 to generate Rce1fl/flKras2LSL/+Mx1-Cre mice. In those mice (hereafter designated Rce1fl/flKLSLM), Cre expression simultaneously inactivated Rce1 expression and activated the expression of K-RASG12D in bone marrow cells. These cells are designated Rce1Δ/ΔKG12DM. For experimental controls, we used mice harboring a single mutant Rce1 allele (Rce1fl/+KLSLM mice); in those mice, Cre inactivates one Rce1 allele and reduces Rce1 expression by 50%. The mice were maintained on a 129/Sv and C57BL/6 mixed genetic background, and the Rce1fl/flKLSLM and Rce1fl/+KLSLM mice were littermates. Animal procedures were approved by the animal research ethics committee in Gothenburg.
Genotyping
Mice were genotyped by polymerase chain reaction (PCR) amplification of genomic DNA obtained by tail biopsy. The Rce1fl allele was detected with forward primer 5′-GCTTTTGGAAAGAACAGGGGCC-3′ and reverse primer 5′-CTCACCTCCAGTTGCTGCTCATC-3′. This PCR reaction yields a 350-bp fragment from the Rce1fl allele and a 270-bp fragment from the Rce1+ allele. The Mx1-Cre transgene was detected with forward primer 5′-GAGCTCCATTCATGTGTGGT-3′ and reverse primer 5′-CTAGAGCCTGTTTTGCACGTTC-3′; the DNA product measured 1009 bp. The Kras2LSL allele was detected with forward primer 5′-CCTTTACAAGCGCACGCAGACTGTAGA-3′ and reverse primer 5′-AGCTAGCCACCATGGCTTGAGTAAGTCTGCA-3′; the amplified fragment measured 600 bp. The presence of the activated Kras2G12D allele was detected with forward primer 5′-GGGTAGGTGTTGGGATAGCTG-3′ and reverse primer 5′-TCCGAATTCAGTGACTACAGATGTACAGAG-3′; the amplified fragment measured 320 bp.
Quantifying the efficiency of Cre-induced recombination of the Rce1fl allele
DNA was prepared from spleen and bone marrow cells and used for real-time quantitative PCR (QPCR) with Power SYBR Green PCR Master Mix on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). The following primer pairs were used: Rce1, 5′-CGAGTAAATCTGTGGGAGAGG-3′ and 5′-CGGTGCAATAACTTGGTTTC-3′; Pggt1b (used as a control), 5′-CATGTCGTGCTGCTTTCATA-3′ and 5′-CAGTTTACCCATCAGGCACA-3′.
Injection of polyinosinic–polycytidylic acid and monitoring
The Mx1-Cre transgene was activated with intraperitoneal injections of 400 μg polyinosinic–polycytidylic acid (pI-pC; Sigma, St Louis, MO) 21 days after birth (at weaning) once every other day, for a total of 4 injections. Blood was drawn from a tail vein before the first pI-pC injection (time point 0) and once per week for the duration of the experiment. The blood was analyzed with a Hemavet 950FS cell counter (Drew Scientific, Oxford, CT) and by manual differential counts of May-Grünwald-Giemsa–stained smears by 2 trained observers blinded to genotype (200 white blood cells per slide were evaluated). At 3 and 5 weeks, groups of mice were killed and tissues were harvested.
Histology
Tissues were fixed in 4% PBS-buffered formalin, dehydrated in 70% to 100% ethanol, and cleared in xylene. Tissues were embedded in paraffin and 4- to 5-μm sections were stained with hematoxylin and eosin. Sections were viewed and photographed with a light microscope (Axiocam HR digital camera; Axioplan 2; Carl Zeiss, Oberkochen, Germany) and analyzed with Axiovision AC software version 4.3. Cytospin preparations of bone marrow cells were stained with May-Grünwald-Giemsa. Cells of the monocytic and granulocytic lineages were identified by staining for nonspecific and specific esterases, respectively, using α-naphthyl butyrate and naphthol AS-D chloroacetate as substrates.
Colony assays
Spleen cells (105) and bone marrow cells (2 × 104) harvested 5 weeks after pI-pC injections of Rce1fl/flKLSLM, Rce1fl/+KLSLM, and wild-type mice were seeded in duplicate wells in methylcellulose medium (MethoCult M3234; StemCell Technologies, Vancouver, BC, Canada) in the absence of growth factors. Ten days later, numbers and sizes of colonies were scored. Colonies were viewed and photographed with a Leica DMIL light microscope and a Leica DC200 digital camera (Leica, Wetzlar, Germany). Genomic DNA from individual colonies was genotyped by PCR (to detect the activated Kras2G12D allele and the excision of Rce1). Cytospin preparations of cells in individual colonies were stained with May-Grünwald-Giemsa and analyzed by light microscopy (Axioplan 2).
Fluorescence-activated cell sorting
Peripheral blood mononuclear cells, bone marrow cells, and cells from methylcellulose colonies were incubated with antibodies against cell-surface antigens (CD11b [catalog no. 550993], CD14 [catalog no. 553739], CD45 [catalog no. 557695], and CD117 [catalog no. 553355]; PharMingen, San Diego, CA) and were analyzed in a FACSaria (BD Biosciences, San Jose, CA). Data were analyzed with CellQuest software (BD Biosciences).
Isolation of mouse embryonic fibroblasts and cell proliferation assays
Mouse embryonic fibroblasts (MEFs) were isolated from Rce1fl/flKLSL and Rce1fl/+KLSL embryos (lacking the Mx1-Cre transgene) at embryonic day 13.5 (E13.5) to E16.5. Experiments were performed on primary (passages 0-3) and on spontaneously immortalized MEFs (passages 15-35). Cells (5 × 105) were seeded onto 60-mm dishes in the presence of adenovirus encoding Cre or β-gal (109 pfu/mL AdRSVCre and AdRSVnlacZ, respectively). Cre-adenovirus treatment of Rce1fl/flKLSL cells produced Rce1Δ/ΔKG12D cells that expressed endogenous K-RASG12D but lacked Rce1 expression; Cre-adenovirus treatment of Rce1fl/+KLSL cells produced Rce1Δ/+KG12D cells that expressed endogenous K-RASG12D and lacked 50% of Rce1 expression. Cells (2 × 104) of each genotype were then seeded in triplicate wells in 12-well plates. At various time points, cells were trypsinized and counted in a cell counter (NucleoCounter; ChemoMetec, Allerød, Denmark). DNA was extracted from a portion of the cells, and the excision of the Rce1 allele and the appearance of the activated Kras2G12D allele were assessed by PCR.
Western blots
Tissue pieces (50-150 mg) were lysed in ice-cold buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 1% NP-40, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 2 mM orthovanadate, and the Complete Mini protease inhibitor cocktail [Roche, Basel, Switzerland]). Lysates were homogenized, sonicated, and centrifuged at 20 000g for 20 minutes, and equal amounts of total protein of the supernatant were size-fractionated on 10% to 20% sodium dodecylsulfate polyacrylamide gels (Criterion; Bio-Rad, Hercules, CA). The proteins were transferred onto nitrocellulose membranes and incubated with antibodies recognizing phosphorylated ERK1/2 (9106), phosphorylated AKT (9271), total ERK (9102; Cell Signaling, Danvers, MA), and p21CIP1 (sc-6246; Santa Cruz Biotechnology, Santa Cruz, CA). Protein bands were visualized with a horseradish peroxidase–conjugated secondary antibody (sc-2314 and sc-2313; Santa Cruz) and the Enhanced Chemiluminescence kit (Amersham, Little Chalfont, Buckinghamshire, United Kingdom).
Statistical analyses
Data are plotted as mean plus or minus SEM. Differences in the concentrations and percentages of white blood cells, the colony-forming ability of hematopoietic cells, and the proliferation of cells in culture were determined with Student t test; survival was assessed by the Mann-Whitney U test.
Results
Inactivation of Rce1 exacerbates K-RAS–induced myeloproliferative disease
To determine whether Rce1 deficiency would inhibit the development of MPD, we monitored pI-pC–injected Rce1fl/flKLSLM mice and control Rce1fl/+KLSLM mice. The pI-pC–injected Rce1fl/+KLSLM mice developed progressive leukocytosis (Figure 1A) with an increased percentage of myeloid cells (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The pI-pC–injected Rce1fl/flKLSLM mice exhibited an increase in white blood cell counts compared with the Rce1fl/+KLSLM mice; this increase was statistically significant 2 weeks after the pI-pC injections (Figure 1A). At 5 weeks, the mean white blood cell count in Rce1fl/flKLSLM mice was 106 ± 12 × 109/L (n = 15) compared with 39 ± 3 × 109/L (n = 11) in Rce1fl/+KLSLM mice (P < .001). The increased proliferation of white blood cells in the pI-pC–injected Rce1fl/flKLSLM mice was associated with reduced survival (Figure 1B; P =.035). We concluded that Rce1 deficiency accelerated the development of K-RASG12D-induced MPD and reduced survival.
Acceleration of K-RAS–induced MPD in mice with Rce1 deficiency. Groups of Rce1fl/flKLSLM mice and control Rce1fl/+KLSLM mice were injected at weaning with pI-pC to induce MPD. (A) The concentration of white blood cells was elevated in Rce1fl/flKLSLM compared with Rce1fl/+KLSLM mice with MPD. Statistically significant changes at each time point are indicated. *P < .05; **P < .01; and ***P < .001; n = 11-27 per time point, except for the values at 42 days (n = 3 [Rce1fl/flKLSLM] and n = 5 (Rce1fl/+KLSLM). (B) Kaplan-Meier curve demonstrating reduced survival of pI-pC–injected Rce1fl/flKLSLM mice (median survival, 40 days; n = 12) compared with Rce1fl/+KLSLM mice (median survival, 54 days; n = 5).
Acceleration of K-RAS–induced MPD in mice with Rce1 deficiency. Groups of Rce1fl/flKLSLM mice and control Rce1fl/+KLSLM mice were injected at weaning with pI-pC to induce MPD. (A) The concentration of white blood cells was elevated in Rce1fl/flKLSLM compared with Rce1fl/+KLSLM mice with MPD. Statistically significant changes at each time point are indicated. *P < .05; **P < .01; and ***P < .001; n = 11-27 per time point, except for the values at 42 days (n = 3 [Rce1fl/flKLSLM] and n = 5 (Rce1fl/+KLSLM). (B) Kaplan-Meier curve demonstrating reduced survival of pI-pC–injected Rce1fl/flKLSLM mice (median survival, 40 days; n = 12) compared with Rce1fl/+KLSLM mice (median survival, 54 days; n = 5).
K-RAS–induced MPD is associated with accumulated immature myeloid cells and increased tissue infiltration in the setting of Rce1 deficiency
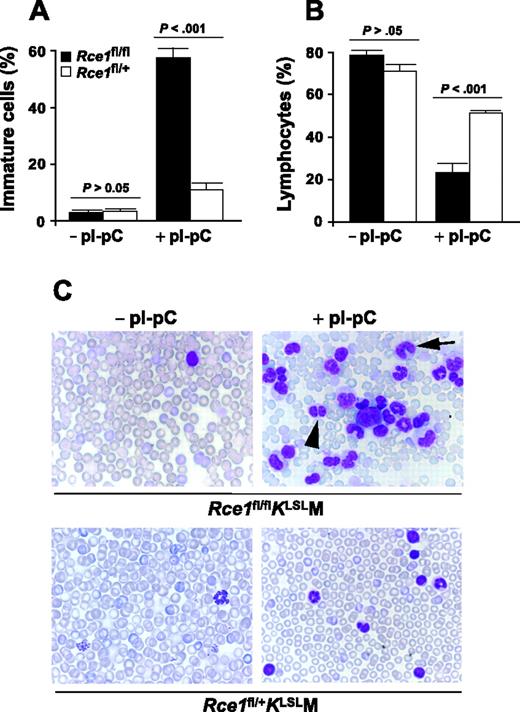
In the pI-pC–treated control Rce1fl/+KLSLM mice with MPD, the percentage of immature myeloid cells was relatively minor (11% vs 3% before pI-pC injections) (Figure 2; Table S1). In contrast, there was a dramatic increase in the percentage of immature myeloid cells in pI-pC–treated Rce1fl/flKLSLM mice (Figure 2A; Table S1). Overall, 58% of white blood cells in these mice were immature. There was a proportionate reduction in the percentage of lymphocytes in the 2 groups of mice (Figure 2B). Thus, the increase in white blood cell counts in pI-pC–treated Rce1fl/flKLSLM mice relative to control mice with MPD was caused by an accumulation of immature myeloid cells.
Increased release of immature K-RASG12D–expressing myeloid cells in the setting of Rce1 deficiency. (A) White blood cells were evaluated in blood smears from Rce1fl/flKLSLM (▪; n = 6) and Rce1fl/+KLSLM mice (□; n = 6) harvested before and after pI-pC injections. Immature cells (myeloblasts, promyelocytes, promonocytes, myelocytes, metamyelocytes, band cells, pelgeroid cells) (A) and lymphocytes (B) are plotted as percentages of total white blood cells. Differences between Rce1fl/flKLSLM and Rce1fl/+KLSLM mice were evident 3 weeks after pI-pC injection, and the proportions did not change over time. Thus, data from 3, 5, and 7 weeks were combined. (C) Photographs of blood smears from before and 5 weeks after pI-pC injections (magnification: 100×/1.30 NA oil-immersion objective lens). (Top right panel) Arrowhead, pelgeroid cell; arrow, band cell; large cell in center, myeloblast.
Increased release of immature K-RASG12D–expressing myeloid cells in the setting of Rce1 deficiency. (A) White blood cells were evaluated in blood smears from Rce1fl/flKLSLM (▪; n = 6) and Rce1fl/+KLSLM mice (□; n = 6) harvested before and after pI-pC injections. Immature cells (myeloblasts, promyelocytes, promonocytes, myelocytes, metamyelocytes, band cells, pelgeroid cells) (A) and lymphocytes (B) are plotted as percentages of total white blood cells. Differences between Rce1fl/flKLSLM and Rce1fl/+KLSLM mice were evident 3 weeks after pI-pC injection, and the proportions did not change over time. Thus, data from 3, 5, and 7 weeks were combined. (C) Photographs of blood smears from before and 5 weeks after pI-pC injections (magnification: 100×/1.30 NA oil-immersion objective lens). (Top right panel) Arrowhead, pelgeroid cell; arrow, band cell; large cell in center, myeloblast.
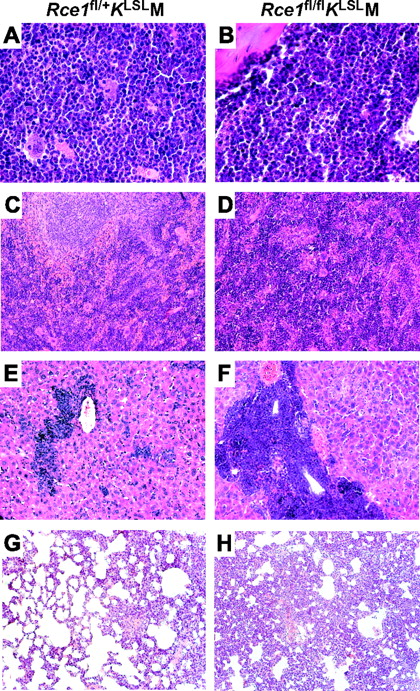
To determine whether the enhanced production of immature white blood cells was associated with increased myeloid infiltration into vital tissues, we conducted histologic studies. Five weeks after pI-pC injections, increased cellularity was noted in the bone marrow in Rce1fl/+KLSLM and Rce1fl/flKLSLM mice (Figure 3A-B). Fluorescence-activated cell sorter (FACS) analyses and double-esterase staining of bone marrow demonstrated a greater proportion of CD45+/CD11b+/CD14− cells expressing specific esterase, consistent with a granulocytic expansion. The proportion of bone marrow cells expressing specific esterase increased from 25.0% ± 3.2% in wild-type mice to 50.8% ± 1.5% in the Rce1fl/+KLSLM mice (P =.007) and further increased to 73.5% ± 1.3% in the Rce1fl/flKLSLM mice (P = .01 vs Rce1fl/+KLSLM mice; P = .001 vs wild-type). The proportion of CD45+/CD117+ bone marrow cells increased from 4.5% in wild-type mice to 8.1% in Rce1fl/+KLSLM mice and 9.3% in Rce1fl/flKLSLM mice.
Tissue infiltration of K-RASG12D–expressing myeloid cells. Hematoxylin and eosin–stained sections of tissues from Rce1fl/+KLSLM (left) and Rce1fl/flKLSLM mice (right) harvested 5 weeks after pI-pC injections. (A-B) Bone marrow. (C-D) Spleen. (E-F) Liver. (G-H) Lung. Magnification: 63×/1.40 (A,B); 20×/0.50 (C-H).
Tissue infiltration of K-RASG12D–expressing myeloid cells. Hematoxylin and eosin–stained sections of tissues from Rce1fl/+KLSLM (left) and Rce1fl/flKLSLM mice (right) harvested 5 weeks after pI-pC injections. (A-B) Bone marrow. (C-D) Spleen. (E-F) Liver. (G-H) Lung. Magnification: 63×/1.40 (A,B); 20×/0.50 (C-H).
The Rce1fl/+KLSLM mice exhibited mild to moderate infiltration of leukocytes into the liver and spleen, effacement of splenic architecture with extramedullary hematopoiesis, and adenoma formation in the lung (Figure 3C,E,G). In the pI-pC–treated Rce1fl/flKLSLM mice, severe infiltration of leukocytes into the liver occurred with congestion of central veins, swelling of hepatocytes, and areas of necrosis (Figure 3F). In addition, complete effacement of splenic architecture and adenoma formation with diffuse hyperplasia in the lung—likely contributing factors in the rapid demise of the Rce1fl/flKLSLM mice—(Figure 3D, H) developed.
To determine the efficiency of Cre-induced recombination in hematopoietic tissues of pI-pC–injected Rce1fl/flKLSLM, we performed quantitative PCR of genomic DNA. Efficiency was 95% ± 1.6% in spleen and 90% ± 2.2% in bone marrow (n = 3). Thus, both alleles of Rce1 were inactivated in most cells in those tissues.
To determine the consequences of inactivating Rce1 in K-RASG12D–expressing tissues on downstream signaling molecules, we performed Western blot analysis. Levels of phosphorylated ERK1/2 in the spleens and livers of pI-pC–injected Rce1fl/+KLSLM and Rce1fl/flKLSLM mice were increased compared with control mice without MPD (Figure S1). Levels of phosphorylated AKT, p21CIP, and total ERK1/2 were not different.
Enhanced colony-forming ability of Rce1-deficient, K-RASG12D–expressing hematopoietic cells
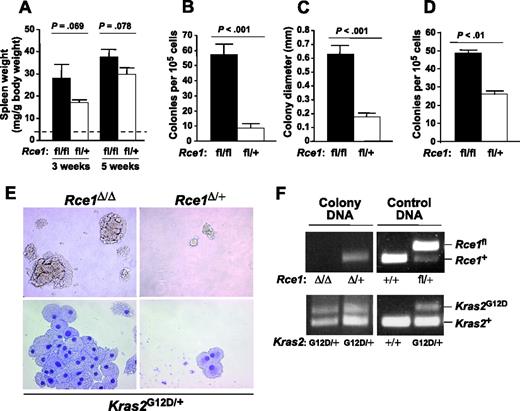
Spleen weights in Rce1fl/flKLSLM and Rce1fl/+KLSLM mice were similar 3 and 5 weeks after pI-pC injections (Figure 4A). However, given the more advanced histologic findings in spleens from Rce1fl/flKLSLM mice (Figure 3D), we assessed the ability of splenocytes from the mice to form colonies in methylcellulose. Rce1fl/+KLSLM splenocytes were capable of forming colonies in the absence of growth factors, though the colonies were small (mean number of colonies, 9 ± 3/105 cells; mean size, 0.19 ± 0.02 mm; Figure 4B-C). In the absence of Rce1, there was a 6.6-fold increase in colony number and a 3.4-fold increase in colony size (Figure 4B-C). With bone marrow cells, there was a 2-fold increase in colony number (Figure 4D). Hematopoietic cells from wild-type mice did not form colonies in the absence of growth factors. Splenocyte colonies from Rce1fl/+KLSLM and Rce1fl/flKLSLM spleens were composed of CD45+/CD11b+/CD13+/CD14− cells that morphologically resembled macrophages (Figure 4E). PCR amplification of genomic DNA from individual colonies demonstrated that the “floxed” Rce1 gene was excised and that the Kras2G12D allele was activated (Figure 4F).
Enhanced colony-forming ability of Rce1-deficient K-RASG12D–expressing hematopoietic cells. (A) Spleen weights of Rce1fl/flKLSLM mice (▪) and Rce1fl/+KLSLM mice (□) 3 and 5 weeks after pI-pC injections (n = 6 and n = 11 for Rce1fl/flKLSLM mice and n = 5 and n = 8 for Rce1fl/+KLSLM mice at 3 and 5 weeks, respectively). Dashed line indicates spleen weights of wild-type mice (4 ± 0.9 mg/g body weight; n =6). (B-C) Growth factor–independent colony growth of splenocytes from Rce1fl/flKLSLM (n = 3) and Rce1fl/+KLSLM (n = 3) mice. (B) Colony number. (C) Colony size. (D) Growth factor–independent colony growth of bone marrow cells (n = 2). (E, top) Photographs showing Rce1Δ/ΔKG12DM and Rce1Δ/+KG12DM splenocyte colonies from a typical experiment in panels B and C (magnification: 20×/0.30 NA objective lens). (Bottom) May-Grünwald-Giemsa–stained cytospins of individual colonies (magnification: 63×/1.40 NA objective lens). (F) PCR amplification of genomic DNA from individual colonies to detect the Rce1fl and Rce1+ alleles (top) and the Kras2+ and Kras2G12D alleles (bottom). Lane 1, Rce1-deficient Kras2G12D colony; lane 2, heterozygous Rce1-deficient Kras2G12D colony; lanes 3-4, control DNA from mouse tails. The same result was found in 5 additional colonies from spleens of Rce1fl/flKLSLM and Rce1fl/+KLSLM mice.
Enhanced colony-forming ability of Rce1-deficient K-RASG12D–expressing hematopoietic cells. (A) Spleen weights of Rce1fl/flKLSLM mice (▪) and Rce1fl/+KLSLM mice (□) 3 and 5 weeks after pI-pC injections (n = 6 and n = 11 for Rce1fl/flKLSLM mice and n = 5 and n = 8 for Rce1fl/+KLSLM mice at 3 and 5 weeks, respectively). Dashed line indicates spleen weights of wild-type mice (4 ± 0.9 mg/g body weight; n =6). (B-C) Growth factor–independent colony growth of splenocytes from Rce1fl/flKLSLM (n = 3) and Rce1fl/+KLSLM (n = 3) mice. (B) Colony number. (C) Colony size. (D) Growth factor–independent colony growth of bone marrow cells (n = 2). (E, top) Photographs showing Rce1Δ/ΔKG12DM and Rce1Δ/+KG12DM splenocyte colonies from a typical experiment in panels B and C (magnification: 20×/0.30 NA objective lens). (Bottom) May-Grünwald-Giemsa–stained cytospins of individual colonies (magnification: 63×/1.40 NA objective lens). (F) PCR amplification of genomic DNA from individual colonies to detect the Rce1fl and Rce1+ alleles (top) and the Kras2+ and Kras2G12D alleles (bottom). Lane 1, Rce1-deficient Kras2G12D colony; lane 2, heterozygous Rce1-deficient Kras2G12D colony; lanes 3-4, control DNA from mouse tails. The same result was found in 5 additional colonies from spleens of Rce1fl/flKLSLM and Rce1fl/+KLSLM mice.
Rce1 deficiency does not enhance the proliferation of K-RASG12D–expressing fibroblasts
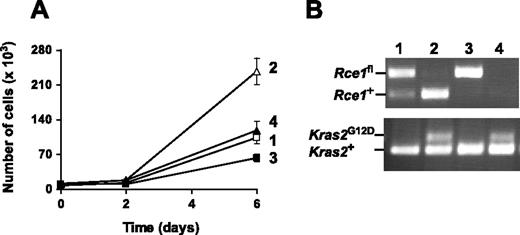
To determine the impact of Rce1 deficiency on the proliferation of other cell types expressing K-RASG12D, we isolated Rce1fl/flKLSL and control Rce1fl/+KLSL MEFs and treated them with a Cre-adenovirus. As expected, Cre-adenovirus treatment of Rce1fl/+KLSL MEFs activated the expression of K-RASG12D and increased cell proliferation (Figure 5A). In experiments with primary and spontaneously immortalized cell lines, the complete inactivation of Rce1 in Rce1fl/flKLSL MEFs attenuated the increased proliferation caused by K-RASG12D. Thus, the inactivation of Rce1 reduced the proliferation of K-RASG12D–expressing MEFs, even though the same genetic intervention increased the proliferation of K-RASG12D–expressing hematopoietic cells.
Inactivation of Rce1 inhibits the growth of K-RASG12D–expressing fibroblasts. (A) Cell proliferation assay of spontaneously immortalized Rce1fl/flKLSL and Rce1fl/+KLSL embryonic fibroblasts treated with Cre and β-gal-adenoviruses. Data are mean of 2 independent cell lines per genotype. The experiment was repeated with a pair of primary cell lines and yielded similar results. (B) PCR amplification of genomic DNA from the cells in panel A demonstrating the activation of the Kras2G12D allele and the inactivation of the Rce1fl allele. Lane 1, β-gal-adenovirus-treated Rce1fl/+KLSL cells; lane 2, Cre-adenovirus-treated Rce1fl/+KLSL cells; lane 3, β-gal-adenovirus–treated Rce1fl/flKLSL cells; lane 4, Cre-adenovirus-treated Rce1fl/flKLSL cells.
Inactivation of Rce1 inhibits the growth of K-RASG12D–expressing fibroblasts. (A) Cell proliferation assay of spontaneously immortalized Rce1fl/flKLSL and Rce1fl/+KLSL embryonic fibroblasts treated with Cre and β-gal-adenoviruses. Data are mean of 2 independent cell lines per genotype. The experiment was repeated with a pair of primary cell lines and yielded similar results. (B) PCR amplification of genomic DNA from the cells in panel A demonstrating the activation of the Kras2G12D allele and the inactivation of the Rce1fl allele. Lane 1, β-gal-adenovirus-treated Rce1fl/+KLSL cells; lane 2, Cre-adenovirus-treated Rce1fl/+KLSL cells; lane 3, β-gal-adenovirus–treated Rce1fl/flKLSL cells; lane 4, Cre-adenovirus-treated Rce1fl/flKLSL cells.
Discussion
We thought the inactivation of Rce1 would inhibit the development of K-RAS–induced MPD in vivo. This hypothesis was not upheld. To our surprise, the inactivation of Rce1 accelerated the development of MPD, increased the release of immature and dysplastic cells into the circulation, and caused mice to die more rapidly, probably because of the infiltration and proliferation of cells in vital tissues. These studies suggested that RCE1 inhibitors would not be useful for the treatment of RAS-induced hematologic malignancies. Indeed, such a strategy could be harmful.
Our experiments involved the use of Cre recombinase to simultaneously inactivate Rce1 and activate K-RASG12D. This strategy worked as planned. PCR analyses of genomic DNA from multiple individual splenocyte colonies confirmed that Rce1 had been fully deleted and that the Kras2G12D allele was activated. Moreover, Rce1 was deleted in more than 90% of the cells in spleen and bone marrow, as judged by quantitative PCR.
Activation of the K-RASG12D allele led to a rapidly progressing MPD, with splenomegaly, increased numbers of mature granulocytes in the circulation, and infiltration of cells in liver and spleen. However, because less than 20% of the myeloid cells in the blood were immature, we could not claim that an acute leukemia syndrome developed in the mice. These findings are entirely consistent with earlier studies on the activation of the latent K-RASG12D allele in hematopoietic cells by Braun et al17 and Chan et al.18,19 In mice with Rce1 deficiency, K-RASG12D–induced MPD progressed to an accelerated-phase MPD with a maturation arrest evident by the more severe leukocytosis and the dramatic increase in the number of immature myeloid cells in the blood. In addition, Rce1-deficient hematopoietic cells displayed an increased capacity to grow in methylcellulose in the absence of growth factors. However, even in mice with Rce1 deficiency, bona fide acute leukemia did not occur because the number of myeloblasts in the blood remained lower than 20%.
The finding of increased numbers of white blood cells in the absence of Rce1 expression is consistent with data reported a few years ago by Aiyagari et al.9 To define the functional importance of Rce1 in normal hematopoiesis, they compared the ability of Rce1−/− and wild-type fetal liver cells to rescue hematopoiesis in lethally irradiated mice. Mice replenished with Rce1−/− cells actually had higher white blood cell counts than mice receiving Rce1+/− or Rce1+/+ cells. At the time, it was unclear whether the higher white blood cell counts were truly significant or simply resulted from chance. In light of the current studies, we suggest that the increased white blood cell counts were in fact caused by Rce1 deficiency, and we further suggest that Rce1 deficiency accelerated the proliferation of normal and K-RASG12D–expressing hematopoietic cells.
We showed previously that the inactivation of Rce1 mislocalizes the RAS proteins away from the plasma membrane and reduces the proliferation of skin carcinoma cells, wild-type fibroblasts, and fibroblasts that overexpress a mutationally activated form of RAS (driven by a strong viral promoter).8 Our current studies reinforced those data: Rce1 deficiency reduced the proliferation of fibroblasts in which the latent K-RASG12D allele was activated (with K-RAS expression driven by the endogenous Kras2 promoter). Thus, our studies indicated that Rce1 deficiency has distinct effects in fibroblasts and hematopoietic cells.
The mechanism for the worsened MPD and the increased proliferation of Rce1-deficient hematopoietic cells is unknown. The simplest potential explanation would be that RCE1 normally processes an isoprenylated CaaX protein that suppresses cell proliferation and that this protein is dysfunctional in the absence of RCE1-mediated endoproteolytic processing. One potential candidate is RAP1, an RCE1 substrate known to be capable of suppressing RAS signaling.20 Regardless of the mechanism, in the future it will be important to determine whether Rce1 deficiency accelerates the development of myeloid leukemia caused by other genetic interventions (eg, Nf1 deficiency21 ). In addition, it will be important to define, with experiments similar to those described here, the in vivo importance of the other CaaX protein–processing enzymes (ie, FTase, GGTase-I, and ICMT) on the development of MPD and leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: A.M.W. designed and performed the research; B.A.C., K.M.E.A., M.L., and A.K.M.S. performed the research; C.K. analyzed the data; B.S. designed the research and analyzed the data; S.G.Y. designed the research; and M.O.B. designed the research and wrote the paper.
We thank Drs Kevin Shannon and Tyler Jacks for providing the Kras2LSL mice, Dr Rosie Perkins for editorial assistance, and the University of Iowa Gene Transfer Vector Core for viral vector preparations.
This work was supported by the Royal Swedish Academy of Sciences, the Swedish Medical Research Council, the Swedish Cancer Society, the Swedish Children's Cancer Fund, and a local clinical grant from Västra Götalandsregionen (M.O.B.) and by National Institutes of Health grants CA099506 and CA103999 (S.G.Y.).

![Figure 1. Acceleration of K-RAS–induced MPD in mice with Rce1 deficiency. Groups of Rce1fl/flKLSLM mice and control Rce1fl/+KLSLM mice were injected at weaning with pI-pC to induce MPD. (A) The concentration of white blood cells was elevated in Rce1fl/flKLSLM compared with Rce1fl/+KLSLM mice with MPD. Statistically significant changes at each time point are indicated. *P < .05; **P < .01; and ***P < .001; n = 11-27 per time point, except for the values at 42 days (n = 3 [Rce1fl/flKLSLM] and n = 5 (Rce1fl/+KLSLM). (B) Kaplan-Meier curve demonstrating reduced survival of pI-pC–injected Rce1fl/flKLSLM mice (median survival, 40 days; n = 12) compared with Rce1fl/+KLSLM mice (median survival, 54 days; n = 5).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/2/10.1182_blood-2006-05-024752/4/m_zh80020706230001.jpeg?Expires=1765900452&Signature=CkK4Kq7SfvYtBasc8cdg5h3xdAug2JHSyLC6n1HOcuoYmDNEtODsqHoMMl8FF5KnoxfPHjPWHGjf4RAjfD0riXQprMLgz9ChgCQIGqhZD6aaF1hfFdUSixp30mpdti3nvgG4pDURLQ-ESgzLXmS80W0MrNJQcsd2zbgf7x3pfrmFssxyoPRhiuU8uc408gJOYwdsVm16lxqiTN656wgL4SSIlGCuF2ZWeJK2zWLhtxILsx-1csTEZW1pWe~OuE98FYgi7~RYpY71p2xeLcdbhmB4OgFAbwsnX9Q3zOqdfHyq-~DC38diNaMLHa17iXO2b5a5d5t6epag7oD7zjcwSg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal