Abstract
Livin, a member of the inhibitor of apoptosis proteins, has been considered to be a poor prognostic marker in malignancies. However, little is known about the clinical relevance of Livin expression in childhood acute lymphoblastic leukemia (ALL). In this study, the expression of Livin was analyzed in 222 patients with childhood ALL using quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) to investigate a possible association with the clinical features at diagnosis and treatment outcomes. Both Livin expression rates and expression levels were higher in patients with favorable prognostic factors. The expression rate was also higher in patients with a favorable day 7 bone marrow response to induction chemotherapy (P < .001). The Livin expression was related to the absence of relapse (P < .001). Similarly, the relapse-free survival rate (± 95% CI) was higher in patients with Livin expression than in patients without Livin expression (97.9% ± 4.0% versus 64.9% ± 11.8%, P < .001). Multivariate analysis for relapse-free survival demonstrated that Livin expression was an independent favorable prognostic factor in childhood ALL (P = .049). This study suggests that Livin expression is a novel prognostic marker in childhood ALL and thus needs to be incorporated into the patient stratification and treatment protocols.
Introduction
Acute lymphoblastic leukemia (ALL) is the most common malignancy in children. It accounts for one fourth of all childhood cancers and approximately 75% of all cases of childhood leukemia. The parameters of initial leukocyte count and age at diagnosis have traditionally provided the most reliable basis for patient stratification; this is because these parameters are readily available and are relatively independent predictors of prognosis.1 The immunophenotype and cytogenetic abnormalities of leukemic cells are also important prognostic factors and are used in the design and analysis of modern therapeutic trials for childhood ALL.2-4
As a result of the accumulation of knowledge on the molecular biology of malignancies, new diagnostic modalities are beginning to be incorporated into diagnostic and therapeutic strategies.5-7 One of these modalities is the quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) that allows the determination of messenger RNA expression levels and, therefore, allows researchers to examine the expression patterns of a large number of genes at the RNA level. If specific patterns of gene expression can be correlated with clinical features in childhood ALL, a refinement of current prognosis-based stratification systems would be possible.
Apoptosis is an active biologic mechanism leading to programmed cell death. A tight regulation is required in biologic systems to ensure a delicate balance between life and death. The loss of apoptosis might result in the development of a wide variety of diseases, including cancers. Cellular defects that halt apoptosis have been shown to be frequently involved in cancer development and progression.8 The up-regulation of antiapoptotic proteins would certainly be advantageous for tumor survival.9-12 During the last decade, a complex network of proapoptotic and antiapoptotic proteins, which strictly regulate the apoptosis pathways, have been identified.13-16 Studies investigating the expression of these molecules in acute leukemia have demonstrated that the expression of proapoptotic or antiapoptotic regulatory molecules varies depending on the types of leukemias and individual patients' characteristics.9,17-25 These differences can be potentially important for the prediction of the response to treatment.23-25
A group of proteins known as the inhibitor of apoptosis proteins (IAP) have been identified. As their name implies, the IAP family proteins are known to inhibit apoptosis induced by a variety of stimuli.15,26 They are the only cellular factors that act on both initiator and effector caspases. Livin (MIM no. 605737; baculoviral IAP repeat-containing 7, BIRC7; aliases, ML-IAP or KIAP) is a member of the IAP family proteins; the gene has been localized to the long arm of chromosome 20 on 20q13.3, a region frequently amplified in melanomas and other malignancies.27 Two splicing variants of Livin (α- and β-isoform) have been identified.15,27 Livin has been shown to antagonize both the death receptor and mitochondria-based apoptotic pathways through the inhibition of caspases 3, 7, and 9, as well as the participation of JNK1.27-29 Therefore, Livin expression has been regarded as a poor prognostic marker in malignancies. However, only a limited number of studies are available to date; thus, the clinical relevance of Livin expression is still controversial in different types of malignancies. For example, Livin expression was associated with poor clinical outcomes in superficial bladder cancer,30 melanoma,31 and neuroblastoma,32 whereas the Livin expression level was not related to the survival of patients with nasopharyngeal cancer.33 No study has been conducted on the clinical relevance of Livin expression in childhood ALL.
In this study, therefore, we analyzed the expression of Livin in 222 patients with childhood ALL using quantitative RT-PCR to investigate a possible relation between Livin expression and the clinical features at diagnosis and treatment outcomes. As a result, Livin expression in childhood ALL was found to be strongly associated with both favorable prognostic factors at diagnosis and better treatment outcomes. This is the first study demonstrating the prognostic implication of Livin expression in childhood ALL.
Patients, materials, and methods
Patients and treatment protocol
From September 1998 to March 2006, a total of 222 patients with newly diagnosed childhood ALL who were younger than 15 years at initial diagnosis were recruited from 2 institutions, the Samsung Medical Center and the Seoul National University Hospital in Seoul, South Korea. The diagnosis of ALL was made based on the morphologic findings from Wright-Giemsa–stained smears of bone marrow aspirates and immunophenotyping analyses of leukemic cells by flow cytometry. Conventional cytogenetic analyses, including fluorescent in situ hybridization (FISH) for t(9;22), 11q23 rearrangement, and t(12;21), were also performed as part of the routine workup. The ploidy was determined directly by the classic method of counting the modal number of chromosomes on metaphase karyotype preparations.
Patients were assigned to the standard-risk group if the leukocyte count was less than 50 × 109/L and the age was 1 to 9 years at diagnosis. Otherwise, patients were assigned to a high-risk group. In the standard-risk patients, the treatment protocols were modified from the Children's Cancer Group (CCG)–189134 or CCG-1952 protocols.35 The regimen used was switched to a modified one from the CCG-1882 protocol36 from consolidation chemotherapy, if the percentage of bone marrow leukemic blasts on day 7 was greater than 25% during induction chemotherapy. If the percentage of leukemic blasts on day 14 was still greater than 25%, the protocol used for the high-risk patients was restarted. In the high-risk patients, the treatment protocols were modified from the CCG-1882, CCG-106B,37 or CCG-1901 protocols.38 If a patient had one or more of the following: a leukocyte count of at least 100 × 109/L, age younger than 1 year, presence of t(9;22), or the 11q23 rearrangement, the patients proceeded to undergo hematopoietic stem cell transplantation when an appropriate donor was available. The protocols used were approved by the Institutional Review Board of each institution. Informed consent was obtained from parents or guardians for both laboratory studies and treatment according to the guidelines of the Korean Food and Drug Administration.
RNA isolation and real-time quantitative RT-PCR
Mononuclear cells (MNCs) were isolated from 2 mL bone marrow aspirate at diagnosis by Ficoll density gradient centrifugation. Total RNA was extracted from MNCs using a QIAamp RNA Blood kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. After treatment with DNA-free (Ambion, Austin, TX) to remove chromosomal DNA, complementary DNA (cDNA) was synthesized using oligo (dT) 15-mer primer by SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) and stored at −20°C until use.
The mRNA expression levels of Livin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were measured by quantitative RT-PCR using an ABI PRISM 7000 Sequence Detector System (Applied Biosystems, Foster City, CA). The quantitative RT-PCR amplification was performed using the predeveloped Assays-on-demand Gene Expression Set for the Livin gene (Hs00223384_m1; GenBank accession no. NM_139317; Applied Biosystems), and TaqMan GAPDH Control Reagents (Applied Biosystems) for the GAPDH gene in combination with the TaqMan Universal PCR Master Mix (Applied Biosystems).
All reactions were performed in triplicate using 20-μL samples containing 50 ng cDNA. The reaction protocol used involved heating for 2 minutes at 50°C and 10 minutes at 95°C, followed by 40 cycles of amplification (15 seconds at 95°C and 1 minute at 60°C). Analysis was performed using ABI PRISM 7000 Sequence Detection software (Applied Biosystems).
The expression levels of the Livin gene in unknown samples were normalized and analyzed by the ΔΔCt method [ΔΔCt = (CtLivin − CtGAPDH)sample − (CtLivin − CtGAPDH)calibrator]. After all reactions, the average ΔΔCt (CtLivin − CtGAPDH) from all samples combined was defined as 1.0, as calibrator. We previously determined that the amplification efficiencies for Livin and GAPDH showed the same slopes. A negative control without template was included in each experiment.
Cytotoxicity assay
Cell proliferation was assessed 24 hours, 48 hours, and 72 hours after the exposure to methylprednisolone (50 μg/mL) by measuring the conversion of the trazolium salt WST-8 to formazan according to the manufacturer's instructions (Cell Counting Kit-8; Dojindo, Kumamoto, Japan).39,40 Briefly, cells (1.5 × 105 cells/well) in RPMI-1640 media supplemented with 10% fetal bovine serum and penicillin were plated onto 96-well plates. The cells were cultured in triplicate wells with 50 μg/mL methylprednisolone for 24, 48, and 72 hours in a humidified atmosphere containing 5% CO2 at 37°C. At the end of each time point, 10 μL WST-8 was added to each well, and the plates were incubated for an additional 4 hours at 37°C to convert WST-8 into formazan. The absorbance of each plate was measured at 450 nm and 600 nm. The absorbance at 450 nm represents a direct correlation with the cell number in this analysis. The results were expressed as the percentage of the absorbance of control (untreated and serial diluted) cells. The control cell number was assessed by trypan blue exclusion (final concentration, 0.2%) for 5 minutes using a hemocytometer.
Western blotting
MNCs from patients' bone marrow were obtained by treatment of lysis buffer (0.8% ammonium chloride solution; StemCell Technology, Vancouver, Canada) to remove red blood cells and were washed 3 times in phosphate-buffered saline. MNCs were resuspended at a final concentration of 1 × 107/mL in RPMI-1640 supplemented with 10% fetal bovine serum and penicillin. Five milliliters of the suspension was then plated onto a T-25 culture flask (Becton Dickinson, Franklin Lakes, NJ) with methylprednisolone (50 μg/mL). A total of 1 × 107 cells was harvested at 0, 24, 48, and 72 hours. Total proteins were immediately extracted by the Pro-Prep solution (Intron, Seoul, South Korea).
The proteins extracted were assessed by the Bradford assay (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. Briefly, the samples were resolved on a 10% Bis-Tris precast gel, following the manufacturer's instructions (Invitrogen). After transferring of the gel to polyvinylidene difluoride membrane (Millipore, Bedford, MA), the membrane was blocked with 5% skim milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) for 1 hour at room temperature. Monoclonal antibodies against Livin (clone 88C570; Abcam, Cambridge, United Kingdom), caspase 3 (Cell Signaling Technology, Beverly, MA), and poly (ADP-ribose) polymerase (PARP; Cell Signaling Technology) were diluted 1:1000, and anti–β-tubulin (Santa Cruz Biotechnology, Santa Cruz, CA) was diluted 1:5000 in blocking solution for immunoblotting. The membrane was exposed to the primary antibodies in a blocking buffer overnight at 4°C, followed by three 5-minute washes with TBST. After washing with TBST 3 times, the blots were incubated with anti–rabbit or anti–mouse IgG horseradish peroxidase–linked antibody (Cell Signaling Technology) for 1 hour at room temperature. Enhanced chemiluminescence (ECL) reaction was performed by ECL detection kit (Amersham Bioscience, Piscataway, NJ) according to the manufacturer's instructions. The full-length (α- and β-isoform) and cleaved forms of Livin were determined by the Western blot assay based on the method by Nachmias et al.31 Densitometric quantification of the Livin protein was performed with a GS-800 imaging densitometer (Bio-Rad, Taiwan). The protein levels of Livin and β-tubulin from the same patient were quantified by using Quantity one 4.2.2 software (Bio-Rad, Hercules, CA), and the ratio of Livin to β-tubulin was then defined as the Livin protein level.
Statistical analysis
Differences in the expression rate of Livin with respect to common prognostic factors (ie, sex, age, leukocyte count, risk group, immunophenotype, numerical or structural cytogenetic abnormalities, cerebrospinal fluid [CSF] involvement, and mediastinal mass) and treatment outcomes (day 7 bone marrow response to induction chemotherapy and occurrence of relapse) were analyzed using the Pearson chi-square test. Differences in expression levels of Livin with respect to the prognostic factors were analyzed using the Mann-Whitney U test. The percentage of survival of leukemic cells in the cytotoxicity assay was also compared using the Mann-Whitney U test. The expression rates of Livin were recorded as a percentage (%) of patients with Livin expression among total patients regardless of the expression level, and the expression levels were represented as the median values along with the ranges. The patients were categorized into 2 groups according to the presence or absence of the Livin expression. Overall survival (OS) rate, relapse-free survival (RFS) rate, and event-free survival (EFS) rate along with 95% confidence interval (CI) were estimated using the Kaplan-Meier method. An event was defined as the occurrence of a relapse or treatment-related death. The differences in survival rates between the 2 groups were compared using the log-rank test. Univariate and multivariate analyses comprising common prognostic factors for relapse-free or event-free survival were performed using the Cox regression analysis. The correlation between the Livin mRNA expression level and protein level was estimated using the Spearman test. Statistical significance was accepted when the P values were less than .05.
Results
Patient characteristics
Clinical characteristics of the patients at diagnosis are presented in Table 1. The median age at diagnosis among the 222 patients (138 boys and 84 girls) was 65.5 months (range, 3-190 months), and their median leukocyte count at diagnosis was 9.00 × 109/L (range, 0.31 × 109–925.1 × 109/L). The median proportion of bone marrow leukemic blasts at diagnosis was 92.0% (range, 31%-100%). The proportion of patients with at least 75% of bone marrow leukemic blasts was 84.2%. The numbers of standard-risk and high-risk patients were 129 and 93, respectively, and 171 patients (77.7%) had the precursor B-cell immunophenotype. The number of patients with hyperdiploidy (≥ 50) and hypodiploidy (≤ 44) were 26 (12.8%) and 9 (4.4%), respectively. Frequent translocations identified by conventional chromosomal analysis, FISH examination, or both were t(12;21) in 35 patients, t(9;22) in 16 patients, and 11q23 rearrangement in 17 patients. Thirty-seven (16.7%) patients received allogeneic hematopoietic stem cell transplant from 11 related and 26 unrelated donors (20 bone marrow, 15 cord blood, and 2 peripheral blood stem cells). Twenty-nine patients received a transplant at first CR, 7 at second CR, and 1 at third CR.
Patient characteristics and the Livin expression rates
| Characteristic . | No. of patients . | Livin expression, no. (%) . | P . |
|---|---|---|---|
| Sex | |||
| Female | 84 | 28 (33.3) | |
| Male | 138 | 29 (21.0) | .042 |
| Age | |||
| 1 y to less than 10 y | 161 | 49 (30.4) | |
| At least 10 y or younger than 1 y | 61 | 8 (13.1) | .008 |
| Leukocyte count | |||
| Less than 50 × 109/L | 166 | 49 (29.5) | |
| At least 50 × 109/L | 56 | 8 (14.3) | .024 |
| Risk group | |||
| Standard-risk patients | 129 | 42 (32.6) | |
| High-risk patients | 93 | 15 (16.1) | .006 |
| Immunophenotype | |||
| Precursor B cell | 171 | 45 (26.3) | |
| Others | 49 | 12 (24.5) | .797 |
| Number of chromosomes | |||
| 50 or more | 26 | 1 (3.8) | |
| 45-49 | 168 | 48 (28.6) | |
| 44 or fewer | 9 | 3 (33.3) | .023 |
| Structural abnormalities of chromosome | |||
| t(12;21) | 35 | 29 (82.9) | |
| Absent or others | 155 | 27 (17.4) | |
| t(9;22) or 11q23 rearrangement | 32 | 1 (3.1) | < .001 |
| CSF involvement | |||
| Absent | 193 | 49 (25.4) | |
| Present | 12 | 1 (8.3) | .182 |
| Mediastinal involvement | |||
| Absent | 215 | 57 (26.5) | |
| Present | 7 | 0 (0) | .114 |
| Day 7 BM response to treatment | |||
| Leukemic blasts less than 25% | 143 | 50 (35.0) | |
| Leukemic blasts at least 25% | 69 | 6 (8.7) | < .001 |
| Relapse | |||
| No | 185 | 56 (30.3) | |
| Yes | 37 | 1 (2.7) | < .001 |
| Characteristic . | No. of patients . | Livin expression, no. (%) . | P . |
|---|---|---|---|
| Sex | |||
| Female | 84 | 28 (33.3) | |
| Male | 138 | 29 (21.0) | .042 |
| Age | |||
| 1 y to less than 10 y | 161 | 49 (30.4) | |
| At least 10 y or younger than 1 y | 61 | 8 (13.1) | .008 |
| Leukocyte count | |||
| Less than 50 × 109/L | 166 | 49 (29.5) | |
| At least 50 × 109/L | 56 | 8 (14.3) | .024 |
| Risk group | |||
| Standard-risk patients | 129 | 42 (32.6) | |
| High-risk patients | 93 | 15 (16.1) | .006 |
| Immunophenotype | |||
| Precursor B cell | 171 | 45 (26.3) | |
| Others | 49 | 12 (24.5) | .797 |
| Number of chromosomes | |||
| 50 or more | 26 | 1 (3.8) | |
| 45-49 | 168 | 48 (28.6) | |
| 44 or fewer | 9 | 3 (33.3) | .023 |
| Structural abnormalities of chromosome | |||
| t(12;21) | 35 | 29 (82.9) | |
| Absent or others | 155 | 27 (17.4) | |
| t(9;22) or 11q23 rearrangement | 32 | 1 (3.1) | < .001 |
| CSF involvement | |||
| Absent | 193 | 49 (25.4) | |
| Present | 12 | 1 (8.3) | .182 |
| Mediastinal involvement | |||
| Absent | 215 | 57 (26.5) | |
| Present | 7 | 0 (0) | .114 |
| Day 7 BM response to treatment | |||
| Leukemic blasts less than 25% | 143 | 50 (35.0) | |
| Leukemic blasts at least 25% | 69 | 6 (8.7) | < .001 |
| Relapse | |||
| No | 185 | 56 (30.3) | |
| Yes | 37 | 1 (2.7) | < .001 |
Differences in Livin expression rates were analyzed using the Pearson chi-square test. Statistical significance was accepted when P values were less than .05.
Both expression rates and expression levels of Livin were higher in patients with favorable prognostic factors
The expression rates of Livin with respect to the common prognostic factors are presented in Table 1. Livin mRNA was expressed in 57 (25.7%) of 222 patients. The expression rate of Livin was higher in female patients (P = .042), patients with an age range of 1 to 9 years (P = .008), patients with a leukocyte number of less than 50 × 109/L (P = .024), standard-risk patients (P = .006), and patients with t(12;21) (P < .001), compared with male patients, patients at least 10 years old or younger than 1 year, patients with a leukocyte count of at least 50 × 109/L, high-risk patients, and children with structural chromosomal abnormalities other than t(12;21). Livin expression rates were also higher in patients without CSF involvement or a mediastinal mass than in those with CSF involvement or a mediastinal mass, albeit without statistical significance. There was no difference in the Livin expression rate with respect to the immunophenotype. The Livin expression rate was lower in patients with hyperdiploidy (≥ 50) than in patients with hypodiploidy (≤ 44) or others (P = .023).
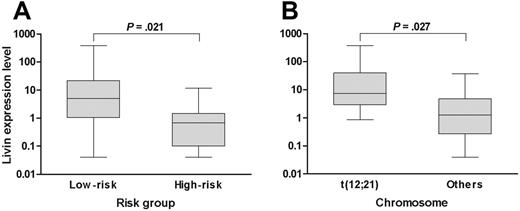
When the analysis was confined to only the patients who exhibited Livin expression, the median expression level of Livin was 3.28 (range, 0.04-382.68). Of note, the expression level of Livin was higher in patients with favorable prognostic factors than in patients with unfavorable prognostic factors when the analysis was confined to only the patients with Livin expression. For example, the expression level of Livin in the standard-risk patients (median, 5.01; range, 0.04-382.68) was higher than in the high-risk patients (median, 0.68; range, 0.04-11.71; P = .021; Figure 1A). Similarly, the Livin expression level was also higher in patients with t(12;21) (median, 7.39; range, 0.86-382.68) than in patients without t(12;21) (median, 1.26; range, 0.04-37.75; P = .027; Figure 1B). The expression level in the only patient with t(9;22) who exhibited Livin expression was very low (0.18). The Livin protein level (median, 0.165; range, 0.029-0.819) measured by Western blot analysis in 7 patients with Livin expression showed a good correlation with the mRNA level (median, 0.207; range, 0.001-3.784; P = .044).
Livin expression level with respect to risk factors. (A) The Livin expression level in low-risk patients was higher than in high-risk patients. (B) Similarly, the Livin expression level in patients with t(12;21) was higher than in patients without t(12;21). The Livin expression levels are presented in box plots showing the median and 90th percentiles of Livin expression. Maximum and minimum values are represented by bars. Statistical analysis was performed using the Mann-Whitney U test.
Livin expression level with respect to risk factors. (A) The Livin expression level in low-risk patients was higher than in high-risk patients. (B) Similarly, the Livin expression level in patients with t(12;21) was higher than in patients without t(12;21). The Livin expression levels are presented in box plots showing the median and 90th percentiles of Livin expression. Maximum and minimum values are represented by bars. Statistical analysis was performed using the Mann-Whitney U test.
Livin expression was associated with a favorable early response to chemotherapy
Livin expression was associated with a favorable early response to induction chemotherapy. The Livin expression rate was higher in patients with a favorable (leukemic blast < 25%) day 7 bone marrow response than in patients with an unfavorable (leukemic blast ≥ 25%) response (35.0% versus 8.7%, P < .001; Table 1). Similarly, the proportion of patients with an unfavorable day 7 bone marrow response was significantly lower in patients with Livin expression than in patients without Livin expression (10.7% versus 40.4%, P < .001; Figure 2A). From the cytotoxicity assay that was used to evaluate the ex vivo susceptibility of leukemic blasts to apoptotic stimuli provided by chemotherapeutic agents, the percentage of surviving leukemic blasts after 24, 48, and 72 hours of cultures with methylprednisolone were significantly lower when the leukemic cells expressed Livin mRNA (Figure 2B). During the cytotoxicity assay, we observed a persistent presence of both full-length Livin protein (α- and β-isoform) and a cleaved form thereof in the leukemic cells with Livin expression.31 The intensities of caspase 3 and PARP proteins increased as the apoptotic process continued (Figure 2C).
Different apoptotic responses to chemotherapeutic agents according to Livin expression. (A) The proportion of patients with unfavorable day 7 bone marrow response (leukemic blasts ≥ 25%) was significantly lower in patients with Livin expression compared with patients without Livin expression (P < .001). (B) In the cytotoxicity assay to evaluate the ex vivo susceptibility of leukemic blasts to apoptotic stimuli provided by methylprednisolone, the percentages of surviving leukemic blasts after 24, 48, and 72 hours of culture were significantly lower when the leukemic cells expressed Livin mRNA. Graph shows means and SEM. (C) In the cytotoxicity assay, both full-length Livin protein (α- and β-isoform) and the cleaved form of Livin protein were persistently observed in the leukemic cells with Livin expression. The intensities of caspase 3 and PARP proteins increased as the apoptotic process continued.
Different apoptotic responses to chemotherapeutic agents according to Livin expression. (A) The proportion of patients with unfavorable day 7 bone marrow response (leukemic blasts ≥ 25%) was significantly lower in patients with Livin expression compared with patients without Livin expression (P < .001). (B) In the cytotoxicity assay to evaluate the ex vivo susceptibility of leukemic blasts to apoptotic stimuli provided by methylprednisolone, the percentages of surviving leukemic blasts after 24, 48, and 72 hours of culture were significantly lower when the leukemic cells expressed Livin mRNA. Graph shows means and SEM. (C) In the cytotoxicity assay, both full-length Livin protein (α- and β-isoform) and the cleaved form of Livin protein were persistently observed in the leukemic cells with Livin expression. The intensities of caspase 3 and PARP proteins increased as the apoptotic process continued.
Livin expression was an independent favorable prognostic factor
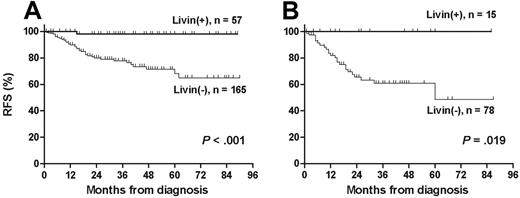
The median follow-up duration among 193 live patients was 37 months (range, 1-90 months). The leukemia relapsed in 37 patients and treatment-related mortality occurred in 7 patients. The 5-year OS, RFS, and EFS rates (± 95% CI) in 222 patients were 82.2% ± 7.0%, 78.6% ± 6.8%, and 76.0% ± 6.8%, respectively. Livin expression was significantly associated with the absence of relapse (P < .001; Table 1). Similarly, RFS rate was higher in patients with Livin expression than in patients without Livin expression (97.9% ± 4.0% versus 64.9% ± 11.8%, P < .001; Figure 3A). In the only relapsed patient with Livin expression, the scheduled chemotherapy could not be delivered prior to relapse because of unexplained severe neurotoxicity during maintenance chemotherapy. Otherwise, all patients who showed Livin expression, including high-risk patients with unfavorable prognostic factors at diagnosis, are currently relapse free (Figure 3B). Among 29 patients who underwent allogeneic stem cell transplantation at first CR, 5 patients with Livin expression are all alive without relapse, whereas 5-year RFS rate of 24 patients without Livin expression was 50.0% ± 22.5% (P = .067).
Relapse-free survival according to Livin expression. (A) Relapse-free survival was significantly longer in patients with Livin expression than in patients without Livin expression (97.9% ± 4.0% versus 64.9% ± 11.8%). (B) All patients in the high-risk group were relapse free when they exhibited Livin expression (100.0% versus 48.7% ± 23.7%).
Relapse-free survival according to Livin expression. (A) Relapse-free survival was significantly longer in patients with Livin expression than in patients without Livin expression (97.9% ± 4.0% versus 64.9% ± 11.8%). (B) All patients in the high-risk group were relapse free when they exhibited Livin expression (100.0% versus 48.7% ± 23.7%).
According to the univariate analyses, age at diagnosis of 1 to 9 years, leukocyte count less than 50 × 109/L at diagnosis, precursor B-cell immunophenotype, the presence of t(12;21), and Livin expression were statistically significant favorable prognostic factors. The presence of t(9;22) or 11q23 rearrangement was also an unfavorable prognostic factor. According to the multivariate analyses for relapse-free survival, Livin expression and structural chromosomal abnormalities were independent favorable prognostic factors with a statistical significance in childhood ALL (hazard ratio of Livin− patients compared with Livin+ patients, 8.844; 95% CI, 1.010-77.479; P = .049; Table 2). The age at diagnosis also had a borderline statistical significance as an independent prognostic factor. Leukocyte count at diagnosis, immunophenotype, and number of chromosomes were not independent prognostic factors.
Univariate and multivariate analyses with prognostic factors for relapse-free survival
| Variable . | No. . | Relapse, no. (%) . | 5-y RFS, ± 95% CI . | Univariate analysis . | Multivariate analysis . | ||||
|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | ||||
| Sex | |||||||||
| Female | 84 | 9 (10.7) | 87.4 ± 7.7 | — | — | — | — | ||
| Male | 138 | 28 (20.3) | 73.9 ± 9.3 | 1.864 | 0.879-3.950 | .104 | 1.367 | 0.599-3.120 | .458 |
| Age | |||||||||
| 1 y to less than 10 y | 161 | 20 (12.4) | 83.4 ± 7.3 | — | — | — | — | ||
| At least 10 y or younger than 1 y | 61 | 17 (27.8) | 65.3 ± 14.2 | 3.425 | 1.778-6.598 | < .001 | 2.095 | 0.996-4.406 | .051 |
| Leukocyte count | |||||||||
| Less than 50 × 109/L | 166 | 20 (12.0) | 83.6 ± 8.9 | — | — | — | — | ||
| At least 50 × 109/L | 56 | 17 (30.4) | 61.7 ± 15.2 | 3.457 | 1.806-6.617 | < .001 | 1.537 | 0.617-3.827 | .356 |
| Immunophenotype | |||||||||
| Precursor B cell | 171 | 24 (14.0) | 80.8 ± 7.6 | — | — | — | — | ||
| Others | 49 | 13 (26.5) | 70.5 ± 14.2 | 2.139 | 1.088-4.204 | .027 | 1.186 | 0.491-2.868 | .705 |
| No. of chromosomes | .805 | .425 | |||||||
| At least 50 | 26 | 5 (19.2) | 65.4 ± 29.0 | — | — | — | — | ||
| 45-49 | 168 | 27 (16.1) | 81.6 ± 6.6 | 0.730 | 0.280-1.899 | .518 | 0.499 | 0.171-1.455 | .203 |
| 44 or fewer | 9 | 2 (22.2) | 75.0 ± 30.0 | 0.839 | 0.162-4.352 | .834 | 0.689 | 0.118-4.038 | .680 |
| Structural abnormality of chromosome | < .001 | .033 | |||||||
| t(12;21) | 35 | 2 (5.7) | 93.6 ± 8.6 | — | — | — | — | ||
| Absent or others | 155 | 19 (12.3) | 83.1 ± 7.7 | 2.342 | 0.545-10.054 | .252 | 0.630 | 0.131-3.036 | .565 |
| t(9;22) or 11g23 rearrangement | 32 | 16 (50.0) | 39.7 ± 23.0 | 12.742 | 2.923-55.550 | .001 | 1.923 | 0.344-10.754 | .457 |
| Livin expression | |||||||||
| Expressed | 57 | 1 (1.8) | 97.9 ± 4.0 | — | — | — | — | ||
| Not expressed | 165 | 36 (21.8) | 71.4 ± 9.0 | 15.098 | 2.069-110.195 | .007 | 8.844 | 1.010-77.479 | .049 |
| Variable . | No. . | Relapse, no. (%) . | 5-y RFS, ± 95% CI . | Univariate analysis . | Multivariate analysis . | ||||
|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | ||||
| Sex | |||||||||
| Female | 84 | 9 (10.7) | 87.4 ± 7.7 | — | — | — | — | ||
| Male | 138 | 28 (20.3) | 73.9 ± 9.3 | 1.864 | 0.879-3.950 | .104 | 1.367 | 0.599-3.120 | .458 |
| Age | |||||||||
| 1 y to less than 10 y | 161 | 20 (12.4) | 83.4 ± 7.3 | — | — | — | — | ||
| At least 10 y or younger than 1 y | 61 | 17 (27.8) | 65.3 ± 14.2 | 3.425 | 1.778-6.598 | < .001 | 2.095 | 0.996-4.406 | .051 |
| Leukocyte count | |||||||||
| Less than 50 × 109/L | 166 | 20 (12.0) | 83.6 ± 8.9 | — | — | — | — | ||
| At least 50 × 109/L | 56 | 17 (30.4) | 61.7 ± 15.2 | 3.457 | 1.806-6.617 | < .001 | 1.537 | 0.617-3.827 | .356 |
| Immunophenotype | |||||||||
| Precursor B cell | 171 | 24 (14.0) | 80.8 ± 7.6 | — | — | — | — | ||
| Others | 49 | 13 (26.5) | 70.5 ± 14.2 | 2.139 | 1.088-4.204 | .027 | 1.186 | 0.491-2.868 | .705 |
| No. of chromosomes | .805 | .425 | |||||||
| At least 50 | 26 | 5 (19.2) | 65.4 ± 29.0 | — | — | — | — | ||
| 45-49 | 168 | 27 (16.1) | 81.6 ± 6.6 | 0.730 | 0.280-1.899 | .518 | 0.499 | 0.171-1.455 | .203 |
| 44 or fewer | 9 | 2 (22.2) | 75.0 ± 30.0 | 0.839 | 0.162-4.352 | .834 | 0.689 | 0.118-4.038 | .680 |
| Structural abnormality of chromosome | < .001 | .033 | |||||||
| t(12;21) | 35 | 2 (5.7) | 93.6 ± 8.6 | — | — | — | — | ||
| Absent or others | 155 | 19 (12.3) | 83.1 ± 7.7 | 2.342 | 0.545-10.054 | .252 | 0.630 | 0.131-3.036 | .565 |
| t(9;22) or 11g23 rearrangement | 32 | 16 (50.0) | 39.7 ± 23.0 | 12.742 | 2.923-55.550 | .001 | 1.923 | 0.344-10.754 | .457 |
| Livin expression | |||||||||
| Expressed | 57 | 1 (1.8) | 97.9 ± 4.0 | — | — | — | — | ||
| Not expressed | 165 | 36 (21.8) | 71.4 ± 9.0 | 15.098 | 2.069-110.195 | .007 | 8.844 | 1.010-77.479 | .049 |
Prognostic effect of each variable was estimated using Cox regression analysis. Statistical significance was accepted when P values were less than .05.
HR indicates hazard ratio; CI, confidence interval; —, not applicable (reference category).
During the study period, 14 bone marrow samples were collected from relapsed patients or from patients who were refractory to chemotherapy after relapse. Only 1 (7.1%) of 14 patients exhibited Livin expression. In this patient, who had been transferred from another hospital after relapse, the day 7 bone marrow response was M1 during reinduction chemotherapy after relapse, and this patient is currently disease free at 22 months after relapse.
Discussion
This is the first study evaluating the expression of Livin, a member of the IAP family proteins, in association with clinical features at diagnosis and treatment outcomes in childhood ALL. Unexpectedly, this study showed that Livin expression in childhood ALL was, albeit not always, associated with the presence of favorable prognostic factors at diagnosis. In addition, Livin expression was associated with a favorable early response to chemotherapy and eventually a higher long-term relapse-free survival.
In this study, the Livin expression rate was significantly higher in patients with favorable prognostic factors. It is particularly interesting to note that the Livin expression rate was very high in t(12;21) and was very low in t(9;22)/11q23 rearrangement, because these translocations are known to be strongly associated with the best41 and worst outcomes,42-44 respectively, in childhood ALL. In this context, it was also interesting to note that the Livin expression rate was lower in cases of hyperdiploidy (≥ 50) than in cases of hypodiploidy (≤ 44) or others. Although the absolute number of chromosomes chosen as the “cutoff point” for analysis may vary slightly between studies, children with hyperdiploidy and hypodiploidy have a better or poorer prognosis, respectively.45-47 The lower rate of Livin expression in our patients with hyperdiploidy may explain their relatively poorer RFS rates (65.4% ± 29.0% at 5 years) than have been generally expected. However, it is not clear why the Livin expression rate was lower in our patients with hyperdiploidy, whereas it was higher in patients with other favorable prognostic factors.
It is also interesting to note that not only the Livin expression rates but also the Livin expression levels were higher in patients with favorable clinical features compared with those in patients with unfavorable clinical features. However, a higher Livin expression level was not associated with a better RFS rate compared with a lower Livin expression level; this was because all but one patient who exhibited Livin expression are currently relapse free regardless of the Livin expression level. These findings suggest that Livin expression itself, not the expression level, is an important prognostic determinant in patients with childhood ALL.
Livin expression in this study was strongly associated with better RFS, EFS, and OS rates in patients with childhood ALL. The better early bone marrow response to induction chemotherapy and, similarly, the better ex vivo response to methylprednisolone in the cytotoxicity assay suggest that Livin expression is associated with a faster apoptotic response of leukemic cells to apoptotic stimuli provided by chemotherapeutic agents and eventually associated with a better relapse-free survival.39 According to the multivariate analyses, Livin expression was an independent favorable prognostic factor harboring statistical significance.
Risk stratification in childhood ALL has been traditionally based on the initial leukocyte count and age at diagnosis.1 Most current treatment protocols for childhood ALL differ in their intensities of treatment according to the risk stratification based on these 2 factors. The immunophenotype and cytogenetic abnormalities of leukemic cells are also important factors considered for prognostic stratification and are thus used in the design and evaluation of modern therapeutic trials in childhood ALL.2-4 However, this study has demonstrated that Livin expression is a new independent favorable prognostic factor in childhood ALL and, therefore, suggests that a new risk stratification system is needed to incorporate this new molecular biologic factor with the traditional prognostic factors. On the basis of the new therapeutic algorithm, patients could be offered a less-intensive therapeutic modality if the leukemic cells exhibit Livin expression. Admitting that the number of cases and the follow-up period in this study were limited, we hope this study would prompt other scientists to investigate the implications of Livin expression in childhood ALL to confirm our observations in a larger series of patients.
Most ex vivo studies and studies on the clinical relevance of Livin expression, albeit limited in number, have shown that Livin is an antiapoptotic regulator; therefore, Livin expression has been considered to be a poor prognostic factor in malignancies. Therefore, the observations from this study were quite unexpected and suggest that the role of Livin in the apoptosis system in leukemogenesis or in the maintenance of leukemic cells might be different from what has been previously recognized. The results from this study clearly showed that a better RFS in patients with Livin expression is associated with a faster cell death in response to apoptotic stimuli. However, the molecular biologic mechanisms to explain these unexpected observations are elusive.
One possible explanation is that the cleaved form of Livin may act as a strong proapoptotic regulator in response to apoptotic stimuli in childhood ALL. Several investigators reported that some of the IAP family proteins could be changed to isoforms that have proapoptotic properties.31,48-50 Nachmias et al31 demonstrated that the caspase-mediated cleavage of Livin converts it from an antiapoptotic to proapoptotic factor. In this study, we could demonstrate the presence of the cleaved form of Livin in the leukemic cells with Livin expression. It seems plausible that this cleaved form might have had proapoptotic activities and thereby partly explains our observations. However, the presence of the cleaved Livin variant has not been investigated in other types of malignancies. Further studies are needed on the expression and implications of the Livin variant protein and the regulatory mechanisms thereof in childhood ALL and other malignant diseases.
Another possible explanation is that the role of Livin expression might be related to the age of patients. Children, not fully matured, have a variety of different biologic features distinguishing them from adults. The regulation of apoptosis, the function of apoptotic regulators, or both in children appear to be different from that in adults. For example, Wuchter et al21 reported that the expression patterns of apoptosis-related molecules were different between children and adults with de novo acute myeloid leukemia. Another example is that the overexpression of Bcl-2, a well-known antiapoptotic protein, was significantly associated with a better prognosis in childhood ALL,23 whereas it was not associated with distinct clinical or biologic characteristics in adult ALL.22 These findings suggest that Livin expression might have a different biologic significance in childhood ALL compared with adult ALL and may also explain the better outcomes in the former than in the latter.
In summary, this is the first study that investigated the relation between Livin expression and the clinical features at diagnosis and treatment outcomes in childhood ALL. We have shown that Livin expression is associated not only with the presence of favorable prognostic factors at diagnosis but also with a significantly better RFS in childhood ALL. A better RFS appears to be associated with a faster death of leukemic cells in patients with Livin expression in response to apoptotic stimuli provided by chemotherapeutic agents. Therefore, Livin expression can be a potential independent prognostic factor in childhood ALL, and new investigational approaches, as part of well-controlled trials, would be needed to develop a modified risk stratification system based on the status of this new molecular biologic factor.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: J.C. performed the research and wrote the paper; Y.K.H. and H.-J.K. performed the research and analyzed the data; K.W.S. designed the research, analyzed the data, and wrote the paper; S.H.L. performed the research and collected the data; K.H.Y., H.L.J., H.H.K., H.J.K., and H.Y.S. designed the research and collected the data; and H.S.A. designed the research and analyzed the data.
K.W.S. and H.H.K. contributed equally to this work.
Acknowledgments
We would like to thank Dr Sun Woo Kim (Samsung Biomedical Research Institute) for critical reading of this manuscript.
This work was supported by the Samsung Biomedical Research Institute (grant SBRI C-A6-319-1) and the National R&D Program of the Ministry of Health and Welfare, Republic of Korea (grant 0520290-1).


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal