Abstract
Resistance to transforming growth factor (TGF)–β1–mediated growth suppression in tumor cells is often associated with the functional loss of TGF-β receptors. Here we describe two B-cell lymphoma cell lines (DB and RL) that differ in their sensitivity to TGF-β1–mediated growth suppression. The TGF-β1–resistant cell line DB lacked functional TGF-β receptor II (TβRII) in contrast to the TGF-β–responsive cell line RL, whereas both cell lines had comparable levels of receptor I (TβRI). Lack of functional TβRII was correlated with the lack of TGF-β1–induced nuclear translocation of phospho-Smad3 and phospho-Smad2, the lack of nuclear expression of p21Cip1/WAF1, and the down-regulation of c-Myc in DB cells. Transfection of wild-type, but not a C-terminal–truncated, form of TβRII rendered the DB cell line responsive to TGF-β1–mediated growth suppression. Analysis of the TβRII gene in DB cells revealed the absence of TβRII message, which was reversed upon 5′-azacytidine treatment, indicating that the promoter methylation might be the cause of gene silencing. Promoter analysis revealed CpG methylations at −25 and −140 that correlated with the gene silencing. These data suggest that promoter methylation plays an important role in TβRII gene silencing and subsequent development of a TGF-β1–resistant phenotype by some B-cell lymphoma cells.
Introduction
Transforming growth factor (TGF)–β1 is a member of the TGF-β superfamily that regulates cell growth and differentiation in a variety of cell types. TGF-β inhibits cell proliferation by arresting cells in the G1 phase of the cell cycle. The mechanisms of the cell-cycle inhibition depend on the cell type.1 Some of the critical regulators in this process include p15INK4b, p21Cip1/WAF1, p27, and c-Myc.1 Activation of the TGF-β receptors (TβRs) occurs via ligand-induced heteromeric complex formation of type I and type II serine/threonine kinase receptors. The constitutively active type II receptor then phosphorylates and activates the type I receptor, which in turn propagates the TGF-β signal by phosphorylating and activating Smad proteins Smad2 and Smad3. Receptor-activated Smads (R-Smads) then hetero-oligomerize with a common partner, Smad4, and translocate to the nucleus where, in association with transcriptional coactivators or repressors, they regulate transcription of TGF-β; target genes. The termination of TGF-β signaling is achieved, in part, by a feedback mechanism through the induction of inhibitory Smads (I-Smads) such as Smad6 and Smad7, which then terminate the activation of R-Smads.2–4
Resistance to TGF-β–mediated growth suppression in tumor cells is often associated with the functional loss of TGF-β receptors and Smads. A frameshift mutation within exon 5 of TβRI has been found in approximately one-third of ovarian cancers and is associated with reduced or absent expression of TβRI.5 Inactivating mutations in TβRI and TβRII have also been reported in human lymphoma,6,7 and loss of TβRII transcripts and protein has been reported in murine lymphoma.8
Mutations in TβRII have been reported in colon and gastric cancers with or without microsatellite instability.9–13 Burkitt lymphoma cell lines and Epstein-Barr virus–transformed B lymphoblastoid cell lines have been shown to harbor reduced expression of TβRII, which correlates with the resistance of these cell lines to TGF-β1.14 Although Smad2 and Smad4 genes have been shown to be affected in different cancers, no mutation in the Smad3 gene has been found in tumors.15,16
In the present study, we examined how B-cell lymphoma cells evade TGF-β–mediated growth suppression. Compared with the TGF-β–responsive B-cell lymphoma cell line RL, the TGF-β–resistant cell line DB lacked functional TβRII on the cell surface, whereas both cell lines had comparable levels of receptor I. Promoter analysis revealed CpG methylations at −25 and −140 that correlated with the gene silencing. The role of promoter methylation in silencing TβRII gene was also observed in another B-cell lymphoma cell line, Akata. These data demonstrated that the nonresponsiveness of some B-cell lymphoma cells to TGF-β was due to the promoter methylation of the TβRII gene.
Materials and methods
Reagents
For Western blot analysis and immunoprecipitation, rabbit polyclonal anti-TβRI antibody (V-22), anti-TβRII (C-16), and mouse monoclonal anti–c-Myc antibody (sc-40) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA); rabbit polyclonal phospho-Smad2 antibody and phospho-Smad3 were purchased from Cell Signaling Technology (Beverly, MA); rabbit polyclonal anti-Smad2 antibody and anti-Smad3 were obtained from Zymed Laboratories (South San Francisco, CA); mouse monoclonal anti-p21Cip1/WAF1 antibody was from Upstate USA (Charlottesville, VA); mouse monoclonal anti-nucleoporin p62 was from BD Transduction Laboratories (San Diego, CA); mouse monoclonal anti-HA (clone 12CA5) was from Roche Applied Science (Indianapolis, IN). All horseradish peroxidase (HRP)–conjugated secondary antibodies were from GE Healthcare (Piscataway, NJ). Recombinant TGF-β1 (100-21R) was from PeproTech (Rocky Hill, NJ); phorbol 12-myristate 13-acetate (PMA) and 5′-azacytidine were from Sigma Chemical (St Louis, MO).
Cell lines and culture conditions
The DB and RL cell lines were derived from tumors of patients with diffuse large-cell lymphoma.17 According to the classification of diffuse large B-cell lymphoma (DLBCL),18,19 the RL line was considered to be germinal center B-cell–like based on the t(14:18) chromosomal translocation and higher expression of LMO2. In contrast, no t(14:18) chromosomal translocation and an undetectable level of LMO2 expression were detected in DB cells. As a proliferation signature, c-Myc expression was highest in DB cells. Based on these and other signature gene analyses, DB appears to have the “activated B-cell” phenotype. RL and DB cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 1000 U penicillin/mL, and 100 μg streptomycin/mL. No exogenous growth factors were added. Cells were grown at 37°C in 5% CO2. Fresh growth medium was added to cells every 3 to 4 days until confluent. In all experiments cells were stimulated with either 0.15 ng/mL PMA and/or 10 ng/mL recombinant TGF-β1 in RPMI 1640 with 5% FBS.
Cell lysis and Western blot analysis
Cytoplasmic and nuclear extracts were prepared according to the procedure described previously.20 Samples were boiled with NuPAGE LDS sample buffer (Invitrogen, Carlsbad, CA), resolved on 4% to 12% NuPAGE Novex Bis-Tris Gels (Invitrogen), and transferred to PVDF membrane. Detection was carried out with ECL-Plus detection reagent (GE Healthcare).
Membrane fraction and immunoprecipitation
Membrane preparations were carried out according to the procedure described by Woods et al.21 Briefly, cells were resuspended in 1 mL hypotonic buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM MgCl2, 5 μg/mL E-64, 1 mM PMSF, 1 mM NaVO4, and 1 mM NaF. Cell suspensions were incubated on ice for 30 minutes before homogenization. NaCl was added to the homogenates to bring the final concentration to 150 mM, and the samples were centrifuged at 210g for 10 minutes. The supernatants were diluted with 3 mL isotonic buffer containing 0.6M NaCl, 5 μg/mL E-64, 1 mM PMSF, 1 mM NaVO4, and 1 mM NaF, and were centrifuged at 100 000g for 45 minutes. The pellet was resuspended in hypotonic buffer containing 150 mM NaCl and 0.5% Triton X-100, and sonicated for 1 minute. After centrifugation at 14 000 rpm for 10 minutes, the supernatants were collected as membrane fractions. For immunoprecipitation, membrane fractions were precleared with protein A/G plus–agarose (sc-2003; Santa Cruz Biotechnology) for 30 minutes at 4°C, and were precipitated overnight with either anti-TβRI or anti-TβRII antibody. This incubation was followed by 1.5 hours of incubation with protein A/G plus–agarose. The agarose beads were washed 3 times with extraction buffer containing 25 mM MOPS (pH 7.2), 15 mM MgCl2, 137 mM NaCl, 1 mM PMSF, 15 mM EGTA, 5 μg/mL E-64, 1 mM NaVO4, 1 mM NaF, and 0.1% Triton X-100. The immune complexes were eluted with NuPAGE LDS sample buffer, boiled for 5 minutes, resolved on 4% to 12% NuPAGE Bis-Tris Gels (Invitrogen) under reducing conditions, and transferred to PVDF membrane. Detection was carried out with ECL-Plus detection reagent.
RT-PCR
Complementary DNA was generated from 2 μg RNA using the SuperScript III reverse transcription (RT) kit and random hexamers (Invitrogen) according to the manufacturer's procedure. For polymerase chain reaction (PCR; 20 μL), 1 μL cDNA, 1.25 U Taq DNA polymerase (New England Biolabs, Beverly, MA), 0.25 mM each dNTP (New England Biolabs), and 500 nM of the respective TβRII primers were used. All samples were subjected to RT-PCR for the housekeeping gene GAPDH, which served as an internal standard. RT-PCR products were resolved on 1% agarose gel in 1 × TAE buffer, visualized by ethidium bromide, and photographed using Multiimage system (Alpha Innotech, San Leandro, CA). Primer sequences are as follows: forward, 5′-tcggtctatgacgagcagcg-3′, reverse, 5′-gcagcctctttggacatgcc-3′.
Transient transfection
Cytomegalovirus (CMV) promoter–based expression plasmids for wild-type (WT) TβRII and the C-terminal–truncated form of TβRII (Δcyt) were gifts from Dr Joan Massague (Memorial Sloan-Kettering Cancer Center, New York, NY).22 Exponentially growing DB cells (5 × 106/point) were transfected with 3 μg TβRII (WT) or TβRII (Δcyt) plasmid using a Nucleofector Kit V (Amaxa, Gaithersburg, MD) according to the manufacturer's protocol. Cells were then exposed to electroporation using program T-27. After incubation for 12 hours, cells were transferred to RPMI 1640 with 5% FBS, and stimulated with PMA/TGF-β for various time points.
5′-azacytidine treatment
Cells were seeded at a density of 0.5 × 106/mL. 5-azacytidine was added to the medium to a final concentration of 2 μM for 7 days. At every 24-hour interval, serum-free medium containing 5′-azacytidine was added. Cells were allowed to recover for 24 hours in complete medium without 5′-azacytidine at 37°C in 5% CO2.
Sodium bisulfite modification
Genomic DNAs from lymphoma cells were modified using a method described previously.23,24 Briefly, 1 μg DNA in a volume of 18 μL was denatured by adding 18 μL 0.6 M sodium hydroxide and incubated for 10 minutes at 37°C. To the denatured DNA, 24 μL 10 mM hydroquinone and 416 μL 3.6 M sodium bisulfite at pH 5 were added, and incubation was done under mineral oil at 37°C for 16 hours. Bisulfite-modified DNA was purified using the QIAquick PCR Purification Kit (Qiagen, Valencia, CA). Modification was completed by adding 11 μL of 3 M sodium hydroxide to 50 μL purified DNA solution, and incubation was done at 37°C for 10 minutes, followed by ethanol precipitation. DNA was resuspended in 30 μL of 2 mM Tris-HCl (pH 8.0) and stored at −20°C.
Methylation-specific PCR
In the first round of PCR, DNA was amplified using the universal primers (forward: 5′-atttaatagattggagtat-3′; reverse: 5′-ccaacaactaaacaaaac-3′). PCR reactions were performed in a final reaction volume of 20 μL, containing 25 ng bisulfite-modified DNA and 1 μL DMSO under the following conditions: initial denaturation at 95°C for 3 minutes, followed by 40 cycles at 95°C for 30 seconds, annealing at 45°C for 30 seconds, extension at 72°C for 30 seconds, and final extension at 72°C for 10 minutes. For the −25 site, the methylation-specific primers (forward: 5′-gaaagtcggttaaagttttcgga-3′; reverse: 5′-acaaaacctctctccgcccg-3′) or the unmethylation-specific primers (forward: 5′-ttgaaagttggttaaagtttttgga-3′; reverse: 5′-aaacaaaacctctctccaccca-3′) were applied with 1 μL DMSO under the following conditions: initial denaturation at 95°C for 3 minutes, followed by 40 cycles at 95°C for 30 seconds, annealing at 65°C (for methylated reaction) or 62°C (for unmethylated reaction) for 30 seconds and extension at 72°C for 30 seconds, and a final 10-minutes extension at 72°C. The PCR products obtained were electrophoresed on 1.5% agarose gel. First-round PCR product was diluted 1:10 in water, and 1 μL diluted solution was taken to carry out the nested second-round PCR in 20 μL. For the −140 site, the methylation-specific primers (forward: 5′-aggagtaatttgaagaaagttgaggg-3′; reverse: 5′-actttcaactacccctcaccg-3′) or the unmethylation-specific primers (forward: 5′-aggagtaatttgaagaaagttgaggg-3′; reverse: 5′-actttcaactacccctcacca-3′) were applied with 1 μL DMSO under the following conditions: initial denaturation at 95°C for 3 minutes, followed by 40 cycles at 95°C for 30 seconds, annealing at 66°C (for methylated reaction) or 65°C (for unmethylated reaction) for 30 seconds and extension at 72°C for 30 seconds, and a final 10-minute extension at 72°C.
Results
Effects of TGF-β on proliferation of 2 B-cell lymphoma cell lines, RL and DB
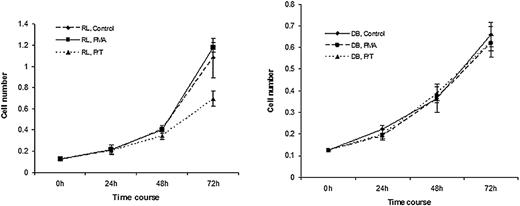
It has been previously shown that RL and DB cells do not express TGF-β receptors on their cell surface, but treatment with a low dose of PMA induced the expression of TGF-β receptors on their cell surface.25 To determine the effect of TGF-β treatment on the proliferative response of these cell lines, RL and DB cells were treated with PMA and TGF-β for various periods of time, and the proliferation was measured by cell counts. As shown in Figure 1, while PMA treatment alone had no effect on the proliferative response of either cell type, addition of TGF-β to the PMA-treated cells caused a significant growth suppression of RL cells. In contrast, TGF-β treatment had no effect on the growth of DB cells.
Treatment with PMA/TGF-β caused growth suppression in RL but not in DB cells. Lymphoma cells were plated at 0.125 × 106 cells/mL and treated with either medium alone, PMA (0.15 ng/mL), or PMA/TGF-β (10 ng/mL) at various time periods. At the end of each time point, cell counts were performed. Results are representative of 3 experiments.
Treatment with PMA/TGF-β caused growth suppression in RL but not in DB cells. Lymphoma cells were plated at 0.125 × 106 cells/mL and treated with either medium alone, PMA (0.15 ng/mL), or PMA/TGF-β (10 ng/mL) at various time periods. At the end of each time point, cell counts were performed. Results are representative of 3 experiments.
Examination of the TGF-β signaling in RL and DB cells
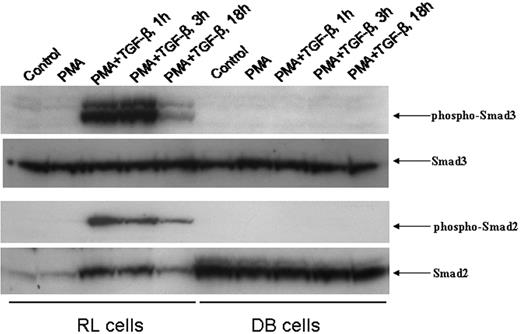
To examine the molecular mechanism underlying the unresponsiveness of DB cells to TGF-β treatment, we first assessed the integrity of the TGF-β signaling pathway in this cell line. We examined the kinetics of nuclear translocation of phospho-Smad3 and phospho-Smad2 upon TGF-β treatment. RL and DB cells were pretreated with PMA for 18 hours, after which TGF-β was added to the culture for various periods of time. While Western blot analysis of the nuclear extracts of RL cells revealed the up-regulation of nuclear phospho-Smad3, no phospho-Smad3 was detected in the nuclear extracts of DB cells (Figure 2). Similarly, no nuclear translocation of phospho-Smad2 was observed in DB cells, whereas the TGF-β–induced nuclear translocation of phospho-Smad2 was quite significant in RL cells (Figure 2). When the membranes were stripped and reprobed with either anti-Smad3 or anti-Smad2 antiserum, basal levels of both Smad3 and Smad2 were observed in the nuclear extracts of DB cells. These data suggested that the lack of TGF-β signaling could be responsible for the unresponsiveness of DB cells to TGF-β treatment.
Time course of nuclear translocation of phospho-Smad3 and phospho-Smad2 induced by PMA/TGF-β treatment. An equal amount of nuclear lysates (30 μg) from RL and DB cells treated with PMA/TGF-β for the time points indicated were analyzed by Western blot analysis. The membranes were probed and reprobed after stripping with different antisera as indicated above.
Time course of nuclear translocation of phospho-Smad3 and phospho-Smad2 induced by PMA/TGF-β treatment. An equal amount of nuclear lysates (30 μg) from RL and DB cells treated with PMA/TGF-β for the time points indicated were analyzed by Western blot analysis. The membranes were probed and reprobed after stripping with different antisera as indicated above.
Status of nuclear p21Cip1/WAF1 and c-Myc in DB and RL cells
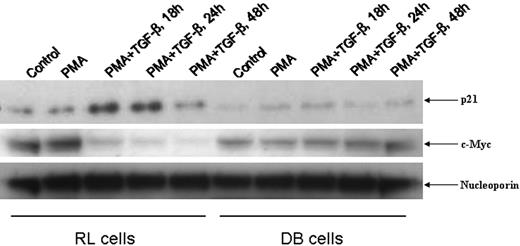
To further investigate whether lack of nuclear translocation of phospho-Smads could account for the unresponsiveness of DB cells to TGF-β–mediated growth suppression, we checked the status of nuclear p21Cip1/WAF1 (hereafter referred to as p21) and c-Myc, 2 TGF-β–responsive genes.26 As shown in Figure 3, PMA/TGF-β treatment up-regulated nuclear p21 and down-regulated c-Myc expression in RL cells in a time-dependent manner, whereas no inducible nuclear p21 and down-regulation of c-Myc were observed in DB cells at any time point. These data strongly suggested a deficit in TGF-β signaling in DB cells.
Status of nuclear p21Cip1/WAF1 and c-Myc in RL and DB cells. An equal amount of nuclear lysates (30 μg) from RL and DB cells treated with PMA/TGF-β for the time points indicated were analyzed by Western blot analysis. The membranes were probed and reprobed after stripping with different antisera as indicated above.
Status of nuclear p21Cip1/WAF1 and c-Myc in RL and DB cells. An equal amount of nuclear lysates (30 μg) from RL and DB cells treated with PMA/TGF-β for the time points indicated were analyzed by Western blot analysis. The membranes were probed and reprobed after stripping with different antisera as indicated above.
Status of TβRI and TβRII in DB and RL cells
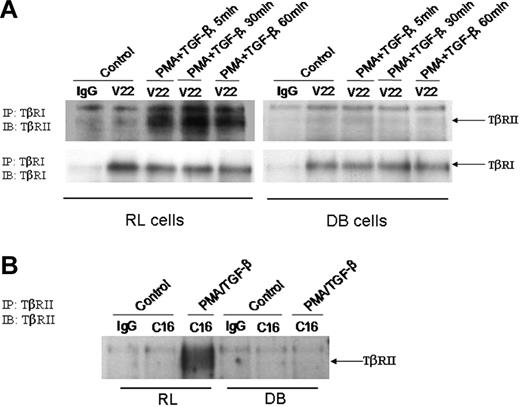
Lack of TGF-β signaling prompted us to investigate the status of TβRI and TβRII in DB cells. To determine the TGF-β–induced interaction between TβRI and TβRII, DB cells were treated with PMA/TGF-β for various periods of time. Membrane fractions were prepared and subjected to immunoprecipitation with anti-TβRI antibody, after which the immunoprecipitated samples were analyzed by Western blot analysis. As shown in Figure 4A, treatment of RL cells with PMA/TGF-β–induced interaction between receptors I and II as early as 5 minutes, and the interaction persisted throughout the time points tested. In contrast, we did not observe any TGF-β–induced interaction between receptors I and II in the membrane fractions of DB cells (Figure 4A). The lack of interaction was not due to inefficient immunoprecipitation, since the anti-TβRI antibody brought down comparable levels of TβRI from both RL and DB samples. These data suggested that the lack of TGF-β signaling in DB cells could be due to an alteration in TGF-β–induced interaction between receptors I and II.
Status of TβRI and TβRII in RL and DB cells. (A) An equal amount of membrane proteins (150 μg) were used for immunoprecipitation with anti-TβRI antibody. The immunoprecipitated (IP) complexes were analyzed by Western blot analysis. The membranes were first probed with anti-TβRII antibody, and then stripped and reprobed with anti-TβRI antibody. (B) An equal amount of membrane fractions (150 μg) were used for immunoprecipitation with anti-TβRII antibody. The immunoprecipitated samples were analyzed by Western blot analysis with the anti-TβRII antibody. IB indicates immunoblot.
Status of TβRI and TβRII in RL and DB cells. (A) An equal amount of membrane proteins (150 μg) were used for immunoprecipitation with anti-TβRI antibody. The immunoprecipitated (IP) complexes were analyzed by Western blot analysis. The membranes were first probed with anti-TβRII antibody, and then stripped and reprobed with anti-TβRI antibody. (B) An equal amount of membrane fractions (150 μg) were used for immunoprecipitation with anti-TβRII antibody. The immunoprecipitated samples were analyzed by Western blot analysis with the anti-TβRII antibody. IB indicates immunoblot.
The absence of interaction between TβRI and TβRII in DB cells could be due to modification in either receptor I or receptor II resulting in the lack of interaction, or total absence of receptor II protein in DB cells. To investigate the status of TβRII, we used membrane fractions from RL and DB cells for immunoprecipitation with anti-TβRII antibody, and the immunoprecipitated samples were tested for the presence of TβRII protein by Western blot analysis. As shown in Figure 4B, while TβRII was present in RL cells, no receptor II was detected in the membrane fractions of DB cells. These data suggested that the unresponsiveness of DB cells toward TGF-β treatment could be due to the lack of functional TβRII at the cell surface.
Effect of ectopically expressed TβRII in DB cells
If the lack of TβRII is responsible for the unresponsiveness of DB cells to TGF-β–mediated growth suppression, reintroduction of TβRII should render DB cells responsive to TGF-β treatment, unless a truncated version of the receptor is acting in a dominant-negative fashion. To investigate the ability of TβRII to restore TGF-β responsiveness, we expressed either WT or a C-terminal–truncated version of receptor II that lacked kinase activity in DB cells. As shown in Figure 5A, ectopically expressed WT, but not c-terminal–truncated receptor II, rendered DB cells responsive to PMA/TGF-β–mediated growth suppression. To monitor whether responsiveness of TβRII-transfected DB cells to PMA/TGF-β treatment was due to the regeneration of TGF-β signaling, we tested the nuclear appearance of phospho-Smad3 and phospho-Smad2 upon PMA/TGF-β treatment. As shown in Figure 5B, both phospho-Smad3 and phospho-Smad2 appeared in the nucleus upon PMA/TGF-β treatment only in the WT-transfected, but not in the c-terminal–truncated receptor II–transfected DB cells. As a consequence of successful TGF-β signaling, we also observed nuclear expression of p21Cip1/WAF1 and down regulation of c-Myc, 2 TGF-β–responsive genes, in the WT receptor II–transfected DB cells only (Figure 5C). Collectively, these data strongly suggested that the lack of TβRII at the cell surface might be responsible for the unresponsiveness of DB cells to TGF-β treatment.
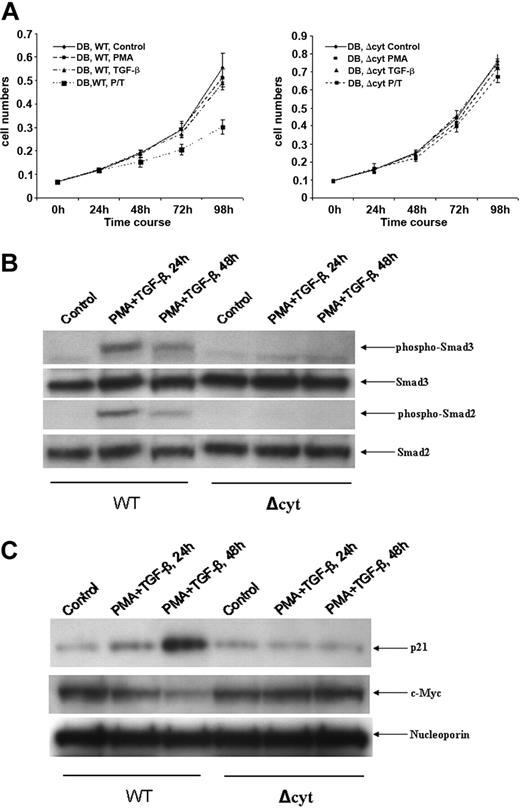
Effect of ectopically expressed TβRII on the responsiveness of DB cells to TGF-β treatment. (A) DB cells were transfected with either WT or Δcyt of TβRII. After transfection, cells (0.1 × 106 cells/mL) were treated with either medium alone, PMA (0.15 ng/mL), TGF-β (10 ng/mL), or PMA/TGF-β (10 ng/mL) for various periods of time. At the end of each time point, cell counts were performed. (B) An equal amount of nuclear lysates (30 μg) from WT- and Δcyt-transfected DB cells treated with PMA/TGF-β for the time points indicated were analyzed by Western blot analysis. The membranes were probed and reprobed after stripping with different antisera as indicated in (A). (C) An equal amount of nuclear lysates (30 μg) from WT- and Δcyt-transfected DB cells treated with PMA/TGF-β for the time points indicated were analyzed by Western blot analysis. The membranes were probed and reprobed after stripping with different antisera as indicated above.
Effect of ectopically expressed TβRII on the responsiveness of DB cells to TGF-β treatment. (A) DB cells were transfected with either WT or Δcyt of TβRII. After transfection, cells (0.1 × 106 cells/mL) were treated with either medium alone, PMA (0.15 ng/mL), TGF-β (10 ng/mL), or PMA/TGF-β (10 ng/mL) for various periods of time. At the end of each time point, cell counts were performed. (B) An equal amount of nuclear lysates (30 μg) from WT- and Δcyt-transfected DB cells treated with PMA/TGF-β for the time points indicated were analyzed by Western blot analysis. The membranes were probed and reprobed after stripping with different antisera as indicated in (A). (C) An equal amount of nuclear lysates (30 μg) from WT- and Δcyt-transfected DB cells treated with PMA/TGF-β for the time points indicated were analyzed by Western blot analysis. The membranes were probed and reprobed after stripping with different antisera as indicated above.
Examination of the TβRII gene in B-cell lymphoma cell lines
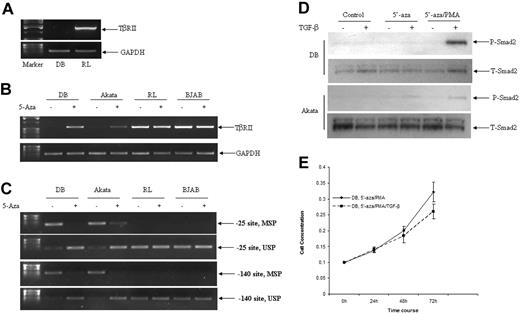
In order to determine whether DB cells completely lacked TβRII message, or carried a modified form of TβRII message, attempts were made to amplify full-length TβRII cDNA by RT-PCR using total RNAs from DB and RL cells. Although we were able to amplify the full-length TβRII cDNA from RL cells, no PCR product was obtained from DB cells (Figure 6A). To determine whether there is any mutation in the TβRII gene in DB cells, all 7 exons of the TβRII gene were amplified, and DNA sequencing was performed. No alteration was observed in any of the exons from the TβRII gene (data not shown). To investigate whether promoter methylation is involved in the suppression of TβRII gene expression, DB cells were treated with the demethylating agent, 5′-azacytidine, according to the protocol described in Materials and methods. As shown in Figure 6B, treatment with 5′-azacytidine restored TβRII gene expression in DB cells. To assess whether the suppression of TβRII gene expression due to the promoter methylation was unique in DB cells, we tested another B-cell lymphoma cell line, Akata, which has been shown to be resistant to TGF-β–mediated growth suppression and lacks TβRII protein.7 Treatment with 5′-azacytidine also resulted in the expression of TβRII message in Akata cells (Figure 6B). RL and BJAB, 2 B-cell lymphoma cell lines that are responsive to TGF-β–mediated growth suppression and have functional TβRII, were used as a positive control (Figure 6B). To determine the status of promoter methylation, CpG sites at −140 and −25 of the TβRII gene were analyzed by methylation-specific and unmethylation-specific primers using bisulfite-modified DNA. As shown in Figure 6C, both DB and Akata cells were positive for methylation-specific primers corresponding to both promoter sites, which were reversed after 5′-azacytidine treatment, indicating that both sites were methylated in DB and Akata cells, and that the methylation status was reversed after 5′-azacytidine treatment. TGF-β–responsive cell lines RL and BJAB, which harbor functional TβRII, were negative for methylation-specific primers, whereas they were positive for unmethylated-specific primers (Figure 6C). Next, we wanted to determine whether 5′-azacytidine–treated DB and Akata cells became responsive to TGF-β treatment. As shown in Figure 6D, both DB and Akata cells were responsive to TGF-β treatment as demonstrated by the inducible phosphorylation of Smad2 after TGF-β treatment. To investigate the functional outcome, 5′-azacytidine–treated DB cells were treated with TGF-β for various periods of time, and the cell numbers were determined to monitor the proliferation. As shown in Figure 6E, 5′-azacytidine–treated DB cells were partially responsive to TGF-β–mediated growth suppression, indicating the reversal of the nonfunctional status of TβRII in DB cells. Collectively, these data indicated that the nonresponsiveness of some B-cell lymphoma cell lines to TGF-β was due to the promoter methylation of the TβRII gene.
Defective expression of TβRII was associated with promoter methylation in B-cell lymphoma cell lines. (A) RT-PCR analysis of TβRII transcription in DB and RL cells. Total RNAs (2 μg) from DB and RL cells were used for cDNA synthesis using a primer pair corresponding to full-length cDNA. (B) RT-PCR analysis of TβRII transcription in B-cell lymphoma cell lines before and after 5′-azacytidine treatment. (C) Methylation status of TβRII gene promoter in B-cell lymphoma cell lines before and after 5′-azacytidine treatment. Methylation-specific PCR (MSP) and unmethylation-specific PCR (USP) corresponding to −25 and −140 promoter regions were performed using bisulphate-modified DNA as template. (D) Status of phospho-Smad2 in DB and Akata cells after 5′-azacytidine. An equal amount of whole-cell lysates (60 μg) from DB and Akata cells before and after 5′-azacytidine treatment were analyzed by Western blot analysis. The membranes were stripped and reprobed with total-Smad2 as indicated above. (E) Treatment with 5′-azacytidine rendered DB cells partially responsive to TGF-β. Lymphoma cells were treated with 5′-azacytidine and PMA as described in Materials and methods. After 1 week, cells were plated at 0.1 × 106/mL and treated with TGF-β (10 ng/mL) for various time periods. At the end of each time point, cell counts were performed.
Defective expression of TβRII was associated with promoter methylation in B-cell lymphoma cell lines. (A) RT-PCR analysis of TβRII transcription in DB and RL cells. Total RNAs (2 μg) from DB and RL cells were used for cDNA synthesis using a primer pair corresponding to full-length cDNA. (B) RT-PCR analysis of TβRII transcription in B-cell lymphoma cell lines before and after 5′-azacytidine treatment. (C) Methylation status of TβRII gene promoter in B-cell lymphoma cell lines before and after 5′-azacytidine treatment. Methylation-specific PCR (MSP) and unmethylation-specific PCR (USP) corresponding to −25 and −140 promoter regions were performed using bisulphate-modified DNA as template. (D) Status of phospho-Smad2 in DB and Akata cells after 5′-azacytidine. An equal amount of whole-cell lysates (60 μg) from DB and Akata cells before and after 5′-azacytidine treatment were analyzed by Western blot analysis. The membranes were stripped and reprobed with total-Smad2 as indicated above. (E) Treatment with 5′-azacytidine rendered DB cells partially responsive to TGF-β. Lymphoma cells were treated with 5′-azacytidine and PMA as described in Materials and methods. After 1 week, cells were plated at 0.1 × 106/mL and treated with TGF-β (10 ng/mL) for various time periods. At the end of each time point, cell counts were performed.
Discussion
In this report, we describe a mechanism by which some tumor cells can avoid TGF-β–mediated growth suppression. A diffuse large B-cell lymphoma cell line, DB, which is refractory to TGF-β–mediated growth suppression, lacks TβRII on its cell surface while maintaining a normal level of TβRI. On the other hand, another diffuse large B-cell lymphoma cell line, RL, which is susceptible to TGF-β–mediated growth suppression, has a normal level of both TβRI and TβRII. The finding that DB cells lack TβRII at its cell surface is in apparent contradiction with our previous finding, where we observed the comparable levels of 125I-labeled TGF-β binding to RL and DB cells after chemical cross-linking.25 One possible explanation for the apparent discrepancy may be due to the fact that an altered form of TβRII is present in DB cells, which demonstrates a normal binding capability to TGF-β but lacks other functions. Another possibility is that DB cells carry a low level of TβRII on their cell surface, which is enough to produce a 125I-labeled band on gel electrophoresis, but is not sufficient to produce any functional outcome. Our preliminary effort to amplify full-length TβRII cDNA by RT-PCR using total RNAs from DB cells was unsuccessful, suggesting a low or undetectable level of TβRII message present in DB cells. Treatment with a demethylating agent, 5′-azacytidine, restored TβRII gene expression in DB cells. In accordance, promoter analysis revealed CpG methylations at −25 and −140 that were correlated with the gene silencing. Present data clearly demonstrate that DB cells lack functional TGF-β receptor II, and this functional deficit can be overcome either by ectopic expression of the WT receptor or demethylation of the TβRII gene promoter.
The TβRII gene is a frequent target for mutational inactivation in colorectal cancers with microsatellite instability.11,12 Microsatellite stable colon cancers have also been shown to harbor mutations in the TβRII gene.10 Mutations were found within a polyadenine tract in colorectal cancers with microsatellite instability that affected both alleles of the TβRII gene.11 Human colon cancer cell lines with high rates of microsatellite instability have also been shown to carry mutated TβRII, resulting in the absence of cell-surface receptor II, and small amounts of RII transcripts.27 Missense mutations in the TβRII gene have also been found in 2 human squamous head and neck carcinoma cell lines.28 Both mutations were located within the conserved serine-threonine kinase domain.28 Expression of the TβRII gene has also been shown to be the target of the EWSR1-FLI1 fusion gene product in Ewing sarcoma.29 Both fusion-positive Ewing sarcoma cell lines and primary tumors have been shown to express low or undetectable levels of TβRII mRNA and protein product.29 Mutations at the TGF-β receptor complex have been reported to be involved in escape mutant responsible for progression of lymphomatoid papulosis to systemic lymphoma.30 Promoter methylation has also been shown to be responsible for the aberrant expression of the TβRII gene in primary non–small-cell lung cancer and prostate cancer cells,31,32 and prostate and lung cancer cell lines.33,34
An examination of the study by Alizadeh et al18 identified TβRII as one of the hundred discriminatory markers for subclassification of DLBCL. While the germinal center–like DLBCL class showed a relative decrease in TβRII expression, the activated B-cell–like DLBCL subclass showed a relative increase in TβRII expression. Heterogeneity in relative expression of TβRII within subclasses was observed. However, this heterogeneity was consistent with observations and statements by the authors18 regarding heterogeneity seen in the complete gene set used for classification. In an alternative classification scheme, Monti et al35 observed that the relative level of transcript for TβRII was increased in the host response cluster of DLBCL compared with the oxidative phosphorylation (OxPhos) cluster and B-cell receptor (BCR)/proliferation cluster. In addition, a comparison of DLBCLs with follicular lymphomas by Goy et al36 revealed a relative increase in TβRII transcript in follicular lymphomas compared with DLBCLs. Altered levels of TβRII expression observed in our study are supported by differences in TβRII expression in subclasses of spontaneously occurring DLBCL. These analyses of the heterogeneity of TβRII expression observed in spontaneous lymphomas may provide useful information on the regulation of the proliferation of these lymphomas as well as the molecular mechanism for regulation of TβRII expression.
It has been shown previously that the resistance of Burkitt lymphoma and Epstein-Barr virus (EBV)–transformed B lymphoblastoid cell lines to TGF-β correlates with a reduced expression of TβRII.14 Interestingly, both RL and DB are EBV-negative cell lines,14,17 and our data indicate that DB cells lack expression of TβRII due to the promoter methylation. The promoter methylation–mediated silencing of TβRII gene expression was not unique in DB cells, since we demonstrate similar suppression of TβRII gene expression in another TGF-β nonresponsive B-cell lymphoma cell line, Akata. Treatment with 5′-azacytidine restored TβRII expression in both cell lines and rendered these cell lines responsive to TGF-β treatment. Identification of the mechanisms underlying the promoter methylation and the subsequent effect of the methylation on the TβRII gene transcription will help us understand in more detail the mechanism by which tumor cells become resistant to TGF-β1–mediated growth suppression.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We would like to thank Dr Joan Massague for providing us with the TβRII plasmids.
This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
National Institutes of Health
Authorship
Contribution: G.C., H.O., L.R., and J.Y. performed research; P.G. wrote the paper; C.Y.S. contributed intellectually; T.J.O. helped in techniques; D.L.L. contributed intellectually and supervised the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paritosh Ghosh, Lymphocyte Cell Biology Section, Laboratory of Immunology, Gerontology Research Center, National Institute on Aging, National Institutes of Health, 5600 Nathan Shock Dr, Baltimore, MD 21224; e-mail: ghoshp@grc.nia.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal