Abstract
We recently identified a disease-specific gene CLLU1 in chronic lymphocytic leukemia (CLL) and also demonstrated that high CLLU1 expression levels predict poor clinical outcome. To validate this finding, we measured CLLU1 mRNA expression levels by real-time reverse transcriptase–polymerase chain reaction (RT-PCR) in 175 patients with CLL. Analyses of IgVH mutational status, ZAP-70 expression, CD38 expression, and chromosomal aberrations were also performed. High levels of CLLU1 expression were associated with shorter overall survival (P < .001), with a 7% increase in risk of early death by each doubling of the CLLU1 expression level. Stratification for age at diagnosis demonstrated a strong prognostic significance of CLLU1 expression in patients younger than 70 years (P < .001), but not in patients aged 70 or older (P = .61). The prognostic significance of IgVH mutational status and ZAP-70 expression had a similar age-dependent variation. Multivariate analysis in the younger age group showed that CLLU1 expression analysis added further prognostic information within all prognostic subgroups, with the exception of patients with unmutated IgVH CLL. Only CLLU1 expression and IgVH mutational status had independent predictive power. Thus, analysis of CLLU1 expression is highly applicable in risk prediction in CLL for patients of an age eligible for risk stratification.
Introduction
We recently demonstrated unique overexpression in chronic lymphocytic leukemia (CLL) of a novel CLL-specific gene CLL Up-regulated gene 1 (CLLU1) located at chromosome 12q22.1 In a small cohort we furthermore demonstrated that CLLU1 mRNA expression levels in patients with CLL predict time to initiation of therapy and overall survival and that CLLU1 is highly up-regulated in any poor-risk group defined by commonly used predictors in CLL.2
Chronic lymphocytic leukemia has long been known to have a variable clinical course.3 The discovery that patients with CLL have varying degrees of somatic hypermutations in the variable region of immunoglobulin heavy chain (IgVH) genes4 led to the first demonstration of a CLL-specific genetic marker (ie, IgVH mutational status) related to clinical course and disease outcome.5–7 These findings raised the possibility that patients who might benefit from early or aggressive treatment could be identified by biologic risk prediction. Subsequent studies have identified a multitude of new biologic risk predictors that can recognize patients with CLL with poor prognosis, even in early stage of disease. Apart from mutational status, the most well-investigated and commonly used biologic prognostic markers are a hierarchy of cytogenetic aberrations defined by del(17)(p13), del(11)(q22), trisomy 12, or del(13)(q14)8,9 ; expression of the intracellular 70-kDa ζ-associated protein (ZAP-70)10–13 ; and expression of the transmembrane glycoprotein CD38.6,14,15
Far from resulting in a straightforward risk prediction in CLL, the introduction of new prognostic markers has exposed a more complex heterogeneity of the disease. Few patients have CLL with only prognostically favorable or unfavorable characteristics; many have CLL with various combinations of favorable and unfavorable traits. Moreover, investigations of the prognostic information offered by these new biomarkers have mainly been performed in retrospective study cohorts and most importantly do not include comparisons of all variables. On this background the use of new prognostic markers has not yet been introduced as the basis for treatment decision,16 which is still essentially directed by clinical staging17,18 and current treatment guidelines.19
The fact that the novel gene CLLU1 is disease specific, albeit the function is unknown, might imply that CLLU1 expression holds some prognostic information not offered by other biologic markers in CLL. We investigated CLLU1 mRNA expression in a large series of patients along with the most commonly used biologic markers to assess the role of this novel gene in risk prediction in CLL.
Patients, materials, and methods
Patient selection and clinical database
Patients with a diagnosis of CLL referred to the Hematological Department of Rigshospitalet, University Hospital of Copenhagen, Denmark, during the period 1991 to 1999 were selected for this investigation. Criteria for inclusion were untreated disease at time of referral, diagnosis made less than 6 months prior to referral, and confirmation of diagnosis by standard immunophenotypic and morphologic criteria, including absolute lymphocytosis above 5 × 109/L.19 A total of 281 patients met the inclusion criteria. Frozen cell samples were available in 176 cases. Clinical data were obtained from the patients' medical records. Sixteen of these patients had already been investigated as part of our initial prognostic study. Assessment of prognostic factors in this subset was independently repeated in this study. All statistical analyses were performed with and without these 16 patients, giving no significant difference in results. Handling of samples adhered to the local research governance procedures and was approved by The Regional Committee for Scientific Ethics (KF 01-021/97, KF 01-021/03, and KF 01-021/03) and conducted in accordance with the Declaration of Helsinki.
Samples and sample preparation
Analyses were performed on samples of either peripheral blood or bone marrow, predominantly consisting of Ficoll-separated mononuclear cells, which had been stored at −80°C or in liquid nitrogen, almost exclusively deriving from time of diagnosis. In 10 cases fresh blood samples were collected later from untreated patients. Total RNA was procured from samples by use of QIAamp RNA Blood mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Contaminating DNA was removed with RNase-free DNase Set (Qiagen). RNA yield was quantified by Nano Drop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE). Viable mononuclear cells were isolated for immunophenotypic analysis by Lymfoprep (Nycomed Pharma, Oslo, Norway) separation. Normal B cells from healthy donors were isolated by magnetic cell sorting (MACS) of Lymfoprep separated buffy coats using MACS CD19 Microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). One hundred 5 frozen samples from the time of diagnosis, already prepared for cytogenetic analysis, were available for fluorescence in situ hybridization (FISH) analysis. In 21 additional cases excessive cells after preparation for flow cytometric analysis were stored in Carnoy fixation and also made available for FISH analysis.
Quantitative reverse transcriptase–polymerase chain reactions (QRT-PCRs)
Quantification of CLLU1 expression was performed on an ABIPrism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) with the use of TaqMan One-Step RT-PCR Master Mix Reagents (Applied Biosystems) as previously described.2 Amplification of the cDNA1 sequence was performed by the comparative Ct method of relative quantification using β2-microglobulin as an internal standard and a pool of isolated normal B cells as calibrator.20 The probes and primers used were as previously described.2 CLLU1 expression levels were measured as fold up-regulation in relation to normal B cells. Satisfactory results of CLLU1 expression were obtained in all but one case (n = 175), which was dismissed because of poor RNA quality.
IgVH sequence analysis
First-strand complementary DNA (cDNA) was synthesized from 1 μg RNA using Superscript III (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Amplification of cDNA was performed using a set of 6 variable region (VH) family-specific upstream primers annealing to sequences in the leader region21 along with a downstream primer specific to the framework I region and the heavy-chain joining region (JH).22 In cases of failed amplification, downstream primers complementary to the constant regions Cμ and Cδ were used.23 PCR products were purified with Qiaquick PCR Purification Kit (Qiagen) or in some cases by gel extraction using E.Z.N.A gel extraction kit (Omega Bio-tek, Doranville, GA). Products were sequenced from both strands with BigDye Terminator v.1.1 (Applied Biosystems) following the manufacturer's instructions and precipitated with ethanol before evaluation by ABIPrism 3730 Gene sequencer (Applied Biosystems). Nucleotide sequences were analyzed using the VBASE24 and IgBLAST25 databases. Mutational status was assessed using 98% homology to germ line as cut-off. IgVH in frame rearrangements and hence mutational status were obtained in 136 cases.
Flow cytometry
All flow cytometric analyses were performed on a FACSCalibur (Becton Dickinson, San Jose, CA). Analysis of intracellular ZAP-70 expression was performed according to the indirect method12 with minor modifications26 using unconjugated anti-ZAP70 monoclonal antibody (mAb) 2F3.2 (Upstate, Waltham, MA). The lymphocyte population was selected based on forward scatter and side scatter characteristics and then gated on B cells and T and natural killer cells. For each sample at least 500 ZAP-70–positive T lymphocytes were acquired to enable setting of the gate corresponding to 98% ZAP-70–positive T cells. This gate was then applied to assess the ZAP-70 expression level within the B-cell population. ZAP-70 status was achieved in 132 cases. A cut-off level of 20% was used for statistical analysis. For analysis of CD38 expression anti-CD38 mAb HB-7 (Becton Dickinson) was used, and CD38 expression level was assessed as the percentage of CD38+ cells of the gated B-cell population. Flow cytometric measurement of CD38 expression was obtained in 130 cases. A cut-off level of 20% was used for statistical analysis of CD38 expression.14
FISH
FISH analysis of interphase cells was performed with the use of a CLL probe panel (Vysis, Downers Grove, IL) according to the manufacturer's instructions. Signals were visualized by use of an epifluorescence microscope (Zeiss Axioscop, Oberkochen, Germany), and images were captured by the Quips Smart Capture FISH Imaging Software (Vysis). On the basis of FISH analyses of healthy controls, cases were classified as deletions if more than 10% of interphase cells harbored less than 2 signals of 13q14.3 or one signal of either p53 or ATM2, whereas cases were classified as trisomy 12 if more than 10% of interphase cells presented more than 2 signals of CEP 12. In cases with more than one type of genetic aberration, patients were assigned to that associated with the highest risk according to a hierarchic model.8 A total of 200 interphase cells were analyzed with each probe. Cytogenetic assessment was performed in a total of 126 cases. For subsequent statistical analyses chromosomal aberrations were divided into 2 risk groups: lower risk with del(13q), normal cytogenetics, or trisomy 12 or higher risk with del(17p) or del(11q).
Statistical analysis
CLLU1 mRNA expression levels are a continuum.2 To asses the influence on overall survival of CLLU1 expression levels without assumptions concerning the functional form, a Cox model27 was used modeling the effect of CLLU1 using penalized splines. To obtain a more interpretable presentation of the association the logarithm of the CLLU1 expression was used instead of the spline model. The influences of further predictors were also modeled in the Cox model.
All analyses were performed using CLLU1 as a continuous variable, but for ease of presentation, we defined a cut-off value to separate high- from low-expression levels. To ensure that conclusions were not substantially affected by the choice of cut-off value, various values were used and compared, including the median value, values based on receiver-operating characteristic plots, and the value giving the optimal segregation of Kaplan-Meier curves. On the basis of these considerations, we chose a 40-fold CLLU1 expression level as cut-off. Group comparisons were done using Kaplan-Meier plots and log-rank tests.28 P values of Cox analyses were score test based to make them comparable with standard log-rank tests.
Overall survival was used as an end-point and defined as time from diagnosis to death or end of follow-up, the latter considered censored observations. For univariate CLLU1 analysis, time to treatment was also considered an end-point, defined as time from diagnosis to first treatment or end of follow-up, the latter being censored observations. Statistical analyses were performed with the use of GraphPad Prism version 4.03 (GraphPad Software, San Diego, CA), SAS version 8.02 (The SAS Institute, Cary, NC) and the R Package version 2.2.0 (The R Foundation for Statistical Computing, Vienna, Austria).
Results
Characteristics of the study population
The clinical characteristics of the study cohort are presented in Table 1. The median age at diagnosis was 69.9 years. Of the 175 patients included, 77 (44%) were women and 98 (56%) were men. At time of diagnosis 115 patients (66%) were Binet stage A, 33 (19%) were stage B, and 27 (15%) were stage C. The median follow-up was 6.4 years and the median overall survival 7.0 years. At the time of last follow-up 107 (61%) patients had started treatment and 110 (63%) patients had died.
Patient characteristics
| Characteristic . | Value . |
|---|---|
| Sex, no. (%) | |
| Female | 77 (44) |
| Male | 98 (56) |
| Age at diagnosis | |
| Median, y | 69.9 |
| Range, y | 29.5-92.3 |
| Younger than 70 y, no. (%) | 92 (53) |
| 70 y or older, no. (%) | 83 (47) |
| Overall survival | |
| Median overall survival, y | 7.0 |
| No. of deaths (%) | 110 (63) |
| Censored events, no. (%) | 65 (37) |
| Median follow-up time, y | 6.4 |
| Range, y | 0.01-14.9 |
| Time to treatment | |
| Median time to first treatment, y | 2.3 |
| No. treated (%) | 107 (61) |
| Censored events, no. (%) | 68 (39) |
| Clinical stage, no. (%) | |
| Binet A | 115 (66) |
| Binet B | 33 (19) |
| Binet C | 27 (15) |
| Characteristic . | Value . |
|---|---|
| Sex, no. (%) | |
| Female | 77 (44) |
| Male | 98 (56) |
| Age at diagnosis | |
| Median, y | 69.9 |
| Range, y | 29.5-92.3 |
| Younger than 70 y, no. (%) | 92 (53) |
| 70 y or older, no. (%) | 83 (47) |
| Overall survival | |
| Median overall survival, y | 7.0 |
| No. of deaths (%) | 110 (63) |
| Censored events, no. (%) | 65 (37) |
| Median follow-up time, y | 6.4 |
| Range, y | 0.01-14.9 |
| Time to treatment | |
| Median time to first treatment, y | 2.3 |
| No. treated (%) | 107 (61) |
| Censored events, no. (%) | 68 (39) |
| Clinical stage, no. (%) | |
| Binet A | 115 (66) |
| Binet B | 33 (19) |
| Binet C | 27 (15) |
CLLU1 expression levels predict overall survival and time to first treatment
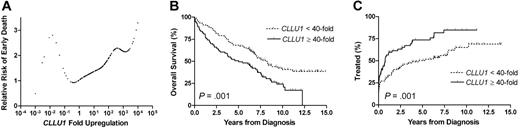
CLLU1 expression levels in the cohort represented a continuum ranging from 0.0005- to 10.000-fold up-regulation compared with that of normal B cells, with a median of 22.9-fold up-regulation. Unlike the other biomarkers investigated, CLLU1 expression levels were not naturally segregated into groups with low and high levels. Applying a Cox proportional hazards model, the CLLU1 expression level was shown to be significantly associated with shorter overall survival, with a hazard ratio (HR) of 1.07 (95% CI, 1.02-1.12; P = .004), signifying a relative increase in risk of dying of 7% by each doubling of the CLLU1 expression level. Although the proportional relation between increased levels of CLLU1 expression and mortality is evident for the vast majority of patients (n = 164), it should be noted that the model does not apply for CLLU1 expression levels much below that of normal B cells (n = 11) (Figure 1A).
CLLU1 expression and risk prediction in 175 patients with CLL. (A) Estimates of relative risk of early death as a continuous function of the CLLU1 mRNA expression level in CLL cells based on a Cox model. An expression level corresponding to that of normal B cells is denoted 1. Most observations are in the range between 0.5- to 1000-fold CLLU1 up-regulation, in which a proportional relationship between risk and CLLU1 fold up-regulation is indicated. The relationship was estimated to correspond to an increased risk of 7% for early death associated with each doubling of the CLLU1 expression level in this range. For CLLU1 expression levels less than 0.5-fold this relationship did not hold. (B) Overall survival according to low or high levels of CLLU1 expression used 40-fold up-regulation as cut-off. The median overall survival was 8.1 years for patients with low CLLU1 expression levels and 5.0 years for patients with high CLLU1 expression levels. (C) Time to first treatment according to low or high levels of CLLU1 expression. The median time to first treatment was 4.6 years in patients with low CLLU1 expression levels and only 9.0 months for patients with high CLLU1 expression levels.
CLLU1 expression and risk prediction in 175 patients with CLL. (A) Estimates of relative risk of early death as a continuous function of the CLLU1 mRNA expression level in CLL cells based on a Cox model. An expression level corresponding to that of normal B cells is denoted 1. Most observations are in the range between 0.5- to 1000-fold CLLU1 up-regulation, in which a proportional relationship between risk and CLLU1 fold up-regulation is indicated. The relationship was estimated to correspond to an increased risk of 7% for early death associated with each doubling of the CLLU1 expression level in this range. For CLLU1 expression levels less than 0.5-fold this relationship did not hold. (B) Overall survival according to low or high levels of CLLU1 expression used 40-fold up-regulation as cut-off. The median overall survival was 8.1 years for patients with low CLLU1 expression levels and 5.0 years for patients with high CLLU1 expression levels. (C) Time to first treatment according to low or high levels of CLLU1 expression. The median time to first treatment was 4.6 years in patients with low CLLU1 expression levels and only 9.0 months for patients with high CLLU1 expression levels.
Analysis of CLLU1 expression as a categorical variable using 40-fold up-regulation as cut-off, showed that patients with CLLU1 expression levels at 40-fold or more had a median overall survival of 5.0 years (95% CI, 2.9-7.1 years) as opposed to 8.1 years (95% CI, 6.7-11.1 years) for patients with lower expression levels (P < .001) (Figure 1B). Similarly, patients with high CLLU1 expression levels had a median time to first treatment of 9.0 months (95% CI, 3.9-26.3 months), whereas the group with lower CLLU1 expression levels had a median time to first treatment of 4.6 years (95% CI, 2.3-8.7 months, P < .001) (Figure 1C).
Age is an important factor in risk prediction in CLL
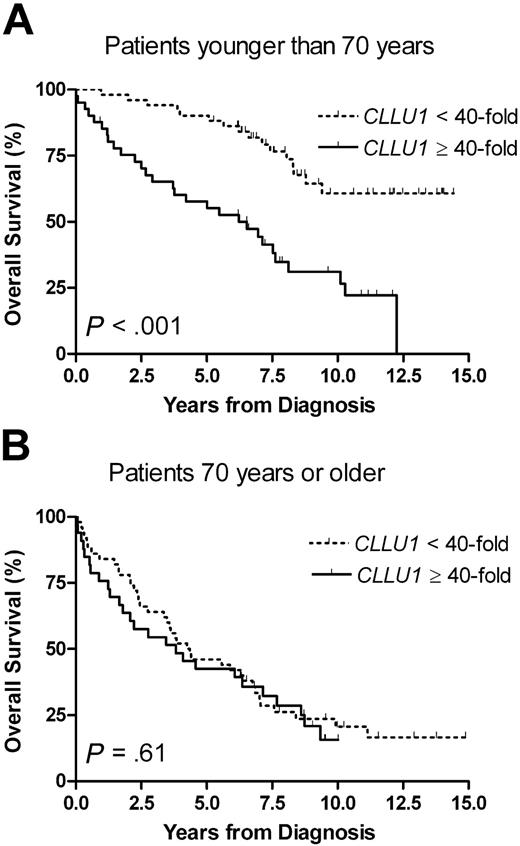
To assess the influence of age-related interaction of non-CLL diseases, we stratified the subsequent analyses by age, using 70 years of age at diagnosis and a 40-fold CLLU1 expression level as cut-off values. Although there was no significant difference in CLLU1 expression levels between age groups, the predictive power of CLLU1 was markedly different. For patients younger than 70 years with high CLLU1 expression levels, the median overall survival was 6.5 years (95% CI, 3.7-8.1 years), whereas it was not reached after 14.9 years for patients with lower CLLU1 levels (P < .001). The prognostic significance of CLLU1 expression disappeared in patients aged 70 or older (P = .61) (Figure 2). This age-dependent variation in predictive power was reproduced when CLLU1 expression was analyzed as a continuous variable: a 17% (HR = 1.17; 95% CI, 1.08-1.27; P < .001) increase in risk of early death by each doubling of CLLU1 expression level was seen for patients younger than 70 years, and no significant increase in risk was seen for patients of 70 years or older (HR = 1.02; 95% CI, 0.96-1.07; P = .59).
Overall survival according to CLLU1 expression levels in different age groups. (A) In patients with CLL younger than 70 years at diagnosis (n = 92) the median overall survival was 6.5 years for patients with high CLLU1 expression levels (n = 41; 45%) but not reached after 14.9 years in patients with low CLLU1 expression levels (n = 51; 55%). (B) In patients 70 years of age or older at diagnosis (n = 83) there was no significant difference in overall survival between groups with high (n = 33; 40%) and low (n = 50; 60%) CLLU1 expression levels.
Overall survival according to CLLU1 expression levels in different age groups. (A) In patients with CLL younger than 70 years at diagnosis (n = 92) the median overall survival was 6.5 years for patients with high CLLU1 expression levels (n = 41; 45%) but not reached after 14.9 years in patients with low CLLU1 expression levels (n = 51; 55%). (B) In patients 70 years of age or older at diagnosis (n = 83) there was no significant difference in overall survival between groups with high (n = 33; 40%) and low (n = 50; 60%) CLLU1 expression levels.
In our cohort the prognostically unfavorable parameters of unmutated IgVH, high expression of ZAP-70 or CD38, and high-risk cytogenetic profile were all associated with a significantly shorter survival in univariate analyses (Table 2). Stratification for age at diagnosis for these markers showed a marked decrease in prognostic significance in older patients for IgVH mutational status and ZAP-70 expression as well, whereas such age dependency was not found for the predictive power of CD38 expression and cytogenetic aberrations (Table 3)
Median overall survival (OS) in investigated prognostic subgroups
| Variable . | No. of patients (%) . | Median OS (95% CI), y . |
|---|---|---|
| CLLU1 expression | 175 (100) | |
| CLLU1 less than 40-fold | 101 (58) | 8.1 (6.7-11.1) |
| CLLU1 at least 40-fold | 74 (42) | 5.0 (2.9-7.1) |
| Clinical stage, Binet | 175 (100) | |
| Stage A | 115 (66) | 8.1 (7.1-10.3) |
| Stage B or C | 60 (34) | 3.8 (2.2-6.5) |
| Mutational status | 136 (77) | |
| IgVH homology less than 98% | 88 (65) | 10.3 (8.4-∞) |
| IgVH homology at least 98% | 48 (35) | 3.6 (2.3-6.2) |
| ZAP-70 expression | 132 (75) | |
| ZAP-70 less than 20% | 51 (39) | Not reached (7.0-∞) |
| ZAP-70 at least 20% | 81 (61) | 4.6 (3.8-7.10) |
| CD38 expression | 130 (74) | |
| CD38 less than 20% | 91 (70) | 9.3 (6.7-∞) |
| CD38 at least 20% | 39 (30) | 4.0 (2.9-7.1) |
| Cytogenetics | 126 (72) | |
| del(13q)/normal/trisomy 12 | 92 (73) | 9.4 (7.1-∞) |
| del(11q)/del(17p) | 34 (27) | 2.3 (1.2-6.2) |
| Variable . | No. of patients (%) . | Median OS (95% CI), y . |
|---|---|---|
| CLLU1 expression | 175 (100) | |
| CLLU1 less than 40-fold | 101 (58) | 8.1 (6.7-11.1) |
| CLLU1 at least 40-fold | 74 (42) | 5.0 (2.9-7.1) |
| Clinical stage, Binet | 175 (100) | |
| Stage A | 115 (66) | 8.1 (7.1-10.3) |
| Stage B or C | 60 (34) | 3.8 (2.2-6.5) |
| Mutational status | 136 (77) | |
| IgVH homology less than 98% | 88 (65) | 10.3 (8.4-∞) |
| IgVH homology at least 98% | 48 (35) | 3.6 (2.3-6.2) |
| ZAP-70 expression | 132 (75) | |
| ZAP-70 less than 20% | 51 (39) | Not reached (7.0-∞) |
| ZAP-70 at least 20% | 81 (61) | 4.6 (3.8-7.10) |
| CD38 expression | 130 (74) | |
| CD38 less than 20% | 91 (70) | 9.3 (6.7-∞) |
| CD38 at least 20% | 39 (30) | 4.0 (2.9-7.1) |
| Cytogenetics | 126 (72) | |
| del(13q)/normal/trisomy 12 | 92 (73) | 9.4 (7.1-∞) |
| del(11q)/del(17p) | 34 (27) | 2.3 (1.2-6.2) |
P values were all less than .001.
Age-stratified Cox proportional hazards analysis of overall survival in poor prognostic subgroups
| Variable . | Younger than 70 y . | 70 y or older . | All ages . | |||
|---|---|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | HR (95% CI) . | P . | |
| CLLU1 at least 40-fold | 3.63 (1.96-6.73) | <.001 | 1.14 (0.69-1.88) | .61 | 1.81 (1.25-2.64) | .002 |
| Binet stage B or C | 2.53 (1.40-4.57) | .002 | 1.52 (0.93-2.48) | .094 | 2.09 (2.08-3.04) | <.001 |
| Unmutated IgVH | 6.04 (2.99-12.20) | <.001 | 2.20 (1.22-3.97) | .007 | 3.53 (2.26-5.51) | <.001 |
| ZAP-70 at least 20% | 4.02 (1.76-9.20) | <.001 | 1.36 (0.72-2.58) | .35 | 2.31 (1.40-3.81) | .001 |
| CD38 at least 20% | 1.91 (0.93-3.90) | .072 | 2.35 (1.27-4.34) | .005 | 2.20 (1.38-3.49) | .001 |
| del(17p)/del(11q) | 3.01 (1.34-6.74) | .005 | 3.43 (1.78-6.62) | <.001 | 3.55 (2.14-5.87) | <.001 |
| Variable . | Younger than 70 y . | 70 y or older . | All ages . | |||
|---|---|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | HR (95% CI) . | P . | |
| CLLU1 at least 40-fold | 3.63 (1.96-6.73) | <.001 | 1.14 (0.69-1.88) | .61 | 1.81 (1.25-2.64) | .002 |
| Binet stage B or C | 2.53 (1.40-4.57) | .002 | 1.52 (0.93-2.48) | .094 | 2.09 (2.08-3.04) | <.001 |
| Unmutated IgVH | 6.04 (2.99-12.20) | <.001 | 2.20 (1.22-3.97) | .007 | 3.53 (2.26-5.51) | <.001 |
| ZAP-70 at least 20% | 4.02 (1.76-9.20) | <.001 | 1.36 (0.72-2.58) | .35 | 2.31 (1.40-3.81) | .001 |
| CD38 at least 20% | 1.91 (0.93-3.90) | .072 | 2.35 (1.27-4.34) | .005 | 2.20 (1.38-3.49) | .001 |
| del(17p)/del(11q) | 3.01 (1.34-6.74) | .005 | 3.43 (1.78-6.62) | <.001 | 3.55 (2.14-5.87) | <.001 |
Only CLLU1 expression and IgVH mutational status have strong independent prognostic significance in patients with CLL presenting before the age of 70
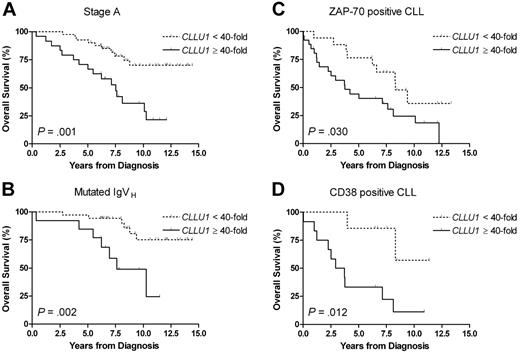
Analysis of high versus low CLLU1 expression in patients younger than 70 years, stratifying for other prognostic factors, revealed a much shorter overall survival for patients with high levels of CLLU1 expression within all the investigated prognostically favorable subgroups (Table 4). In patients in clinical stage A, high CLLU1 expression levels increased the risk of early death by 3.73 times (P < .001) as compared with lower CLLU1 levels (Figure 3A). In patients with mutated IgVH CLL, high CLLU1 expression levels increased the risk of early death 4.81 times (P = .002) (Figure 3B). Also, in the prognostically unfavorable groups of ZAP-70–positive CLL (Figure 3C), CD38+ CLL (Figure 3D), and patients with high-risk cytogenetic profile [ie, del(11q) or del(17p)], high CLLU1 expression levels were associated with significantly shorter overall survival. Only in patients with CLL with unmutated IgVH did CLLU1 expression not add prognostic information (Table 4).
Relative risk of early death associated with high CLLU1 expression levels in prognostic subgroups for patients younger than 70 years
| Variable . | No. of patients . | HR* (95% CI) . | P . |
|---|---|---|---|
| Binet stage A | 65 | 3.73 (1.69-8.24) | <.001 |
| Binet stage B or C | 27 | 2.43 (0.90-6.54) | .071 |
| Mutated | 48 | 4.81 (1.59-14.57) | .002 |
| Unmutated | 25 | 0.92 (0.33-2.53) | .86 |
| ZAP-70 less than 20% | 31 | 7.41 (1.65-33.28) | .002 |
| ZAP-70 at least 20% | 43 | 2.35 (1.06-5.19) | .030 |
| CD38 less than 20% | 51 | 4.37 (1.83-10.45) | <.001 |
| CD38 at least 20% | 19 | 5.88 (1.26-27.50) | .012 |
| del(13q)/normal/trisomy 12 | 56 | 3.24 (1.41-7.44) | .004 |
| del(11q)/del(17p) | 15 | 8.28 (1.01-67.53) | .020 |
| Variable . | No. of patients . | HR* (95% CI) . | P . |
|---|---|---|---|
| Binet stage A | 65 | 3.73 (1.69-8.24) | <.001 |
| Binet stage B or C | 27 | 2.43 (0.90-6.54) | .071 |
| Mutated | 48 | 4.81 (1.59-14.57) | .002 |
| Unmutated | 25 | 0.92 (0.33-2.53) | .86 |
| ZAP-70 less than 20% | 31 | 7.41 (1.65-33.28) | .002 |
| ZAP-70 at least 20% | 43 | 2.35 (1.06-5.19) | .030 |
| CD38 less than 20% | 51 | 4.37 (1.83-10.45) | <.001 |
| CD38 at least 20% | 19 | 5.88 (1.26-27.50) | .012 |
| del(13q)/normal/trisomy 12 | 56 | 3.24 (1.41-7.44) | .004 |
| del(11q)/del(17p) | 15 | 8.28 (1.01-67.53) | .020 |
Hazard ratio for CLLU1 at least 40-fold versus CLLU1 less than 40-fold.
Overall survival according to CLLU1 expression levels in different prognostic subgroups for patients younger than 70 years at diagnosis. (A) In 65 patients in stage A, the median overall survival was 7.5 years for patients with high CLLU1 expression levels and not reached after 14.4 years for patients with low CLLU1 expression levels. (B) In 48 patients with mutated IgVH CLL the median overall survival was 7.6 years for patients with high CLLU1 expression levels and not reached after 14.4 years for patients with low CLLU1 expression levels. (C) In 43 patients with ZAP-70–positive CLL the median overall survival was 3.8 years for patients with high CLLU1 expression levels and 8.3 years for patients with low CLLU1 expression levels. (D) In 19 patients with CD38+ CLL the median overall survival was 3.3 years for patients with high CLLU1 expression levels and not reached after 11.3 years for patients with low CLLU1 expression levels.
Overall survival according to CLLU1 expression levels in different prognostic subgroups for patients younger than 70 years at diagnosis. (A) In 65 patients in stage A, the median overall survival was 7.5 years for patients with high CLLU1 expression levels and not reached after 14.4 years for patients with low CLLU1 expression levels. (B) In 48 patients with mutated IgVH CLL the median overall survival was 7.6 years for patients with high CLLU1 expression levels and not reached after 14.4 years for patients with low CLLU1 expression levels. (C) In 43 patients with ZAP-70–positive CLL the median overall survival was 3.8 years for patients with high CLLU1 expression levels and 8.3 years for patients with low CLLU1 expression levels. (D) In 19 patients with CD38+ CLL the median overall survival was 3.3 years for patients with high CLLU1 expression levels and not reached after 11.3 years for patients with low CLLU1 expression levels.
Because of the difference in prognostic significance of CLLU1 expression in patients with unmutated IgVH versus mutated IgVH CLL, the possibility of interaction between CLLU1 expression and mutational status was assessed in a bivariate Cox proportional hazards model (n = 73). This analysis confirmed that high CLLU1 expression levels in patients with mutated IgVH CLL was associated with a relative risk of 4.52 (95% CI, 1.51-13.54; P = .007) of early death, whereas it did not add significant prognostic information in patients with unmutated IgVH (HR = 0.87; 95% CI, 0.10-7.54). Conversely, the added relative risk of shorter overall survival associated with unmutated IgVH was 11.75 (95% CI, 3.50-39.42; P < .001) in patients with CLLU1 expression less than 40-fold and only 2.26 (95% CI, 0.29-17.47) in patients with CLLU1 expression at 40-fold or above (data not shown).
To take this interaction between CLLU1 expression and IgVH mutational status into account, the different combinations of these 2 markers were incorporated in further multivariate analysis. The prognostic contribution of other biologic markers to this bivariate model in younger patients with known mutational and CLLU1 status was first tested with the separate addition of one variable at a time. No single marker could add significant prognostic information to the model (data not shown). A multivariate Cox analysis comprising all investigated prognostic variables in the 51 CLL patients younger than 70 years at diagnosis for whom complete data were available is shown in Table 5. High CLLU1 expression levels or unmutated IgVH CLL will increase risk of early death significantly, even when adjusting for all other prognostic markers. Of the other markers investigated, only high-risk cytogenetics contributed with borderline significant prognostic information in this model. Regardless of CLLU1 expression level, IgVH mutational status, ZAP-70, and CD38 expression, a high-risk cytogenetic profile increased the risk of early death 3.44 times (95% CI, 0.97-12.13; P = .055).
Multivariate Cox proportional hazards analysis of overall survival for patients with CLL younger than 70 years at diagnosis
| Variable . | HR* (95% CI) . | P . |
|---|---|---|
| Low CLLU1 and mutated IgVH | 1.00 (reference) | — |
| High CLLU1 and mutated IgVH | 8.00 (2.00-32.04) | .003 |
| Low CLLU1 and unmutated IgVH | 23.69 (4.44-126.36) | <.001 |
| High CLLU1 and unmutated IgVH | 7.08 (1.93-25.94) | .003 |
| ZAP-70 positive versus ZAP-70 negative | 2.65 (0.85-8.31) | .094 |
| CD38+ versus CD38− | 0.38 (0.11-1.25) | .11 |
| del(17p)/del(11q) versus trisomy 12/normal/del(13q) | 3.44 (0.97-12.13) | .055 |
| Variable . | HR* (95% CI) . | P . |
|---|---|---|
| Low CLLU1 and mutated IgVH | 1.00 (reference) | — |
| High CLLU1 and mutated IgVH | 8.00 (2.00-32.04) | .003 |
| Low CLLU1 and unmutated IgVH | 23.69 (4.44-126.36) | <.001 |
| High CLLU1 and unmutated IgVH | 7.08 (1.93-25.94) | .003 |
| ZAP-70 positive versus ZAP-70 negative | 2.65 (0.85-8.31) | .094 |
| CD38+ versus CD38− | 0.38 (0.11-1.25) | .11 |
| del(17p)/del(11q) versus trisomy 12/normal/del(13q) | 3.44 (0.97-12.13) | .055 |
— indicates not applicable.
Based on a model including ZAP-70, CD38, cytogenetics, and interaction between CLLU1 expression and mutational status.
Discussion
In this single-center retrospective study we have confirmed that expression of CLLU1, a recently identified CLL-specific gene, has prognostic significance in CLL. Age-stratified analysis revealed that the prognostic significance was particularly strong in patients younger than 70 years of age at diagnosis, and absent in patients aged 70 or older. This age-dependent variation of prognostic impact was seen for IgVH mutational status and ZAP-70 expression as well. We have furthermore demonstrated that CLLU1 expression status can add complementary prognostic information to all subgroups of prognostic biomarkers currently in use, except for patients with unmutated IgVH CLL.
This study confirmed the finding of our initial study2 that CLLU1 expression has strong prognostic significance for overall survival in CLL. In the present, much larger patient cohort we demonstrate a continuous proportional relationship between the expression level of CLLU1 at time of diagnosis and the relative risk of early death. Because the expression level of CLLU1 is highly variable within different CLL cases, the linear relation suggests that CLLU1 can improve risk prediction in CLL with more differentiated information. This continuous model, however, does not apply in the few CLL cases with CLLU1 expression levels much below that of normal B cells (n = 11) (Figure 1A). Taking into account that the very few observations in this range (10−4- to 10−1-fold expression level) do not allow for a proper modeling in this area, it is interesting, however, that a disproportionately high number of these individual cases had cytogenetic aberrations (3 of 11 patients had 17p deletions), suggesting that these patients may represent a subgroup with a particular pathogenesis.
In the evaluation of new prognostic markers it is essential to have a long follow-up time. Given a median age at diagnosis in CLL close to 70 years,29 the increase of interacting diseases and deaths unrelated to CLL will shorten follow-up time unproportionally in the elderly. Thus, in our study population the median overall survival was 8.8 years for patients younger than 70 years at diagnosis and only 4.1 years for patients aged 70 or older at time of diagnosis. To take this into account, we chose to perform our analyses by age stratification. We found a striking change in the predictive power of CLLU1 expression as well as for IgVH mutational status and ZAP-70 expression, when analyzed separately in younger and older patients. In contrast, the prognostic significance of cytogenetic aberrations was preserved in the elderly. In our opinion, a loss or decrease in the predictive power of some markers in the elderly can in part be explained by the accumulation of deaths unrelated to CLL in this age group, masking the disease-specific effect of these markers. To explain the discrepancy in age-dependent variation between different prognostic markers, we suggest that CLLU1 is expressed from very early CLL onset but may exert its clinical effect over a long time. A similar interpretation could be given for mutational status and ZAP-70 expression, whereas some prognostic factors [ie, high-risk cytogenetics such as del(17p) and del(11q)] might be markers of very advanced disease, contributing with molecular changes that affect overall survival more rapidly.
Biologic prognostic markers have clinical value when they can lead to early identification of patients at high risk for developing progressive disease; the earlier the more useful for interventional purposes. For younger patients when such strategies are relevant, we have here shown that CLLU1 expression adds significant prognostic information within all commonly used prognostic subgroups, whether favorable or unfavorable, except in patients with IgVH unmutated CLL. Moreover, in a multivariate analysis, only CLLU1 expression and IgVH mutational status were found to have independent predictive power.
In conclusion we find that analysis of CLLU1 expression is highly applicable in risk prediction in CLL for patients of an age well suited for risk-adapted treatment strategies. CLLU1 might identify patients at time of diagnosis likely to have CLL requiring aggressive treatment, despite an otherwise favorable prognostic marker profile. Equally important CLLU1 might identify cases of CLL presenting before the age of 70 with otherwise unfavorable prognostic markers, where aggressive treatment is not beneficial. Because of the lack of clear conclusive data, new biologic biomarkers are not yet recommended as basis for treatment-related decisions in CLL.16 The potential value of these markers for use in risk-adapted treatment strategies must be evaluated in prospective clinical trials. On the basis of this study, we suggest that the prognostic information offered by new biologic markers should be evaluated in context with age and propose that analysis of CLLU1 expression is included in such clinical trials.
Presented in part at the 47th annual meeting of the American Society of Hematology, Atlanta, GA, December 13, 2005.30
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Lone Bredo Pedersen for assistance with CLLU1 expression analysis, Lone Bach for assistance with flow cytometric analysis, Inge-Lise Frost Andersen for assistance with FISH analysis, Brian V. Hansen for assistance with RNA preparation, and the Department of Clinical Biochemistry, Rigshospitalet, for assistance with DNA sequencing.
This work was supported by the University of Copenhagen, the Carla Thiel Krag Legacy, the Danish Cancer Society, the Danish Cancer Research Foundation, the John and Birthe Meyer Foundation, the Anders Hasselbalch Foundation, the Fraenkel Foundation, and the Alfred Benzon Foundation.
Authorship
Contribution: P.J. performed research, statistical analysis, and wrote the manuscript; C.H.G. participated in designing the project; H.L. contributed to analysis of CLLU1 expression; J.H.P. assisted in statistical analysis; M.K.A. was responsible for FISH analyses, and J.J. and A.M.B. designed and supervised the project. All authors participated in interpretation of data and checked the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pär Josefsson, The Leukemia Laboratory 4041, Department of Hematology, Rigshospitalet, Blegdamsvej 9, DK-2100 Copenhagen, Denmark; e-mail: par.josefsson@pc.dk.