Abstract
Monoclonal TCRαβ+/CD4+ T-large granular lymphocyte (T-LGL) lymphocytosis is a T-cell disorder with a restricted TCR-Vβ repertoire. In the present study we explored the potential association between the expanded TCR-Vβ families, the CDR3 sequences of the TCR-Vβ gene, and the HLA genotype of patients with monoclonal TCRαβ+/CD4+ T-LGL lymphocytosis. For that purpose, 36 patients with monoclonal TCRαβ+/CD4+ T-LGL lymphocytosis (15 TCR-Vβ13.1 versus 21 non–TCR-Vβ13.1) were selected. For each patient, both the HLA (class I and II) genotype and the DNA sequences of the VDJ-rearranged TCR-Vβ were analyzed. Our results show a clear association between the TCR-Vβ repertoire and the HLA genotype, all TCR-Vβ13.1+ cases being HLA-DRB1*0701 (P = .004). Interestingly, the HLA-DR7/TCR-Vβ13.1–restricted T-cell expansions displayed a highly homogeneous and strikingly similar TCR arising from the use of common TCR-Vβ gene segments, which shared (1) unique CDR3 structural features with a constantly short length, (2) similar combinatorial gene rearrangements with frequent usage of the Jβ1.1 gene, and (3) a homolog consensus protein sequence at recombination junctions. Overall, these findings strongly support the existence of a common antigen-driven origin for monoclonal CD4+ T-LGL lymphocytosis, with the identification of the exact peptides presented to the expanded T cells deserving further investigations.
Introduction
Monoclonal chronic T-cell lymphocytosis and T-cell leukemias/lymphomas are a heterogeneous group of disorders whose diagnosis and classification have been hampered by their relatively low frequency and variable clinical and histopathological behavior,1–4 the lack of easily applicable clonality markers for T cells, and the substantial clinical overlap with nonmalignant inflammatory disorders.1,5,6 Although the pathogenetic mechanisms involved in the development of clonal T-cell disorders remain largely unknown, in recent years significant advances have been made in this regard.7–9 Among other observations, an association between chronic inflammatory and infectious processes and the occurrence of (mono)clonal expansions of lymphoid cells has recurrently been reported, particularly for chronic B-cell malignancies10 but also for mature T-cell neoplasias.1,8–9 Accordingly, different viruses (eg, human T-cell lymphotropic virus type I [HTLV-I], Epstein-Barr virus [EBV], and cytomegalovirus [CMV]) and bacterial superantigens (ie, staphylococci-derived superantigens) have been associated with the pathogenesis of specific mature T-cell malignancies, either because they infect tumor cells11–14 or because they could induce an antigen-driven expansion of neoplastic T cells.9,15,16 In line with the latter hypothesis, recent reports suggest that T-cell receptor (TCR)–associated signals could contribute to tumor development, particularly in T-cell large granular lymphocyte (T-LGL) leukemia.5,9 In these cases, antigen-driven expansions of cytotoxic T lymphocyte (CTL) clones could precede the occurrence of oncogenic events leading to neoplastic transformation and/or dysregulation of growth/apoptosis resulting in T-LGL leukemia9 with a restricted TCR-Vβ/Vα usage.17 This notion is supported by the observation that TCRαβ+/CD8+ T-LGL leukemia often occurs in the context of specific autoimmune diseases9,16 and that in about one third of cases TCRαβ+/CD4+ T-LGL leukemia/lymphocytosis is associated with neoplasias other than the T-LGL and a preferential usage of the TCR-Vβ13.1 family.18 In addition, the reactive versus neoplastic nature of some (mono)clonal expansions of T-LGL remains a matter of debate,15 particularly in cases where it is associated with viral infection, severe immune disturbances, or in the elderly.19,20 In this regard, the search for the potential involvement of common antigens in driving the development of monoclonal T-cell disorders through the analysis of complementary determining region 3 (CDR3) sequences of TCR genes has provided controversial findings. Accordingly, while CDR3 sequences from CD8+ T-LGL leukemia did not show any apparent structural homology,21,22 in nearly half of all TCRγδ+ T-LGL neoplasias, clonal T cells express the same TCR-Vδ/Vγ family members (TCR-Vγ9/Vδ2) and share common TCR sequences, as reflected by the systematic presence of the antigen-selected invariant T nucleotide in the first codon of the Vδ2-Jδ1 junctional region from all patients.23 Such apparent discrepancy could be related to the fact that antigen-driven TCRαβ+/CD8+ T-LGL leukemias would depend not only on the complementary sequences and specific binding of the TCR to the antigen, but also on the individual HLA haplotypes, while for TCRγδ+ T cells this HLA restriction would not apply.
In the present study, we have analyzed a large series of 36 patients with monoclonal TCRαβ+/CD4+ T-LGL lymphocytosis grouped according to TCR-Vβ13.1+ usage versus other TCR-Vβ families. Our aim was to explore the potential existence of an association in these patients between the expanded TCR-Vβ families, the CDR3 sequences of the TCR-Vβ gene, and the HLA genotype. Our results indicate that all patients with monoclonal expansions of TCR-Vβ13.1+/CD4+ T cells display a common HLA-DRB0701+ genotype and express identical motifs in the CDR3-TCR-Vβ sequence, suggesting a common antigen-driven origin.
Patients, materials, and methods
Peripheral blood (PB) samples from human patients were obtained after informed consent was given by the patients (all of them more than 18 years old), in accordance with the local ethics committee of the University Hospital of Salamanca and the Declaration of Helsinki.
Patients and samples
A total of 161 T-LGL cases were referred to the Cytometry Service of the University Hospital of Salamanca (63 TCRαβ+/CD8++/CD4−, 55 TCRαβ+/CD4+/NKa+/CD8−/+dim, 40 TCRγδ+, and 3 TCRαβ+/CD8−/CD4− cases) between September 1999 and March 2006. From them, 36 individuals with monoclonal TCRαβ+/CD4+/NKa+/CD8−/+dim lymphocytosis (19 males and 17 females; mean age, 63 ± 12 years, ranging from 36 to 81 years) were selected and included in this study. In all these latter cases, PB samples were collected into tubes containing K3-EDTA, according to the local ethics committee's recommendations. The major clinical and laboratory features of this group of patients, according to the type of TCR-Vβ family expressed (TCR-Vβ13.1 versus non–TCR-Vβ13.1), are shown in Table 1 in comparison with those of a randomly selected series of 24 patients with monoclonal TCRαβ+/CD8+/CD4− lymphocytosis. At the close of the study, the median follow-up of the patients with monoclonal TCRαβ+/CD4+/NKa+/CD8−/+dim lymphocytosis was 72 months (range, 10 to 233 months).
Clinical and laboratory characteristics of monoclonal TCRαβ+/CD4+ LGL lymphocytosis according to the type of TCR-Vβ family expressed (Vβ13.1 versus non-Vβ13.1) compared with monoclonal TCRαβ+/CD8+ LGL lymphocytosis
| Characteristic . | Monoclonal TCRαβ+/CD4+ LGL lymphocytosis . | Monoclonal TCRαβ+/CD8+ LGL lymphocytosis . | P . | ||
|---|---|---|---|---|---|
| TCR-Vβ13.1 . | Non-TCR-Vβ13.1 . | Total cases . | |||
| No. of patients | 15 | 21 | 36 | 24 | |
| Mean age ± 1 SD, y (range) | 64 ± 8 (52-80) | 61 ± 13 (36-81) | 63 ± 12 (36-81) | 56 ± 15 (31-79) | NS |
| % male/% female | 25/75 | 61/39 | 47/53 | 37/63 | NS |
| Reason for consulting, %; | |||||
| Routine blood analysis | 100 | 83 | 90 | 83 | NS |
| Skin lesions | 0 | 11 | 7 | 0 | NS |
| Abdominal distension | 0 | 6 | 3 | 0 | NS |
| Fever | 0 | 0 | 0 | 4 | NS |
| General symptoms | 0 | 0 | 0 | 13 | NS |
| Physical examination, %; | |||||
| Adenomegalies | 11 | 6 | 7 | 0 | NS |
| Hepatomegaly | 0 | 0 | 0 | 4 | NS |
| Splenomegaly | 0 | 6 | 3 | 4 | NS |
| Skin lesions | 0 | 11 | 7 | 0 | NS |
| Associated neoplasias, % | 25 | 22 | 23 | 26 | NS |
| Associated autoimmune diseases, % | 18 | 0 | 13 | 33 | .05 |
| Laboratory parameters, %; | |||||
| Leukocytosis, WBC count more than 10 × 109/L | 64 | 61 | 63 | 25 | .01 |
| Lymphocytosis, lymphocyte count more than 5 × 109/L | 70 | 67 | 68 | 42 | .09 |
| Neutropenia, neutrophil count less than 1.5 × 109/L | 14 | 0 | 3 | 61 | < .001 |
| Anemia, hemoglobin level less than 10 g/dL | 0 | 0 | 0 | 29 | .003 |
| Thrombocytopenia, platelet count less than 100 × 109/L | 10 | 0 | 3 | 8 | NS |
| Increased lactic dehydrogenase level, more than 460 U/L | 10 | 0 | 3 | 22 | NS |
| Increased β2-microglobulin level, more than 2 mg/dL | 0 | 7 | 3 | 63 | .001 |
| Cases requiring treatment because of lymphocytosis or the associated autoimmune disease, % | 0 | 0 | 0 | 37 | .001 |
| Outcome: stable disease, % | 100 | 100 | 100 | 87 | NS |
| Total deaths, % | 10 | 18 | 15 | 8 | NS |
| Deaths related to the TCRαβ+ LGL lymphocytosis, % | 0 | 0 | 0 | 4 | NS |
| Mean follow-up ± 1 SD, mo (range) | 68 ± 48 (13-136) | 74 ± 56 (10-233) | 72 ± 53 (10-233) | 44 ± 40 (1-152) | .03 |
| Characteristic . | Monoclonal TCRαβ+/CD4+ LGL lymphocytosis . | Monoclonal TCRαβ+/CD8+ LGL lymphocytosis . | P . | ||
|---|---|---|---|---|---|
| TCR-Vβ13.1 . | Non-TCR-Vβ13.1 . | Total cases . | |||
| No. of patients | 15 | 21 | 36 | 24 | |
| Mean age ± 1 SD, y (range) | 64 ± 8 (52-80) | 61 ± 13 (36-81) | 63 ± 12 (36-81) | 56 ± 15 (31-79) | NS |
| % male/% female | 25/75 | 61/39 | 47/53 | 37/63 | NS |
| Reason for consulting, %; | |||||
| Routine blood analysis | 100 | 83 | 90 | 83 | NS |
| Skin lesions | 0 | 11 | 7 | 0 | NS |
| Abdominal distension | 0 | 6 | 3 | 0 | NS |
| Fever | 0 | 0 | 0 | 4 | NS |
| General symptoms | 0 | 0 | 0 | 13 | NS |
| Physical examination, %; | |||||
| Adenomegalies | 11 | 6 | 7 | 0 | NS |
| Hepatomegaly | 0 | 0 | 0 | 4 | NS |
| Splenomegaly | 0 | 6 | 3 | 4 | NS |
| Skin lesions | 0 | 11 | 7 | 0 | NS |
| Associated neoplasias, % | 25 | 22 | 23 | 26 | NS |
| Associated autoimmune diseases, % | 18 | 0 | 13 | 33 | .05 |
| Laboratory parameters, %; | |||||
| Leukocytosis, WBC count more than 10 × 109/L | 64 | 61 | 63 | 25 | .01 |
| Lymphocytosis, lymphocyte count more than 5 × 109/L | 70 | 67 | 68 | 42 | .09 |
| Neutropenia, neutrophil count less than 1.5 × 109/L | 14 | 0 | 3 | 61 | < .001 |
| Anemia, hemoglobin level less than 10 g/dL | 0 | 0 | 0 | 29 | .003 |
| Thrombocytopenia, platelet count less than 100 × 109/L | 10 | 0 | 3 | 8 | NS |
| Increased lactic dehydrogenase level, more than 460 U/L | 10 | 0 | 3 | 22 | NS |
| Increased β2-microglobulin level, more than 2 mg/dL | 0 | 7 | 3 | 63 | .001 |
| Cases requiring treatment because of lymphocytosis or the associated autoimmune disease, % | 0 | 0 | 0 | 37 | .001 |
| Outcome: stable disease, % | 100 | 100 | 100 | 87 | NS |
| Total deaths, % | 10 | 18 | 15 | 8 | NS |
| Deaths related to the TCRαβ+ LGL lymphocytosis, % | 0 | 0 | 0 | 4 | NS |
| Mean follow-up ± 1 SD, mo (range) | 68 ± 48 (13-136) | 74 ± 56 (10-233) | 72 ± 53 (10-233) | 44 ± 40 (1-152) | .03 |
P value corresponds to comparisons between monoclonal TCRαβ+/CD4+ LGL and TCRαβ+/CD8+ LGL lymphocytosis; NS indicates no statistically significant differences: P > .1. No statistically significant differences (P > .05) were found between TCR-Vβ13.1 and non-TCR-Vβ13.1 clonal CD4+ LGL lymphocytosis. A detailed description of the clinical characteristics of patients with monoclonal TCRαβ+/CD4+ LGL lymphocytosis is provided by Lima et al.18
A total of 930 PB samples from unrelated healthy subjects were used as controls to establish the frequency of the different HLA haplotypes in the healthy population, while PB samples from 15 adult individuals (older than 50 years) were used as controls to establish the TCR-Vβ repertoire usage in TCRαβ+/CD4+ T cells.
Immunophenotypic studies
For the analysis of the TCR-Vβ repertoire of CD4+/CD8−/+dim LGL T lymphocytes, a panel of 24 monoclonal antibodies (MAbs) directed against an identical number of members of 21 different TCR-Vβ families (TCR-Vβ repertoire Kit; Immunotech, Marseille, France) was used in 4-color stainings. Further phenotypic characterization of CD4+/CD8−/+dim LGL T cells was performed using the following 4-color combinations of MAbs: fluorescein isothiocyanate (FITC)/phycoerythrin (PE)/PE–cyanin 5 (PC5) or peridinin chlorophyll protein (PerCP)/allophycocyanin (APC): CD2/CD7/CD4/CD8, CD5/CD7/CD4/CD8, CD38/CD11b/CD4/CD8, CD57/CD11c/CD4/CD8, CD16/CD56/CD4/CD8, CD122/CD25/CD4/CD8, CD45RA/CD45RO/CD4/CD8, CD62L/CD28/CD4/CD8, CD11a/HLA-DR/CD4/CD8, CD16/NKB1/CD4/CD8, CD158a/CD161/CD4/CD8, CD57/CD8/CD56/CD4, and cytoplasmic (Cy) perforin/Cy granzyme B/CD56/CD4. The source and specificity of each MAb reagent used has been previously described in detail.18
Cell staining was performed using a whole blood “stain-and-then-lyse” method (FACS lysing solution; Becton Dickinson Biosciences [BDB], San Jose, CA) and a direct immunofluorescence technique, as previously reported in detail.18 For the cytoplasmic staining, the Fix & Perm reagent kit (Invitrogen, Carlsbad, CA) was used according to the recommendations of the manufacturer.
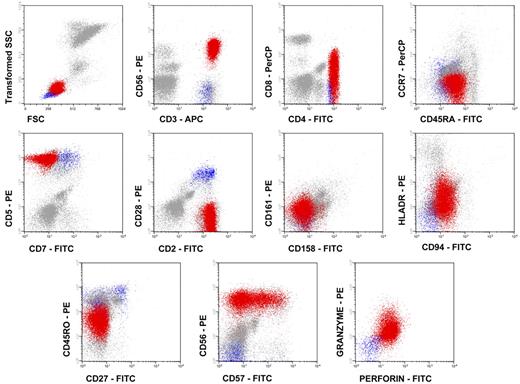
Data acquisition was performed immediately after completion of sample preparation in a FACSCalibur flow cytometer (BDB) using the CellQUEST software program (BDB). The Paint-A-Gate Pro software program (BDB) was used for data analysis. In each case, the aberrant T-cell population was defined as CD4+/CD8−/+dim and/or CD4+/CD56+ large granular—intermediate sideward light scatter (SSCintermediate)—events (Figure 1) for its further phenotypic characterization.
Preparation of DNA and HLA typing
High molecular weight DNA was prepared from 200 μL PB using the QIAGEN bloodmicro kit (Qiagen, Hilden, Germany). HLA genotyping for HLA-ABC and both HLA-DRB1 and HLA-DQB1 was performed by sequence-specific oligonucleotide–polymerase chain reaction (SSPO-PCR) techniques using the Dynal Reli SSO kit (DYNAL Biotech, Bromborough, United Kingdom). Ambiguous results were resolved by sequence-based typing (SBT). DNA samples were amplified by PCR using the Big-Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Sequencing was performed on an ABI 377 DNA sequencer (Applied Biosystems), and the data obtained were analyzed using the Match Tools v 1.0 Sequencing Analysis software program (Applied Biosystems). The ancestral haplotypes are putative because it was not possible to verify their segregation from family studies.
PCR amplification and nucleotide sequence analysis of TCR
High molecular weight DNA was prepared from freshly frozen PB samples using standard protocols including proteinase K treatment. In addition, total RNA was isolated from fluorescence-activated cell sorter (FACS)–sorted CD4+ and CD8+ T-cell populations (purity more than 95%) from pooled PB mononuclear cells (MNCs) of 5 non–HLA-DR*0701 adult healthy donors and from PB MNCs of 1 HLA-DR*0701 healthy individual and reverse transcribed into cDNA. TCR Vβ13.1 gene family–specific PCR was performed using specific primers as previously described.24 PCR products were cloned into pGEM-T easy vector (Promega, Madison WI), and single-colony PCR was performed on positive clones. Single-colony PCR products were directly sequenced.
DNA was amplified using a mixture of sense primers annealing to the TCR-Vβ13 sequence in conjunction with a mixture of antisense primers complementary to the germ-line J regions as previously reported in detail.24 In most samples, clonal products from the Vβ gene PCR were sequenced directly using the BigDye Terminator Cycle Sequencing Reaction Kit. In fact, the amplified sequences exhibited identical rearranged monoclonal TCR sequences, even in the VDJ junctional hypervariable regions, indicating that these expanded regions were clonal. To confirm the validity of the sequences obtained (thereby avoiding the possibility of either contamination or sequencing mistakes), a more detailed analysis of the sequences obtained was performed in some cases. For that purpose, PCR products were inserted into the PCR2.1-TOPO vector (Invitrogen, Barcelona, Spain), which was followed by transformation into competent Escherichia coli cells; on average, 5 colonies were randomly selected for sequencing using the BigDye Terminator Cycle Sequencing Reaction Kit. A total of 98 Vβ13.1 clones (69 from the non-HLA-DR*0701 donors and 29 from the HLA-DR*0701 donor) in the CD4+ T-cell fraction and 53 Vβ13.1 clones (38 from the non–HLA-DR*0701 donors and 15 from the HLA-DR*0701 donor) in the CD8+ T-cell fraction were sequenced and analyzed. All sequence reactions were analyzed using an automated DNA sequencer (ABI 377; Applied Biosystems).
Database searches
Sequences obtained were aligned using the Basic Local Alignment Search Tool (BLAST) (National Center for Biotechnology Information, Bethesda, MD) and ImMunoGeneTics (IMGT) databases.
Statistical methods
For all clinical and laboratory parameters included in Table 1, mean, standard deviation, and range were calculated using the SPSS program (SPSS 12.0, Chicago, IL). To establish the statistical significance of the differences observed between groups, either the Pearson χ2 test or Fisher exact test was used for categorical variables, and the Mann-Whitney U nonparametric test was used (SPSS 12.0) for continuous variables. P values below .05 were considered to be associated with statistical significance.
Results
Immunophenotype of the expanded CD4+ LGL T cells
In all cases studied, expanded CD4+ LGL T cells showed relatively high SSC features as compared with normal PB CD4+ T lymphocytes and common phenotypic characteristics consisting of TCRαβ+/CD4+/CD8−/+dim cells with a typical cytotoxic (granzyme B+, CD56+, CD57+, CD11b+/−) activated, memory/effector T-cell phenotype (CD2+bright, CD7−/+dim, CD11a+bright, CD28−, CD62L−, HLA-DR+) (Figure 1). In about half (47%) of the cases, clonal T cells coexpressed CD45RA and CD45RO, while in the other cases they had a CD45RA+/CD45RO− phenotype. Other NKa markers (CD11c, CD16, CD94, CD158a, CD161, and NKB1) and T-cell activation-related antigens (CD25, CD38, and CD122) were absent on the CD4+ LGL T cells. Overall, these cells represented 47% ± 23% of all PB lymphocytes, with a mean (±1 SD) absolute number of 6.6 × 109 ± 3.3 × 109 PB TCRαβ+/CD4+/NKa+/CD8−/+dim T lymphocytes per liter.
Immunophenotypic features of monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL. Representative dot plots illustrate the phenotypic patterns shown by monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL. Red dots correspond to monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL, blue dots correspond to normal residual non-LGL CD4+ T cells, while gray dots correspond to PB leukocytes other than CD4+ T cells.
Immunophenotypic features of monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL. Representative dot plots illustrate the phenotypic patterns shown by monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL. Red dots correspond to monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL, blue dots correspond to normal residual non-LGL CD4+ T cells, while gray dots correspond to PB leukocytes other than CD4+ T cells.
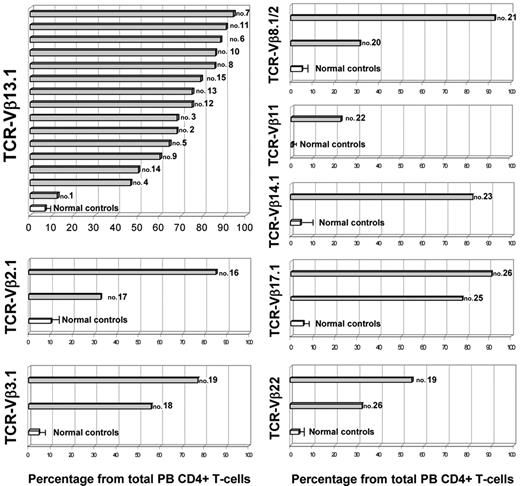
Flow cytometric analysis of the TCR-Vβ repertoire of CD4+/CD8−/+dim LGL T cells was consistent with a (mono)clonal expansion in all cases studied, which accounted for 72% ± 21% of all PB CD4+ T cells. In 27 cases the expanded TCR-Vβ family was identified with the panel of TCR-Vβ reagents used, corresponding to TCR-Vβ13.1 in 15 cases (42%), TCR-Vβ2.1 in 2 (5.6%), TCR-Vβ3.1 in 2 (5.6%), TCR-Vβ8.1 + Vβ8.2 in 2 (5.6%), TCR-Vβ17.1 in 2 (5.6%), TCR-Vβ22 in 2 (5.6%), and TCR-Vβ11 or TCR-Vβ14.1 in 1 case each (2.8%). In the remaining 9 patients, the expanded TCR-Vβ family was not identified (25%) with the panel of MAbs used. Figure 2 shows the nominal size of the TCR-Vβ expansion present in each patient (as percentage of the total PB CD4+ T cells) in comparison with the size of the corresponding TCR-Vβ family observed in a cohort of age-matched healthy subjects; the proportion of TCR-Vβ13.1+ cells represented 6.7% ± 2.5% of total PB CD4+ T cells from healthy subjects, while in the patient group it represented between 12.5% and 94.1% (median, 75%).
Illustrative representation of the size of the actual identifiable TCR-Vβ expansion present in each patient (as percentage of total PB CD4+ T cells) in comparison with the size of the corresponding TCR-Vβ family observed in a cohort of age-matched healthy subjects (n = 15). Gray bars correspond to patients, each one identified by the corresponding case number, while white bars and horizontal lines correspond to the mean value and 1 SD found in healthy controls, respectively.
Illustrative representation of the size of the actual identifiable TCR-Vβ expansion present in each patient (as percentage of total PB CD4+ T cells) in comparison with the size of the corresponding TCR-Vβ family observed in a cohort of age-matched healthy subjects (n = 15). Gray bars correspond to patients, each one identified by the corresponding case number, while white bars and horizontal lines correspond to the mean value and 1 SD found in healthy controls, respectively.
Association between the HLA haplotypes and the TCR-Vβ repertoire of clonal TCRαβ+/CD4+/NKa+/CD8−/+dim T-LGL lymphocytosis
A significant association (P = .004) was found between the TCR-Vβ repertoire and the HLA genotype of the studied cases (Table 2). Accordingly, all 15 patients who showed expansion of TCR-Vβ13.1+ CD4+ T cells were HLA-DRB*0701+ (Figure 3). In turn, the frequency of the HLA-Cw*0401 allele was slightly higher among those patients in whom the expanded TCR-Vβ family was not contained in the panel of MAb reagents used than among both TCR-Vβ13.1+ patients and cases expressing a known TCR-Vβ other than 13.1 (67% versus 33%; P = .1). In addition, the frequency of cases with a HLA-DRB1*03 and HLA-DRB1*04 genotype was lower among CD4+ T-LGL patients than in the control group (14% versus 29% and 6% versus 22%, respectively; P ≤ .04).25 Interestingly, 4 of the 15 HLA-DRB*0701 cases included the 44.2 ancestral haplotype (HLA-A*2902, B*4403, C*1601, DRB*0701, and DQB1*0202) (cases 7, 9, 11, and 14) (Table 2). Another 2 cases (cases 2 and 10) were related to the 57.1 ancestral haplotype (HLA-A*01, B*5701, C*0602, DRB1*0701, and DQB1*0303). The ancestral haplotypes 35.2 (HLA-A*1101, B*3501, C*0401, DRB*0101, DQB1*0501) and 35.1 (HLA-C*0401, B*3501, DRB1*11, DQB1*0301) were also found in 3 (cases 5, 6, and 14) and 1 patients (case 1), respectively; however, among these patients, only case 14 showed a complete 35.2 haplotype, with a 4-locus haplotype being detected in the other 3 cases. Cases 8, 13, and 15 were not considered to be related with ancestral haplotypes, although they showed haplotypes that are frequently present in the Spanish population.26
HLA genotype of patients with clonal TCRαβ+/CD4+ expansions
| Case no. . | Expanded TCR-Vβ family . | HLA-DRB . | HLA-DQB . | HLA-A . | HLA-B . | HLA-C . |
|---|---|---|---|---|---|---|
| 1 | 13.1 | 0701/1103 | 0301/0303 | 2301/3201 | 3501/5002 | 0401/0401 |
| 2 | 13.1 | 0701/1404 | 0202/0503 | 0101/2601 | 1401/5701 | 0602/0802 |
| 3 | 13.1 | 0701/0102 | 0303/0501 | 0201/0201 | 5101/5701 | 0102/0701 |
| 4 | 13.1 | 0701/1401 | 0303/0503 | 0201/2402 | 4402/5701 | 0501/0701 |
| 5 | 13.1 | 0701/0101 | 0202/0501 | 0201/0201 | 3501/4901 | 0401/0701 |
| 6 | 13.1 | 0701/0101 | 0202/0501 | 0301/0301 | 1302/3501 | 0401/0602 |
| 7 | 13.1 | 0701/0401 | 0202/0301 | 2601/2902 | 4402/4403 | 0501/1601 |
| 8 | 13.1 | 0701/0701 | 0202/0202 | 0101/2402 | 1801/5001 | 0501/0602 |
| 9 | 13.1 | 0701/0101 | 0202/0501 | 0101/2902 | 1401/4403 | 0802/1601 |
| 10 | 13.1 | 0701/1501 | 0303/0602 | 0201/0201 | 0702/5701 | 0602/0702 |
| 11 | 13.1 | 0701/0301 | 0201/0202 | 0301/2902 | 1801/4403 | 0501/1601 |
| 12 | 13.1 | 0701/0301 | 0201/0202 | 0101/2301 | 0801/4403 | 0401/0701 |
| 13 | 13.1 | 0701/0301 | 0201/0202 | 2301/3002 | 1801/5801 | 0501/0701 |
| 14 | 13.1 | 0701/0101 | 0201/0501 | 1101/2902 | 3501/4403 | 0401/1601 |
| 15 | 13.1 | 0701/1501 | 0202/0602 | 2902/3002 | 0702/4101 | 0701/1701 |
| 16 | 2.1 | 0301/1317 | 0201/0603 | 2402/2402 | 0801/3508 | 0401/0701 |
| 17 | 2.1 | 0701/1301 | 0202/0609 | 3002/6801 | 4403/5101 | 0701/1402 |
| 18 | 3.1 | 0403/1501 | 0302/0602 | 0301/3202 | 0702/3501 | 0401/0702 |
| 19 | 3.1 | 1501/1602 | 0502/0602 | 0201/0201 | 3701/4402 | 0501/0602 |
| 20 | 8.1 + 8.2 | 1101/1101 | 0301/0301 | 0201/6901 | 1801/3508 | 0701/1203 |
| 21 | 8.1 + 8.2 | 0301/1501 | 0201/0602 | 0201/0201 | 1518/3501 | 0401/0704 |
| 22 | 11 | 1101/1601 | 0301/0502 | 0101/0201 | 3701/4002 | 0602/1204 |
| 23 | 14.1 | 0701/0701 | 0202/0303 | 0103/2902 | 4403/5701 | 0701/1601 |
| 24 | 17.1 | 0802/1101 | 0301/0402 | 0102/2301 | 1401/3501 | 0701/0801 |
| 25 | 17.1 | 0701/1401 | 0202/0503 | 0201/2902 | 4403/5101 | 1502/1601 |
| 26 | 22 | 0102/0801 | 0402/0501 | 2402/3301 | 1402/3503 | 0401/0802 |
| 27 | 22 | 1103/1301 | 0301/0603 | 0201/3201 | 1509/4002 | 0202/0704 |
| 28 | NI | 0701/1104 | 0201/0301 | 1101/2902 | 4001/4403 | 0304/1601 |
| 29 | NI | 0701/1101 | 0201/0301 | 0201/0301 | 3503/5001 | 0401/0501 |
| 30 | NI | 0701/1301 | 0202/0604 | 3201/6801 | 1401/5301 | 0401/0802 |
| 31 | NI | 0701/1602 | 0202/0502 | 0301/2402 | 0702/3801 | 0702/1203 |
| 32 | NI | 1101/1401 | 0301/0503 | 0201/2603 | 4002/3501 | 0401/0501 |
| 33 | NI | 0101/1301 | 0501/0604 | 1101/1101 | 4004/5601 | 0102/0202 |
| 34 | NI | 0101/1101 | 0301/0501 | 0201/2402 | 1801/3501 | 0401/1203 |
| 35 | NI | 0101/1301 | 0501/0604 | 1101/3101 | 3501/5101 | 0401/0501 |
| 36 | NI | 1302/1305 | 0301/0604 | 0201/0205 | 3508/4101 | 0401/0701 |
| Case no. . | Expanded TCR-Vβ family . | HLA-DRB . | HLA-DQB . | HLA-A . | HLA-B . | HLA-C . |
|---|---|---|---|---|---|---|
| 1 | 13.1 | 0701/1103 | 0301/0303 | 2301/3201 | 3501/5002 | 0401/0401 |
| 2 | 13.1 | 0701/1404 | 0202/0503 | 0101/2601 | 1401/5701 | 0602/0802 |
| 3 | 13.1 | 0701/0102 | 0303/0501 | 0201/0201 | 5101/5701 | 0102/0701 |
| 4 | 13.1 | 0701/1401 | 0303/0503 | 0201/2402 | 4402/5701 | 0501/0701 |
| 5 | 13.1 | 0701/0101 | 0202/0501 | 0201/0201 | 3501/4901 | 0401/0701 |
| 6 | 13.1 | 0701/0101 | 0202/0501 | 0301/0301 | 1302/3501 | 0401/0602 |
| 7 | 13.1 | 0701/0401 | 0202/0301 | 2601/2902 | 4402/4403 | 0501/1601 |
| 8 | 13.1 | 0701/0701 | 0202/0202 | 0101/2402 | 1801/5001 | 0501/0602 |
| 9 | 13.1 | 0701/0101 | 0202/0501 | 0101/2902 | 1401/4403 | 0802/1601 |
| 10 | 13.1 | 0701/1501 | 0303/0602 | 0201/0201 | 0702/5701 | 0602/0702 |
| 11 | 13.1 | 0701/0301 | 0201/0202 | 0301/2902 | 1801/4403 | 0501/1601 |
| 12 | 13.1 | 0701/0301 | 0201/0202 | 0101/2301 | 0801/4403 | 0401/0701 |
| 13 | 13.1 | 0701/0301 | 0201/0202 | 2301/3002 | 1801/5801 | 0501/0701 |
| 14 | 13.1 | 0701/0101 | 0201/0501 | 1101/2902 | 3501/4403 | 0401/1601 |
| 15 | 13.1 | 0701/1501 | 0202/0602 | 2902/3002 | 0702/4101 | 0701/1701 |
| 16 | 2.1 | 0301/1317 | 0201/0603 | 2402/2402 | 0801/3508 | 0401/0701 |
| 17 | 2.1 | 0701/1301 | 0202/0609 | 3002/6801 | 4403/5101 | 0701/1402 |
| 18 | 3.1 | 0403/1501 | 0302/0602 | 0301/3202 | 0702/3501 | 0401/0702 |
| 19 | 3.1 | 1501/1602 | 0502/0602 | 0201/0201 | 3701/4402 | 0501/0602 |
| 20 | 8.1 + 8.2 | 1101/1101 | 0301/0301 | 0201/6901 | 1801/3508 | 0701/1203 |
| 21 | 8.1 + 8.2 | 0301/1501 | 0201/0602 | 0201/0201 | 1518/3501 | 0401/0704 |
| 22 | 11 | 1101/1601 | 0301/0502 | 0101/0201 | 3701/4002 | 0602/1204 |
| 23 | 14.1 | 0701/0701 | 0202/0303 | 0103/2902 | 4403/5701 | 0701/1601 |
| 24 | 17.1 | 0802/1101 | 0301/0402 | 0102/2301 | 1401/3501 | 0701/0801 |
| 25 | 17.1 | 0701/1401 | 0202/0503 | 0201/2902 | 4403/5101 | 1502/1601 |
| 26 | 22 | 0102/0801 | 0402/0501 | 2402/3301 | 1402/3503 | 0401/0802 |
| 27 | 22 | 1103/1301 | 0301/0603 | 0201/3201 | 1509/4002 | 0202/0704 |
| 28 | NI | 0701/1104 | 0201/0301 | 1101/2902 | 4001/4403 | 0304/1601 |
| 29 | NI | 0701/1101 | 0201/0301 | 0201/0301 | 3503/5001 | 0401/0501 |
| 30 | NI | 0701/1301 | 0202/0604 | 3201/6801 | 1401/5301 | 0401/0802 |
| 31 | NI | 0701/1602 | 0202/0502 | 0301/2402 | 0702/3801 | 0702/1203 |
| 32 | NI | 1101/1401 | 0301/0503 | 0201/2603 | 4002/3501 | 0401/0501 |
| 33 | NI | 0101/1301 | 0501/0604 | 1101/1101 | 4004/5601 | 0102/0202 |
| 34 | NI | 0101/1101 | 0301/0501 | 0201/2402 | 1801/3501 | 0401/1203 |
| 35 | NI | 0101/1301 | 0501/0604 | 1101/3101 | 3501/5101 | 0401/0501 |
| 36 | NI | 1302/1305 | 0301/0604 | 0201/0205 | 3508/4101 | 0401/0701 |
Extended/ancestral haplotypes are underlined.
NI indicates TCR-Vβ family not identified by immunophenotyping.
Frequency of the HLA-DRB1*0701 genotype in patients with monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL lymphocytosis grouped according to the expanded TCR-Vβ family. NI indicates that the exact TCR-Vβ family expanded was not identified with the panel of anti–TCR-Vβ MAbs used.
Frequency of the HLA-DRB1*0701 genotype in patients with monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL lymphocytosis grouped according to the expanded TCR-Vβ family. NI indicates that the exact TCR-Vβ family expanded was not identified with the panel of anti–TCR-Vβ MAbs used.
Expanded clonal CD4+/CD8−/+dim T cells from HLA-DRB1*0701 patients exhibit conserved TCRβ chain motifs
Molecular analysis of the length of CDR3 of CD4+/CD8−/+dim T cells from those cases expressing TCR-Vβ13.1 showed a pattern consistent with monoclonality based on both TCR-Vβ usage and CDR3 length (Table 3). Further comparison of CDR3 size distribution in clonal CD4+/CD8−/+dim T cells from the same patients showed a highly restricted usage of VβDβJβ gene segments and shared CDR3 configurations. Accordingly, 11 of 13 patients used the same J1.1 and V segments, and they had highly similar CDR3 configurations (Table 3). Although 3 different J segments were used in the clonal expansions derived from CD4+/CD8−/+dim T lymphocytes, similar VDJ junctional region sequences were found (Table 4). Remarkably, in 5 of the 11 Vβ13.1-Dβ1-Jβ1.1 cases, the combinatorial process involved the deletion of 1 to 3 nucleotides from the 5′ end of Vβ13.1 gene and the insertion of a variable number of nontemplate nucleotides (Table 4). Accordingly, in all cases analyzed (n = 14) the mean length of CDR3 was considerably shortened (4 or 5 codons), the distance between the CASS and FG motifs constantly being of 9 codons. Interestingly, shared motifs consisting of at least 2 identical amino acids were found within the VDJ junctional regions of the expanded CD4+ T cells derived from different patients, a consensus XQGX motif being shared by all cases (Table 3). For 7 cases the GGG codon yielded a glycine; in contrast, in 2 other cases the glycine was generated by the GGT and in another case by a GGA codon. In turn, glutamine (Q) was yielded in all cases by the CAG codon. One of the motifs (YQGA) was identical among the clonally expanded CD4+/CD8−/+dim T cells derived from 3 patients, and the XQGA motif was detected in 7 individuals. In contrast, only 2 (2.9%), 8 (11.6%), and 2 (2.9%) of 69 clones of PB CD4+ T cells from non–HLA-DR*0701 healthy adults were found to use the Jβ1.1, Jβ1.2, and Jβ1.5 gene segments, respectively; similarly, from the 29 clones of CD4+ T cells sequenced from the HLA-DR*0701 adult healthy donor, only 1 (3.4%) and 3 (10%) of them were found to use Jβ1.1 and Jβ1.2 gene segments, respectively. Interestingly, of these 16 clones of CD4+ T cells, only 1 of those 3 clones using the Jβ1.1 gene segment was highly similar in the CDR3 configuration to those detected in the patients analyzed (Tables 3–4); this clone was sequenced from the pooled non–HLA-DR*0701 MNCs. In addition, the XQGX configuration could not be detected in other clones of PB CD8+ T cells from healthy adults using the Jβ1.1, Jβ1.2, and Jβ1.5 gene segments (0 of 9 clones from the 38 CD8+ T-cell clones sequenced from non–HLA-DR*0701 donors and 0 of 3 from the 15 CD8+ T-cell clones sequenced from the HLA-DR*0701 donor). Finally, we searched GenBank for VDJ rearrangements with similar XQGX amino acid sequences, but no TCR close matches were found.
VDJ protein sequences of clonally expanded TCRαβ+/CD4+ T-LGL
| Case no. . | V-J usage . | CDR2 . | 3′ end of Vβ . | CDR3: N-Dβ1-N . | 5′ end of Jβ . |
|---|---|---|---|---|---|
| 1 | Vβ13S1-J1.1 | SVGAGI | CASS | KQGV | TEAFFG |
| 2 | Vβ13S1-J1.1 | SVGAGI | CASS | KQGA | TEAFFG |
| 3 | Vβ13S1-J1.1 | SVGAGI | CAS | RKQGA | TEAFFG |
| 4 | Vβ13S1-J1.1 | SVGAGI | CASS | YQGA | TEAFFG |
| 5 | Vβ13S1-J1.1 | SVGAGI | CASS | SQGT | TEAFFG |
| 6 | Vβ13S1-J1.1 | SVGAGI | CAS | RHQGS | TEAFFG |
| 7 | Vβ13S1-J1.2 | SVGAGI | CASS | HQGA | NGYTFG |
| 8 | Vβ13S1-J1.5 | SVGAGI | CASS | YQGA | QPQHFG |
| 9 | Vβ13S1-J1.5 | SVGAGI | CASS | YQGS | QPQHFG |
| 10 | Vβ13S1-J1.1 | SVGAGI | CAS | RRQGY | TEAFFG |
| 12 | Vβ13S1-J1.1 | SVGAGI | CASS | YQGA | TEAFFG |
| 13 | Vβ13S1-J1.1 | SVGAGI | CAS | RRQGA | TEAFFG |
| 14 | Vβ13S1-J1.1 | SVGAGI | CAS | NLQGS | TEAFFG |
| 15 | Vβ13S1-J1.1 | SVGAGI | CASS | YQGSA | EAFFG |
| 10* | Vβ13S1-J1.1 | ND | CASS | Y | NTEAFFG |
| 52* | Vβ13S1-J1.1 | ND | CASS | WQGVD | TEAFFG |
| 29*† | Vβ13S1-J1.1 | ND | CAS | NRGLY | TEAFFG |
| Case no. . | V-J usage . | CDR2 . | 3′ end of Vβ . | CDR3: N-Dβ1-N . | 5′ end of Jβ . |
|---|---|---|---|---|---|
| 1 | Vβ13S1-J1.1 | SVGAGI | CASS | KQGV | TEAFFG |
| 2 | Vβ13S1-J1.1 | SVGAGI | CASS | KQGA | TEAFFG |
| 3 | Vβ13S1-J1.1 | SVGAGI | CAS | RKQGA | TEAFFG |
| 4 | Vβ13S1-J1.1 | SVGAGI | CASS | YQGA | TEAFFG |
| 5 | Vβ13S1-J1.1 | SVGAGI | CASS | SQGT | TEAFFG |
| 6 | Vβ13S1-J1.1 | SVGAGI | CAS | RHQGS | TEAFFG |
| 7 | Vβ13S1-J1.2 | SVGAGI | CASS | HQGA | NGYTFG |
| 8 | Vβ13S1-J1.5 | SVGAGI | CASS | YQGA | QPQHFG |
| 9 | Vβ13S1-J1.5 | SVGAGI | CASS | YQGS | QPQHFG |
| 10 | Vβ13S1-J1.1 | SVGAGI | CAS | RRQGY | TEAFFG |
| 12 | Vβ13S1-J1.1 | SVGAGI | CASS | YQGA | TEAFFG |
| 13 | Vβ13S1-J1.1 | SVGAGI | CAS | RRQGA | TEAFFG |
| 14 | Vβ13S1-J1.1 | SVGAGI | CAS | NLQGS | TEAFFG |
| 15 | Vβ13S1-J1.1 | SVGAGI | CASS | YQGSA | EAFFG |
| 10* | Vβ13S1-J1.1 | ND | CASS | Y | NTEAFFG |
| 52* | Vβ13S1-J1.1 | ND | CASS | WQGVD | TEAFFG |
| 29*† | Vβ13S1-J1.1 | ND | CAS | NRGLY | TEAFFG |
Comparison of the CDR2 and CDR3 of clonal TCR Vβ13S1 gene rearrangements from 14 CD4+ T-LGL cases (this analysis could not be performed in case 11 due to sample shortage). Segments are aligned according to the conserved motifs (CASS for the Vβ and FG for the Jβ segment). Nucleotide sequences were aligned to TCR sequences according to the Basic Local Alignment Search Tool (BLAST) and ImMunoGeneTics (IMGT) databases.
ND indicates not determined. Underlining indicates the motifs shared in common.
VDJ protein sequence of the 3 of 98 clones using the Jβ1.1 segment found among PB Vβ13.1+ CD4+ T cells from adult healthy donors.
This clone corresponds to an HLA-DR*0701 donor.
Sequence of the VDJ junctional TCRβ regions of the clonally expanded TCRαβ+/CD4+ T-LGL showing Vβ13S1-J1.1
| Case no. . | V-J usage . | 3′ end of Vβ13.1 . | N . | Dβ1 . | N . | 5′ end of Jβ . |
|---|---|---|---|---|---|---|
| 1 | Vβ13S1-J1.1 | AGCAGT | AA | gggACAGGGggc | AGT | aaCACTGAAGCTTTC |
| 2 | Vβ13S1-J1.1 | AGCAGT | AA | gggACAGGGGGC | — | aaCACTGAAGCTTTC |
| 3 | Vβ13S1-J1.1 | AGCAG | AAA | gggACAGGGGGC | — | aaCACTGAAGCTTTC |
| 4 | Vβ13S1-J1.1 | AGCAGT | TAC | gggaCAGGGGGc | CG | aacACTGAAGCTTTC |
| 5 | Vβ13S1-J1.1 | AGCAGT | TCC | gggaCAGGGGgc | AC | aaCACTGAAGCTTTC |
| 6 | Vβ13S1-J1.1 | AGCAG | ACAT | gggaCAGGGggc | TAG | aaCACTGAAGCTTTC |
| 10 | Vβ13S1-J1.1 | AGC | CGGC | ggGACAGGGGgc | AAA | aacACTGAAGCTTTC |
| 12 | Vβ13S1-J1.1 | AGCAGT | TAT | gggaCAGGGGGC | — | aCACTGAAGCTTTC |
| 13 | Vβ13S1-J1.1 | AGCAG | GC | ggGACAGGGGGC | — | aacACTGAAGCTTTC |
| 14 | Vβ13S1-J1.1 | AGCA | ATCT | gggACAGGGggc | TAG | aaCACTGAAGCTTTC |
| 15 | Vβ13S1-J1.1 | AGCAGT | — | TACCAAGGCTCGG | — | aacaCTGAAGCTTTC |
| 10* | Vβ13S1-J1.1 | AGCAGTTAC | — | — | — | AACACTGAAGCTTC |
| 52* | Vβ13S1-J1.1 | AGCAGTT | GG | gggaCAGGGGGc | TGG | aACACTGAAGCTTTC |
| 29*† | Vβ13S1-J1.1 | AGCA | — | gggACAGGGGGc | TTGT | aACACTGAAGCTTTC |
| Case no. . | V-J usage . | 3′ end of Vβ13.1 . | N . | Dβ1 . | N . | 5′ end of Jβ . |
|---|---|---|---|---|---|---|
| 1 | Vβ13S1-J1.1 | AGCAGT | AA | gggACAGGGggc | AGT | aaCACTGAAGCTTTC |
| 2 | Vβ13S1-J1.1 | AGCAGT | AA | gggACAGGGGGC | — | aaCACTGAAGCTTTC |
| 3 | Vβ13S1-J1.1 | AGCAG | AAA | gggACAGGGGGC | — | aaCACTGAAGCTTTC |
| 4 | Vβ13S1-J1.1 | AGCAGT | TAC | gggaCAGGGGGc | CG | aacACTGAAGCTTTC |
| 5 | Vβ13S1-J1.1 | AGCAGT | TCC | gggaCAGGGGgc | AC | aaCACTGAAGCTTTC |
| 6 | Vβ13S1-J1.1 | AGCAG | ACAT | gggaCAGGGggc | TAG | aaCACTGAAGCTTTC |
| 10 | Vβ13S1-J1.1 | AGC | CGGC | ggGACAGGGGgc | AAA | aacACTGAAGCTTTC |
| 12 | Vβ13S1-J1.1 | AGCAGT | TAT | gggaCAGGGGGC | — | aCACTGAAGCTTTC |
| 13 | Vβ13S1-J1.1 | AGCAG | GC | ggGACAGGGGGC | — | aacACTGAAGCTTTC |
| 14 | Vβ13S1-J1.1 | AGCA | ATCT | gggACAGGGggc | TAG | aaCACTGAAGCTTTC |
| 15 | Vβ13S1-J1.1 | AGCAGT | — | TACCAAGGCTCGG | — | aacaCTGAAGCTTTC |
| 10* | Vβ13S1-J1.1 | AGCAGTTAC | — | — | — | AACACTGAAGCTTC |
| 52* | Vβ13S1-J1.1 | AGCAGTT | GG | gggaCAGGGGGc | TGG | aACACTGAAGCTTTC |
| 29*† | Vβ13S1-J1.1 | AGCA | — | gggACAGGGGGc | TTGT | aACACTGAAGCTTTC |
— indicates not applicable.
*Sequence of the 3 clones using the Jβ1.1 segment found among PB Vβ13.1+ CD4+ T cells from adult healthy donors.
This clone corresponds to an HLA-DR*0701 donor.
Discussion
Monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL lymphocytosis is a subgroup of monoclonal LGL lymphoproliferative disorders, different from both the CD8+ TCRαβ+ T-LGL, TCRγδ+ T-LGL, and natural killer (NK) cell–type LGL leukemias.18 Noteworthy, in the present study, the former subgroup of clonal T-LGL lymphocytosis was found at a higher frequency than both TCRγδ- and NK-LGL leukemias, whereas it was slightly less common than TCRαβ+ CD8+ T-LGL. In contrast to TCRαβ+ CD8+ T-LGL, monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL cases have been only sporadically reported in the literature, while they were relatively frequent in our series. According to the present study, such discrepancy might be related to the fact that TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL cases usually display a more indolent clinical course—although rare cases associated with aggressive disease have also been reported in the literature—associated with a significantly lower frequency of neutropenia, anemia, and other associated autoimmune diseases, in addition to a lower percentage of cases requiring treatment, in comparison with TCRαβ+/CD8+ T-LGL lymphocytosis. However, the apparently high frequency of CD4 LGL cases found in our series could also be due to the fact that we actively searched for these cases. Recently, we showed that in patients with monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL lymphocytosis the expanded clonal T cells display a restricted usage of a limited number of TCR-Vβ families,18 from which TCR-Vβ13.1 was particularly overrepresented in comparison with its frequency in the PB counterpart of these cells from healthy individuals.27 These observations suggest the potential involvement of a common antigen in driving the expansion of clonal T cells in these patients. In such a situation, shared HLA haplotypes, as well as common motifs in the CDR3 sequences of the TCR-Vβ genes, could be expected. Upon comparing TCR-Vβ13.1+ cases with all non–TCR-Vβ13.1 individuals, a clear association was found between the expanded TCR-Vβ family and the HLA genotype, all TCR-Vβ13.1+ cases displaying an HLA-DRB1*0701 allele. The random chance that both events coincide is about 2%, versus 42% in our patients. In line with these observations, it has recently been reported28 that most CD4+ T cells from an HIV-1+/CMV+–infected patient with lytic granules containing cytotoxic proteins (such as granzymes and perforin) displayed a clear HLA class II– and not class I–restricted lytic activity. Accordingly, after specifically blocking of HLA class II, CMV-specific CD4+ LGL T cells from this patient resulted completely inhibited in their in vitro ability to produce cytokines. In addition to the strong association between the expanded TCR-Vβ and HLA class II, all (unrelated) HLA-DRB1*0701+ patients showing TCR-Vβ13.1 expansions had a common CDR3 amino acid motif (XQGX) in the expanded T lymphocytes. Interestingly, this common “XQGX” CDR3 amino acid motif could not be found among the TCR-VDJβ sequences of T lymphocytes from healthy individuals deposited in GenBank, and it was detected only at very low frequencies among the few clones using the Jβ1.1, Jβ1.2, and Jβ1.5 gene segments identified in both purified CD4+ (1 of 16 clones) and CD8+ (0 of 12 clones) PB T cells from healthy adults. In addition, in a normal T-cell repertoire, different T cells have distinct CDR3 lengths that result in a gaussian distribution, while in our series virtually all expanded monoclonal CD4+ T-LGL cases expressing TCR-Vβ13.1 showed the presence of TCRβ chains characterized by a unique CDR3 length. Altogether, the association between monoclonal expansions of TCR-Vβ13.1 T-LGL, the HLA-DRB1*0701 genotype, and a common XQGX motif in the CDR3 sequence strongly suggests that monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL from these unrelated patients has been selected by a specific common antigen and that they could be the result of a chronic, long-term, antigen-driven process, as previously reported for TCRγδ LGL leukemias23 and B-cell chronic lymphocytic leukemias using the VH3-21 gene, based on their CDR3 homology.29 Because the expanded CD4+/CD8−/+d T-LGL clones expressed TCR-Vβ13.1 with restricted antigen-binding sites in the context of HLA-DR*0701, it could be suggested that they result from an exogenous peptide-driven T-cell stimulation. Furthermore, if this selection involves antigen binding and triggering through the TCR, the antigenic epitope would most likely be restricted in its nature and structure,30 although some differences in the amino acid sequences of the CDR3 region were noted. The overlapping phenotypes of the expanded cells between different (unrelated) patients would further reinforce an underlying common pathogenesis. As previously reported,18 monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL cases show a remarkably uniform cytotoxic T-cell phenotype, as reflected by a common pattern of expression of NKa surface markers and cytotoxic proteins (CD56+, CD57+, Cy granzyme B+) in the absence of expression of other (CD16−, CD94−, CD158a−, CD161−, NKB1−) NK-associated receptors.
Recent reports provide strong accumulating evidence for a role of chronic antigen stimulation in clonal selection and progression of B-cell lymphomas10 as well as T-LGL leukemias.9,22,23 Although identical TCR gene rearrangement are typically identified in LGL leukemia, indicating a (mono)clonal proliferative disease, demonstration of monoclonality does not necessarily imply either neoplastic or malignant transformation.9,23 In fact, in the present study we were unable to demonstrate the presence of any genetic alteration in the patients studied, either by conventional karyotyping or by fluorescence in situ hybridization (FISH) (data not shown). Accordingly, the most probable pathogenetic mechanism leading to an increased survival and/or proliferation of specific T-cell clones in CD4+ T-LGL patients could be more probably related to chronic antigenic stimulation than to a cytogenetic-associated neoplastic transformation. TCRαβ+/CD4+/NKa+/CD8−/+d T cells have been found in increased proportions in humans in different disease conditions where chronic antigen stimulation may occur, such as neoplasias, chronic viral infections, autoimmune disorders, and allografts.28,31–34 Unfortunately, no study has been reported in which CDR3 sequences of the expanded cells have been analyzed in such disease conditions; an exception would be graft versus host disease (GVHD) after allogeneic hematopoietic stem cell transplantation (allo-HSCT), where the expanded CTL clones (including both CD4+ and CD8+ T cells) have been clonotyped.35,36 Accordingly, a variable but frequently high degree of CDR3 homology within a given Vβ family has been reported in patients undergoing allo-HSCT35 whereby the extent of the alteration of the T-cell repertoire is significantly higher in PBMCs from patients with acute GVHD than it is in cases without GHVD.36 Based on these results, it has been hypothesized that such abnormalities could reflect multiple antigen-driven T-cell clonal expansions against alloantigens. Altogether, the evidence of oligoclonal expansions of TCRαβ+/CD4+/NKa+/CD8−/+d T cells in several pathological conditions, interpreted as a specific T-cell response against tumor cells, virus, and autoantigens or alloantigens, clearly suggests that clonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL lymphocytosis represents a dysregulated reaction to exogenous antigens. As a result, a wide and complex spectrum consisting of different clinical entities (from transient immune reaction to LGL leukemia) could be expected, similar to that described for clonal TCRαβ+/CD8++ T-LGL lymphocytosis.9 Although the exact identity of such antigen(s) remains unknown, based on our results we may conclude that monoclonal TCRαβ+/CD4+/NKa+/CD8−/+d T-LGL lymphocytosis cases are not random, because they do not reflect the expected Vβ-J physiological frequencies. In addition, the diverse geographic origin of our patients would suggest that the potential antigen involved in these processes is widely distributed. We can also rule out the involvement of a superantigen, due to the clear major histocompatibility complex (MHC)–TCRVβ–restricted association here observed.37 Finally, the HLA-II restriction found for these clonal expansions of CD4+ T cells supports the involvement of a peptide with an exogenous origin leading to a repetitive and chronic engagement of the TCR of the expanded CD4+ T-LGL.
Another interesting observation is that monoclonal expansions of CD4+ T-LGL have only rarely been reported in the literature18 despite the fact that HLA-DRB1*0701 is frequently observed in the Caucasian population (about 30%).25 These observations further support the role of factors other than the HLA genotype in leading to the dysregulation of the immune response and clonal expansion of CD4+ T-LGL. In this sense, the presence of common extended haplotypes among the TCR-Vβ13.1+ patients suggests that a genetic influence cannot be ruled out. In particular, polymorphisms in genes within the MHC (ie, MICA, cytokines) should be considered with regard to dysregulation of CD4+ cytotoxic T cells.
In summary, in the present study we show that patients with monoclonal expansions of TCR-Vβ13.1+/CD4+ T-LGL display a common HLA-DRB1*0701 genotype and express identical motifs in a constantly shorter-length CDR3-TCR-Vβ sequence, supporting a common antigen-driven origin for these T-cell disorders. Further identification of the short peptides bound to HLA molecules preferentially expressed by clonal TCRαβ+/CD4+ T-LGL would provide new insight into the pathogenesis of the disease; at the same time it could facilitate the identification and establishment of novel preventive and/or therapeutic strategies in individuals with monoclonal CD4+ T-LGL lymphocytosis at risk for transformation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Federico Garrido (Department of Análisis Clínicos, Hospital Universitario Virgen de las Nieves, Granada, Spain) for his helpful discussions and support. We also thank the members of the Hematology Services from the following hospitals for their assistance in the sample and data collection: Hospital Universitario Virgen de las Nieves (Granada, Spain), Hospital Universitario de Salamanca (Spain), Hospital Rio Hortega (Valladolid, Spain), Hospital General Yagüe (Burgos, Spain), Clínica San Miguel (Pamplona, Spain), Hospital de Navarra (Pamplona, Spain), Hospital del Bierzo (Ponferrada, Spain), Hospital General de Segovia (Spain), Hospital de León (Spain), Hospital Virgen de la Concha (Zamora, Spain), Hospital Miguel Servet (Zaragoza, Spain), Hospital Virgen de la Victoria (Málaga, Spain), Hospital Santo António (Porto, Portugal), Centro Hospitalar de Coimbra (Portugal), Instituto Português de Oncologia (Lisbon, Portugal), Hospital São João (Porto, Portugal), and Hospital Egas Moniz (Lisbon, Portugal).
This work has been partially supported by the following grants: FIS 02/1244 and FIS 05/0399, from the Ministerio de Sanidad y Consumo, Madrid, Spain; RETICC RD06/0020/0035 from the Instituto de Salud Carlos III, Ministerio de Sanidad y Consumo, Madrid, Spain; SA 103/03, from the Consejería de Educación y Cultura, Junta de Castillo y León, Valladolid, Spain; and 05/287, from the Consejerra de Salud, Junta de Andalucía, Sevilla, Spain. P.B. is supported by a grant from the University of Salamanca (Reg. N. 430). A.C.G.-M. is supported by a grant from FIS (CP03/00035).
National Institutes of Health
Authorship
Contribution: P.G., Y.S., M.L., A.B., M.G., M.A.L.-N., A.W.L., and A.C.G.-M. performed research and analyzed data; F.R.-C. designed research, analyzed data, and wrote the paper; P.B. performed research and collected and analyzed data; J.C. performed research; and J.A. and A.O. designed research, collected data, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
P.G. and F.R.-C. contributed equally to this work, and J.A. and A.O. contributed equally to this work.
Correspondence: Alberto Orfao, Servicio General de Citometría, Centro de Investigación del Cáncer, Campus Miguel de Unamuno, 37007-Salamanca, Spain; e-mail: orfao@usal.es.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal