Abstract
Although hyperactivation of Ras is a common feature of myeloid malignancy, its role in subverting hematopoiesis is unclear. We have examined the influence of Ras on normal human uncommitted myeloid subsets and show that expression of this oncogene strongly favors monocyte lineage selection in bipotential granulocyte/macrophage progenitors while inhibiting colony formation in other uncommitted subsets. Ras also promoted monocytic differentiation but not the proliferation of these cells. The mechanism through which Ras drives monocyte lineage selection was dependent on PKC activity and Ras was found to promote the expression, membrane translocation, and phosphorylation of conventional and novel PKC isoforms. We further show that Ras promoted the expression of the AGC kinase master regulator, PDK1, which maintains the stability and activity of PKC isoforms. Consistent with this, overexpression of PDK1 itself promoted monocyte colony formation and translocation of PKC. Overexpression of PDK1 was found to be a common feature of acute myeloid leukemia (45% of patients) and was closely associated with hyperphosphorylation of PKC. These data demonstrate that Ras is able to promote monocyte lineage selection via PKC and show for the first time the involvement of the kinase master regulator, PDK1, in both lineage specification and in human leukemia.
Introduction
Understanding of the molecular genetics of leukemia has led to an appreciation that particular molecular abnormalities give rise to specific subtypes of disease. For example, in myeloid leukemogenesis, PML-RARα and BCR-ABL are defining features of acute promyelocytic leukemia and chronic myeloid leukemia, respectively. In addition to these, Ras oncogenes are preferentially associated with disease with monocytic features as in myeloproliferative disorders, such as chronic myelomonocytic leukemia (CMML; 60% of patients)1,2 and monocytic subtypes of acute myeloid leukemia (AML).3 The fundamental basis of myeloid leukemia is a developmental inhibition resulting in accumulation of cells that have a reduced or absent capacity to undergo terminal differentiation. The preferential association of molecular abnormalities with particular developmental subsets of leukemia may therefore reflect the selective disruption of the development of the affected lineage. Alternatively, because it is generally considered that leukemia arises in cells with multipotent developmental capacity, it may also be the case that such associations arise as a result of a disturbance in the process of lineage selection, biasing the production of progeny toward a particular lineage. In this case, the associated abnormality may either play an accessory role in leukemogenesis (specifying the dominant lineage without otherwise influencing its developmental capacity) or it may play a dual role whereby it also perturbs subsequent differentiation, as in the case of PML/RARα.4
Whereas much is known about the transcriptional events that drive developmental programming in hematopoietic cells, comparatively little is known of the primary events leading to the induction of these developmental programs, though evidence supports the importance of environmental factors such as adhesion to stromal cells and the presence of cytokines.5 In this context, Ras may play an important role, because it integrates signals from a variety of membrane-bound receptors including adhesion molecules6 and cytokine receptors.7 Ras activity is frequently dysregulated in myeloid leukemia and myelodysplastic syndrome by both direct and indirect mechanisms,3,8–11 probably contributing to the developmental disruption characteristic of these conditions.
Previously, we have shown that expression of mutant Ras in human primary erythroid cells promotes the activity of protein kinase C (PKC).12 This family of serine/threonine kinases act as lineage discriminators determining cell fate decisions.13,14 Here, we determine the effect of Ras on individual multipotential and bipotential progenitor populations and show that Ras drives monocytic lineage commitment in granulomonocytic bipotential cells by promoting PKC activity. We also report that Ras promotes the expression of the phosphoinositide-dependent kinase-1 (PDK1), which is responsible for maintaining the stability and activity of conventional and novel isoforms of PKC (c/nPKC). PDK1 also acts as a master regulator of AGC kinases, including Akt, p70 ribosomal S6 kinase, p90 ribosomal S6 kinases (Rsk), and serum and glucocorticoid-induced protein kinase.15 Overexpression of PDK1 itself also promoted monocyte colony formation, suggesting that Ras influences lineage commitment via this kinase. Moreover, analysis of a cohort of patients with AML demonstrated frequent overexpression of PDK1 in AML patients and corresponding hyperphosphorylation of PKC, demonstrating the clinical relevance of these data.
Materials and methods
Retroviral constructs
Human H-RasV12, N-RasD12, and myrPDK1 (kindly provided by Phillip Hawkins, Biotechnology and Biological Sciences Research Council (BBSRC), Cambridge, United Kingdom) were subcloned into the retroviral vector, PINCO (a bicistronic vector in which the gene of interest is expressed from the LTR and GFP from an internal cytomegalovirus promoter). Retrovirus was generated in Phoenix-AM cells as previously decribed.12
Cell culture and patient samples
CD34+ cells from neonatal cord blood were isolated and cultured as previously.16 Twenty-four hours following isolation, cells were infected with retrovirus on 2 consecutive days on tissue culture plates preadsorbed with retroviral particles.12 Cells were subsequently cultured in medium containing 5 ng/mL IL-3, granulocyte colony-stimulating factor (G-CSF), granulocyte macrophage colony-stimulating factor (GM-CSF), and stem cell factor (SCF) (R&D Systems, Abingdon, United Kingdom). In some experiments, erythropoietin (Epo; 1 U/mL) was also included. Colony assays (by limiting dilution in 96-well plates) and subsequent analyses were carried out as described previously.17 In PKC inhibitor experiments, cultures were treated with either GF109203X or chelerythrine (LC Labs, Woburn, MA) or vehicle. Where cells were subsequently plated for colony formation this was also carried out in the indicated concentration of inhibitor. Note that each experiment has been carried out with cord blood from a different individual, which contributes to interassay variation.
Patient samples (MRC-AML15 trial), cord blood, and normal marrow were obtained with informed consent with approval from the South East Wales Research Ethics Committee and the Multi-Centre Research Ethics Committee for Wales. AML samples were isolated by Ficoll separation and assessed for blast cell purity by flow cytometry (CD45 versus side-scatter). Samples 70% or less pure were not included in the analysis (≤ 20% of AMLs).
Flow cytometric analysis and sorting
Transduced cultures were analyzed by 4-color cytometric analysis following staining with CD64-APC (Invitrogen, Paisley, United Kingdom) in combination with M-CSFR-biotin (custom preparation, Santa Cruz Biotechnology, Santa Cruz, CA) and one of the following phycoerythrin (PE)–labeled antibodies: CD11b, CD14 (Dako Ely, United Kingdom), or CD163 (BD PharMingen, Oxford, United Kingdom). Biotin was subsequently labeled with SA-PerCP-CY5.5 (BD PharMingen). For single-cell sorting of lineage-uncommitted subpopulations, day-3 transduced cultures were labeled with CD45RA-APC (Invitrogen), CD123-PE (eBioscience, San Diego, CA), and a cocktail of biotinylated lineage markers: CD36 (Ancell, Bayport, MN), CD15 (Sigma, Poole, United Kingdom), and CD11b and CD64 (Serotec, Oxford, United Kingdom). Biotin was subsequently labeled with SA-PerCP-CY5.5. Sorting of lineage-negative (lin−), GFP+ subpopulations was carried out using a MoFlo sorter (DakoCytomation, Glostrop, Denmark) equipped with an argon-ion laser tuned to 488 nm and a 635-nm diode laser. Individual cells were sorted in 96-well plates using CyClone instrumentation and pulse-width discrimination to exclude doublets. Cell deposition efficiency was determined by microscopic examination of the sorted plates. After 7 days, colonies were scored and 2000 latex beads (BD, Oxford, United Kingdom) added to each colony (to control for recovery in determination of colony size). Each colony was then aspirated and individually stained with a cocktail of antibodies to define lineage: CD235a (glycophorin A)–PE (erythroid), CD14-PE (monocytic), CD15-biotin (granulocytic), and CD41-biotin (megakaryocytic) together with SA-PerCP-CY5.5. CD13-APC (Leinco, St Louis, MO) was also included to segregate CD235a-PE (always CD13−) from CD14-PE staining (always CD13+). This approach was similarly applied to discriminate CD15 from CD41 staining (not detected in these experiments).
Western blotting and real-time polymerase chain reaction analysis
Cells were fractionated into cytosolic and membrane fractions as previously.12 Because frequency of retroviral transduction exceeded 60%, enrichment of GFP+ cells was unnecessary. Protein corresponding to 5 × 104 cell equivalents/lane was loaded on a 4% to 12% precast minigel and electroblotted as described previously.12 Blots were probed for the following PKC isoforms: α (Santa Cruz Biotechnology; 208-R), βI (209-R), βII (210-R), δ (937-R), ϵ (214-R), θ (212-R), phospho-ζ (12894R), and ζ (Calbiochem, Nottingham, United Kingdom; no. 538539). Antibodies to phospho-PKC (c- and -isoforms) PDK1 (no. 9371 and no. 3062), Akt (no. 9275), Erk (no. 9101), and panRas (no. 3965) were from Cell Signaling Technology (Danvers, MA). RasVal12 specific antibody (OP38) was from Calbiochem. Signal detection was performed using enhanced chemiluminescence (ECL) Advance substrate (Amersham, Chalfont St Giles, United Kingdom). In the analysis of patient samples, expression levels were quantified by comparison with recombinant standards for PKC and PDK1 (Nventa, San Diego, CA). Band volumes were estimated by densitometric analysis (UVItec, Cambridge, United Kingdom) and were calculated as fold-expression relative to the corresponding control fraction. Data are expressed as the mean-fold differences recorded in the indicated number of independent replicate experiments.
Quantitation of mRNAs was carried out using a LightCycler FastStart DNA Masterplus SYBRGree-I (Roche, Lewes, United Kingdom) using the following primers: 5′-GCACGGCAAGGGCATCATTC-3′ and 5′-CGACATAGCCGGCAGGTAAGC-3′ (for PDK1) and 5′-CGAGGAAGGAAACATGGAACTCAG-3′ and 5′-TTTCCACTACGAACGGCTGTCC-3′ (for PKCα). Quantitative analysis was performed by comparing the crossing points (CTs) of gene expression compared to the CT of the housekeeping gene, S14 (see Roche technical note LC13/2001 for details).
Results
Expression of Ras in human progenitor cells promotes macrophage colony formation and differentiation
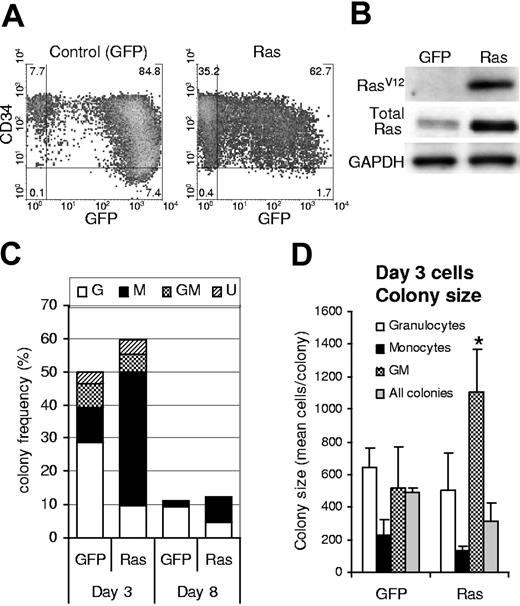
We generated a population of human hematopoietic progenitor cells (CD34+) expressing mutant Ras by retroviral transduction with a vector that coexpressed GFP (Figure 1A). An equivalent population expressing GFP alone acted as a control in these experiments. Ras-transduced cells showed specific expression of RasV12, which was overexpressed 3-fold compared to total endogenous levels (Figure 1B). Following retroviral transduction (day 3), we determined the effect of Ras on myeloid colony formation by limiting dilution in 96-well plates. After 7 days, individual colonies were stained with lineage markers (CD13, CD14, and CD15) and analyzed by flow cytometry to define lineage identity and GFP positivity. Cells expressing Ras generated macrophage colonies at more than 3-fold the efficiency of corresponding control cells (P ≤ .01; Figure 1C; day 3). This occurred at the expense of granulocyte and granulocyte-macrophage (GM) colony formation (approximately 50% of control) and partly as a result of an overall increase in colony-forming efficiency (127% of control). We reasoned that Ras could give rise to these changes by a number of mechanisms. First, Ras may selectively promote an increase in the proliferation of macrophage colonies; however, if this were the case we should also have observed an increase in colony size. This proved not to be the case; only the relatively infrequent GM colonies showed an average increase in size; otherwise colony size not significantly different (Figure 1D). Alternatively, Ras may promote the self-renewal of monocyte progenitors during the period of retroviral transduction (days 1-3) or the survival of these cells under clonal conditions. In either case, this would predict that cells expressing Ras should demonstrate better retention of colony-forming ability; however, cells plated after 8 days of culture showed the same relative decline in colony formation (8-fold) as control cells (Figure 1C; day 8), indicating that Ras did not affect the retention of colony-forming ability under these conditions.
Expression of Ras in CD34+ cells and its effect on colony formation. (A) Expression of GFP and CD34 on retrovirally transduced cells (day 3); quadrants delineate autofluorescence/background staining using isotype-matched controls; figures represent the frequency of cells (%) in each quadrant. (B) Corresponding expression of mutant Ras (using a RasV12-specific antibody) and total Ras (using a pan-specific antibody). (C) Cells were plated for colony-forming assay by limiting dilution on day 3 or day 8 of culture (as indicated) in IL-3, GM-CSF, G-CSF, and SCF. Colonies were scored 7 days later. GFP positivity and lineage identity were determined by flow cytometric analysis (see “Materials and methods”). G indicates granulocytic; M, macrophage; GM, granulocyte/macrophage; U, colonies for which lineage could not be unambiguously defined by cytometric analysis. (D) Average colony size for each colony type (for colonies plated on day 3); error bars represent SD; *P ≤ .05. Each data set (C-D) represents the mean of at least 4 independent replicate experiments (n = 4).
Expression of Ras in CD34+ cells and its effect on colony formation. (A) Expression of GFP and CD34 on retrovirally transduced cells (day 3); quadrants delineate autofluorescence/background staining using isotype-matched controls; figures represent the frequency of cells (%) in each quadrant. (B) Corresponding expression of mutant Ras (using a RasV12-specific antibody) and total Ras (using a pan-specific antibody). (C) Cells were plated for colony-forming assay by limiting dilution on day 3 or day 8 of culture (as indicated) in IL-3, GM-CSF, G-CSF, and SCF. Colonies were scored 7 days later. GFP positivity and lineage identity were determined by flow cytometric analysis (see “Materials and methods”). G indicates granulocytic; M, macrophage; GM, granulocyte/macrophage; U, colonies for which lineage could not be unambiguously defined by cytometric analysis. (D) Average colony size for each colony type (for colonies plated on day 3); error bars represent SD; *P ≤ .05. Each data set (C-D) represents the mean of at least 4 independent replicate experiments (n = 4).
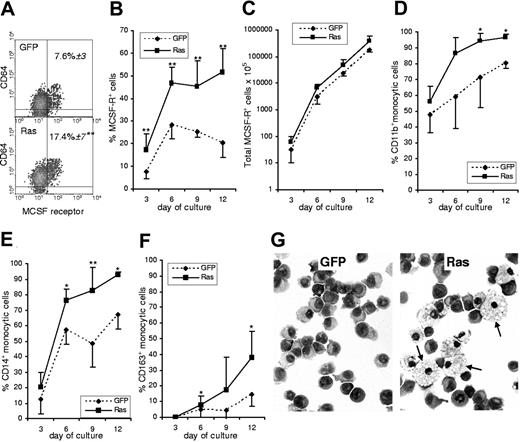
A third possible cause was that Ras affected lineage decision-making in myeloid multipotent or bipotent (GM) cells. The earliest definitive marker for the identification of monocytic cells is expression of the M-CSF receptor (CD115).18,19 We used this marker to establish the frequency of monocytic cells in Ras-expressing and in control cultures. Furthermore, by multiparameter labeling with differentiation-associated cell surface markers we determined the subsequent effect of Ras on the differentiation of M-CSFR+ cells. We found that immediately following transduction, the frequency of cells that had undergone commitment to the monocytic lineage in the Ras expressing population was more than twice that seen in control cells (Figure 2A). Such a difference could arise from Ras promoting monocyte lineage selection or as a result of a pro-proliferative influence of Ras on these cells; however, from day 3 onward there was no further relative change in the proportion of M-CSFR–expressing cells, which remained at 2-fold control levels (Figure 2B). There was also no significant difference between the growth rate of Ras versus control monocytes (Figure 2C), which is consistent with the data on colony size (Figure 1D). We also noted that markers associated with early and intermediate stages of monocytic differentiation (CD11b and CD14) were more strongly up-regulated on monocytic cells expressing Ras, suggesting that this oncogene also promoted their differentiation (Figure 2D-E). This interpretation was borne out by the increased frequency of CD163 expression (Figure 2F), which is considered to be a late marker for monocyte development expressed on the majority of tissue macrophages.20 In support of this, morphologic analysis of purified monocytic cells sorted from these cultures showed that Ras promoted a mature macrophage morphology (Figure 2G arrows).
Effect of Ras on monocyte development. (A) Dual parameter dot plots showing expression of M-CSFR and the myeloid antigen, CD64, on day 3 cells; data are gated on GFP+ cells. Figures indicate the percentage of M-CSFR+ cells ± SD; quadrants delineate background staining using isotype-matched controls. (B) Changes in the frequency of GFP+, M-CSFR+ cells with subsequent culture. (C) Total GFP+, M-CSFR+ cells. (D-F) Differentiation antigen expression on GFP+, M-CSFR+ cells. (G) Morphology of GFP+, M-CSFR+ cells sorted on day 9 (arrows indicate mature macrophages); Wright-Giemsa stain (Leica DM4000B microscope, 40×/0.75 HCL PL objective lens, Leica DC500 digital camera, and Leica IM50 version 4.0 software (Leica, Cambridge, United Kingdom). original magnification ×400). Error bars represent extent of positive or negative SD, shown separately for clarity (n = 6). *P ≤ .05; **P ≤ .01.
Effect of Ras on monocyte development. (A) Dual parameter dot plots showing expression of M-CSFR and the myeloid antigen, CD64, on day 3 cells; data are gated on GFP+ cells. Figures indicate the percentage of M-CSFR+ cells ± SD; quadrants delineate background staining using isotype-matched controls. (B) Changes in the frequency of GFP+, M-CSFR+ cells with subsequent culture. (C) Total GFP+, M-CSFR+ cells. (D-F) Differentiation antigen expression on GFP+, M-CSFR+ cells. (G) Morphology of GFP+, M-CSFR+ cells sorted on day 9 (arrows indicate mature macrophages); Wright-Giemsa stain (Leica DM4000B microscope, 40×/0.75 HCL PL objective lens, Leica DC500 digital camera, and Leica IM50 version 4.0 software (Leica, Cambridge, United Kingdom). original magnification ×400). Error bars represent extent of positive or negative SD, shown separately for clarity (n = 6). *P ≤ .05; **P ≤ .01.
Effect of Ras on lineage specification of uncommitted subpopulations
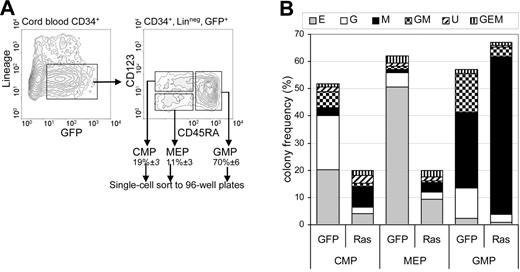
These data suggested that Ras promoted monocyte differentiation and lineage selection, observations that are compatible with previous reports.21–23 We next established how Ras affected lineage commitment for each of the uncommitted myeloid subpopulations. We examined its effect on multipotent and bipotent myeloid progenitors as defined by Manz et al.24 Using single-cell sorting, we cultured individual CD34+, lin−, GFP+ cells representing 3 subpopulations: multipotent myeloid cells (common myeloid progenitors [CMPs]) and the bipotent populations: granulocyte-macrophage progenitors (GMPs) and megakaryocyte-erythrocyte progenitors (MEPs) on the basis of CD123 and CD45RA expression (Figure 3A). Ras had no significant effect on the relative frequencies of these populations (not shown). After 7 days, individual colonies derived from each population were counted, stained with lineage-specific antibodies (see “Materials and methods”), and assigned lineage following flow cytometric analysis. Epo was included in these assays to allow the formation of erythroid colonies.
Effect of Ras on lineage specification. (A) CD34+ GFP+ lin− cells (day 3) were single-cell sorted as 3 subpopulations representing multipotent myeloid cells (CMP) and bipotent populations: GMP and MEP on the basis of CD123 and CD45RA expression. Figures indicate mean ± SD population frequencies. Data illustrated are for Ras-transduced cells. (B) Frequency distribution of colonies derived from each uncommitted subpopulation: E indicates erythroid; G, granulocytic; M, macrophage; GM, granulocyte/macrophage; U, indefinable by surface phenotype. Individual cells were cultured in IL-3, IL-6, GM-CSF, G-CSF, SCF, and Epo and harvested 7 days later.
Effect of Ras on lineage specification. (A) CD34+ GFP+ lin− cells (day 3) were single-cell sorted as 3 subpopulations representing multipotent myeloid cells (CMP) and bipotent populations: GMP and MEP on the basis of CD123 and CD45RA expression. Figures indicate mean ± SD population frequencies. Data illustrated are for Ras-transduced cells. (B) Frequency distribution of colonies derived from each uncommitted subpopulation: E indicates erythroid; G, granulocytic; M, macrophage; GM, granulocyte/macrophage; U, indefinable by surface phenotype. Individual cells were cultured in IL-3, IL-6, GM-CSF, G-CSF, SCF, and Epo and harvested 7 days later.
As expected, control CMP cells gave rise to colonies of each myeloid lineage (Figure 3B), though we were unable to derive megakaryocyte (CD41+) colonies (which may be related to the use of cord blood for these experiments).25 MEP cells predominantly gave rise to erythroid colonies (> 80% of colonies formed) and GMP cells gave rise to a mixture of granulocyte, macrophage, and GM colonies. As would be expected, this lin− subpopulation contained GM colonies at higher frequency than the colony assays derived from unfractionated cells (Figure 1A). Expression of Ras caused considerable distortion to this pattern of colony formation. Within the GMP (bipotential) population, expression of Ras resulted in almost complete abrogation of GM colony formation compared with controls (5% ± 3% versus 26% ± 1% of colonies; P < .05). Instead, nearly all colonies formed were pure macrophage (87% ± 7% versus 49% ± 5%; P < .05). Ras did not significantly affect the colony-forming efficiency of GMP cells (76% ± 3% versus 62% ± 13%). A similar distortion was observed in the CMP (multipotent) population: 38% of colonies were pure macrophage (versus 6% ± 3% in control; P < .05). However, Ras also significantly reduced the colony-forming efficiency of CMP cells (20% ± 5% versus 46% ± 12%; P < .05) and also generated a significant number of colonies for which no lineage could be assigned, due to the absence of differentiation-marker expression (17% of colonies). Ras also reduced colony-forming efficiency of MEP cells (in line with previously published work)12,16 but caused no significant distortion. These data indicate that Ras can grossly perturb lineage selection of granulocyte/macrophage bipotential cells in favor of macrophage colony formation. Ras may also perturb lineage selection of CMP cells; however, the reduction in colony-forming efficiency makes this less clear-cut.
Of the 3 Ras genes associated with myeloid malignancy, N-Ras is the most frequently mutated (H-Ras mutations being relatively rare).3 We therefore repeated these experiments using an identical construct expressing mutant N-Ras. This construct had an almost identical, though slightly attenuated, effect on macrophage colony formation of the GMP subpopulation—doubling the frequency of macrophage colonies (63% ± 2% of colonies versus 36% ± 2% in controls, P < .001). The effects on MEPs were also similar (49% ± 5% reduction in colony-forming capacity) though in contrast to H-Ras, N-Ras had no significant effect on the distribution or frequency of colonies in the CMP population. Overall, these data support the assertion that expression of mutant Ras strongly promotes monocyte lineage selection in bipotential progenitor cells with granulocyte-macrophage colony-forming capacity.
Ras promotes the expression and activation of classical and novel PKC isoforms
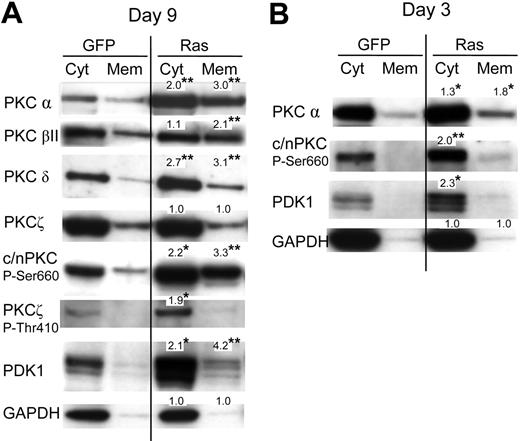
Previously, it has been reported that activation of PKC promotes both monocyte lineage selection in granulocyte-macrophage progenitors13,26 as well as monocyte differentiation.27,28 Activation of PKC is associated with phosphorylation (which promotes PKC maturation and stability) and membrane translocation. To investigate whether Ras promoted the activation of PKC in these cells, we therefore carried out subcellular fractionation to establish the effect of this oncogene on the expression and membrane translocation of PKC. We analyzed its effect on the PKC isotypes: α, βI, βII, δ, ϵ, θ, and ζ. Of these only the α, βII, δ, and ζ isoforms were detected in either Ras or control cells (Figure 4A; data for PKCϵ and PKCθ are not shown). Ras promoted the expression of both the classical PKC isoforms (α and βII) and the novel isoform, PKC δ. In contrast, Ras had no effect on the expression of PKCζ (with the caveat that, in our hands, this antibody also cross-reacted with the PKCι atypical form). Ras also increased translocated levels 2- to 3-fold for conventional and novel PKC isoforms (c/nPKC) indicating that Ras promoted PKC activation as well as expression. To determine whether augmentation of PKC expression by Ras was due to transcriptional activation, we examined the levels of PKCα mRNA in these cells by real-time quantitative polymerase chain reaction (RQ-PCR); however, we found the levels of PKCα mRNA to be identical compared with control cells (data not shown). This suggested that the observed differences were due to posttranscriptional/posttranslational events. In this respect, we noted that Ras also strongly promoted the C-terminal phosphorylation of c/nPKC (Figure 4A) as well as activation loop phosphorylation (at Thr514; not shown), though the concomitant increase in overall expression made it difficult to assess whether the proportion of c/nPKC phosphorylated was increased. Ras additionally promoted the phosphorylation of the atypical form, PKCζ (Figure 4A). Phosphorylation of PKC is initiated via the phosphoinositide-dependent kinase, PDK1.29–31 Interestingly, Ras strongly promoted the expression of PDK1 in these cells (Figure 4A). Because PDK1-mediated phosphorylation of c/nPKC isoforms increases their stability as well as their activity, this suggested that the Ras-induced changes in PKC expression and translocation may have been mediated via PDK1. In support of this interpretation, PDK1 is not apparently required to maintain the stability of atypical PKC isoforms (despite promoting their phosphorylation), which may explain why the expression of PKCζ was unaffected by Ras.29
Effect of Ras on PKC expression and translocation. (A) Representative Western blot data from day 7 cultures, fractionated into cytosolic and membrane components, and probed with the indicated antibody; c/nPKC P-Ser660 binds all conventional and novel isoforms of PKC only when phosphorylated at Ser660 (or equivalent residue). A total of 5 × 104 cell equivalents were loaded in each lane. Note: PDK1 is a multiply phosphorylated protein that gives rise to species of different mobilities on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE); treatment of extracts with phosphatase gives rise to a single fast-migrating band (not shown). (B) Corresponding analysis at day 3 of culture (day 0 following infection). In each case, quantitation was carried out by densitometric analysis of band volumes (from at least 4 replicate experiments); *P ≤ .05; **P ≤ .01. Note that the values are normalized relative expression levels (compared with the equivalent control fraction). These are mean values and therefore may not exactly match the fold-changes shown in the corresponding representative data. To clearly show membrane-translocated species, some cytoplasmic banding is overexposed.
Effect of Ras on PKC expression and translocation. (A) Representative Western blot data from day 7 cultures, fractionated into cytosolic and membrane components, and probed with the indicated antibody; c/nPKC P-Ser660 binds all conventional and novel isoforms of PKC only when phosphorylated at Ser660 (or equivalent residue). A total of 5 × 104 cell equivalents were loaded in each lane. Note: PDK1 is a multiply phosphorylated protein that gives rise to species of different mobilities on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE); treatment of extracts with phosphatase gives rise to a single fast-migrating band (not shown). (B) Corresponding analysis at day 3 of culture (day 0 following infection). In each case, quantitation was carried out by densitometric analysis of band volumes (from at least 4 replicate experiments); *P ≤ .05; **P ≤ .01. Note that the values are normalized relative expression levels (compared with the equivalent control fraction). These are mean values and therefore may not exactly match the fold-changes shown in the corresponding representative data. To clearly show membrane-translocated species, some cytoplasmic banding is overexposed.
If increased PKC and PDK1 expression/translocation is a causative change driving lineage selection, changes in activity should be evident directly following transduction with Ras. We therefore repeated the analysis of c/nPKC and PDK1 taking freshly transduced CD34+ cells. Augmented expression, as well as increased phosphorylation of PKC, was consistently observed in cells expressing Ras with an approximate 2-fold overexpression in each case (Figure 4B). A corresponding increase in the level of translocated PKCα was also observed. These data demonstrate that changes in the activity of PKC occur in synchrony with the period during which lineage selection is taking place and that Ras constitutively promotes the expression of both PKC and PDK1.
The effect of Ras on monocyte lineage selection is dependent on PKC
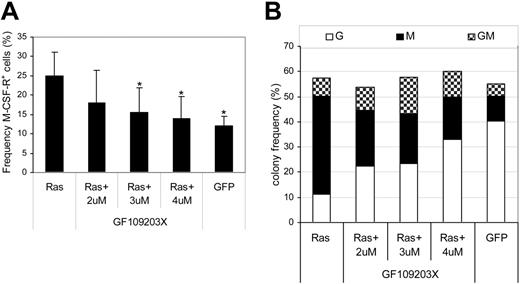
Having established that Ras promoted the activation of PKC isoforms, we further examined whether the PKC activity was indispensable to the effect of Ras on monocyte lineage selection. To this end, we carried out transduction of CD34+ cells with Ras in the presence of the highly selective PKC inhibitor, GF109203X, which is inhibitory for the classical and novel isoforms of PKC. In the untreated Ras-expressing population, we observed the expected 2-fold increase in monocyte progenitor frequency (M-CSFR+) compared with cells expressing GFP alone (Figure 5A). Treatment with inhibitor abrogated this effect across the dose range tested, indicating that the effect of Ras on monocyte lineage commitment was dependent on the activity of PKC. We next examined the subsequent ability of these cells to form colonies. The presence of inhibitor had no significant effect on colony-forming efficiency (Figure 5B); however, it did have a conspicuous effect on the type of colonies formed by CD34+ cells expressing Ras. As expected, cells transduced with Ras produced mainly macrophage colonies (68% ± 6% of colonies formed compared with 18% ± 3% in GFP controls); however, the same cells treated with inhibitor gave rise to a near normal distribution of colony formation, dominated by granulocytic and mixed colonies (GMs) with only 28% pure macrophage colonies. To reduce the likelihood that these results were due to nonspecific inhibition of other protein kinases, we repeated these experiments with chelerythrine chloride, an alternative highly selective PKC inhibitor. This affected colony formation in a similar manner, reducing the proportion of macrophages by greater than 2-fold (55% ± 10% to 22% ± 4%, P < .05 at 1 μM; n = 3) without affecting colony-forming frequency. These data therefore support the hypothesis that activation of PKC by Ras is indispensable for its effect on monocyte lineage selection.
Effect of PKC inhibition on monocyte progenitor frequency and colony formation. Day-1 cells were transduced in the presence of the indicated concentration of GF109203X. (A) GFP+ cells were assessed for M-CSFR expression on day 3 as shown in Figure 2A. Error bars represent SD (n = 3); *P ≤ .05. (B) Frequency distribution of colony formation of day 3 cells was assessed as described in Figure 1 (n = 3). G indicates granulocytic; M, macrophage; GM, granulocyte/macrophage.
Effect of PKC inhibition on monocyte progenitor frequency and colony formation. Day-1 cells were transduced in the presence of the indicated concentration of GF109203X. (A) GFP+ cells were assessed for M-CSFR expression on day 3 as shown in Figure 2A. Error bars represent SD (n = 3); *P ≤ .05. (B) Frequency distribution of colony formation of day 3 cells was assessed as described in Figure 1 (n = 3). G indicates granulocytic; M, macrophage; GM, granulocyte/macrophage.
Activated PDK1 mimics the effect of Ras on colony formation and PKC activation
The correlation of PDK1 overexpression with increased phosphorylation of c/nPKC isoforms suggested that the effect of Ras on PKC may be mediated by overexpression of PDK1. We therefore examined whether Ras promoted PDK1 expression through a transcriptional process; however, we again found that PDK1 mRNA was present at identical levels compared with control cells (data not shown), suggesting that the effect of Ras on the expression of this molecule was mediated via posttranscriptional/posttranslational events.
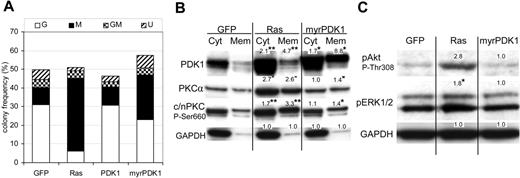
We next examined the effect of overexpressing wild-type PDK1 (wtPDK1) in human CD34+ cells. Despite overexpressing this protein 5-fold, we found it did not significantly affect either the phosphorylation of n/cPKC or monocyte colony formation (Figure 6A). However, previous work has shown that Ras may also promote the phosphorylation of PDK1 and membrane attachment of this kinase.32–34 We also observed increased membrane-associated PDK1 in cells expressing Ras (Figure 4A), though we did not observe nuclear translocation of PDK1 as described by Scheid et al34 (not shown). We therefore repeated this analysis using PDK1 myristolated at its N-terminus to promote membrane association (myrPDK1). In contrast to wtPDK1, myrPDK1 consistently promoted monocyte colony formation 3-fold (P ≤ .05; Figure 6A) and also constitutively promoted the membrane translocation of PKCα, albeit more weakly than Ras (Figure 6B). MyrPDK1 also promoted phosphorylation of membrane-associated c/nPKC (Figure 6B). In contrast to Ras, myrPDK1 did not significantly affect the phosphorylation of Erk (Figure 6C). Similarly, myrPDK1 did not promote the phosphorylation of Akt at Thr308; however, this is consistent with the observation that Akt is a poor substrate for PDK1 unless first phosphorylated at Ser473.35 We were unable to detect the expression of phospho-Rsk1/2/3 in this mixed progenitor population. These data suggest that commitment to the monocytic lineage can occur independently of the hyperactivation of Erk or Akt.
Activated PDK1 mimics the effect of Ras on colony formation and PKC activation. (A) Effect of PDK1 and myrPDK1 on colony formation of CD34+ cells. Conditions as in Figure 1 (n = 4). (B) Western blot analysis of cytosolic and membrane components from the indicated cultures (day 7). Figures indicate mean relative expression (from 4 replicate experiments) compared to equivalent control cells. *P ≤ .05, **P ≤ .01 (see also note in Figure 4). (C) Effect of Ras and myrPDK1 on the phosphorylation of Akt and Erk. Representative Western blot data of total protein from day 7 cultures probed with the indicated antibody. Data are representative of 3 independent experiments.
Activated PDK1 mimics the effect of Ras on colony formation and PKC activation. (A) Effect of PDK1 and myrPDK1 on colony formation of CD34+ cells. Conditions as in Figure 1 (n = 4). (B) Western blot analysis of cytosolic and membrane components from the indicated cultures (day 7). Figures indicate mean relative expression (from 4 replicate experiments) compared to equivalent control cells. *P ≤ .05, **P ≤ .01 (see also note in Figure 4). (C) Effect of Ras and myrPDK1 on the phosphorylation of Akt and Erk. Representative Western blot data of total protein from day 7 cultures probed with the indicated antibody. Data are representative of 3 independent experiments.
Overall, these data indicate that membrane-associated PDK1 is causally linked to the promotion of monocyte colony formation and activation of PKC isoforms.
PDK1 and phospho-PKC are commonly overexpressed in patients with AML
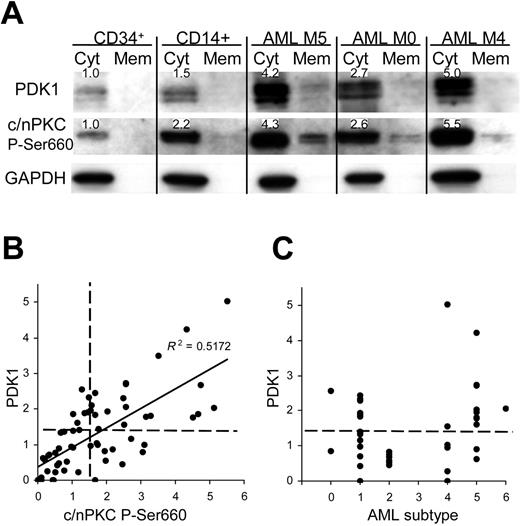
We examined whether similar dysregulation of PDK1 and phospho-PKC was observed in AML compared with normal bone marrow. PDK1 and phospho-PKC were undetectable in unfractionated marrow and in granulocytic cells (not shown); however, low levels were detected in both CD34+ and monocyte (CD14+) subpopulations (Figure 7A). In a cohort of 66 patients with AML, 45% significantly overexpressed both PDK1 (up to 5-fold compared with normal CD34+ cells) and a similar number (46%) overexpressed phospho-PKC (up to 6-fold) with enhanced levels of translocation in each case (Figure 7A). In fact, there was a close correlation between the expression of PDK1 and phospho-PKC (Figure 7B), as observed in the model system. Overexpression of PDK1 occurred across all represented AML subtypes (in the 42 patients where this data were available) except in M2 (Figure 7C). M5 patients displayed the highest levels of this protein, which is consistent with a role in monocytic development and indeed relatively high levels of both PDK1 and phospho-PKC were seen in normal monocytic cells (Figure 7A); however, overexpression was not universal, even within this group. These data demonstrate for the first time the overexpression of PDK1 in leukemia and indicate that its expression is important in regulating PKC phosphorylation.
PDK1 and phospho-PKC are overexpressed in patients with AML. (A) Western blot data from normal bone marrow subpopulations and AML blasts fractionated into cytosolic and membrane components. Figures show expression levels relative to normal CD34+ bone marrow cells for which representative data are shown (n = 4); see also note Figure 4. (B) Quantitative analysis of total PDK1 and phospho-PKC expression in AML blasts (normalized to mean expression in CD34+ cells; n = 5). Dashed lines indicate statistical threshold of normal expression (mean ± 2 SD of normal CD34+ cells). (C) Expression of PDK1 related to AML FAB subtype.
PDK1 and phospho-PKC are overexpressed in patients with AML. (A) Western blot data from normal bone marrow subpopulations and AML blasts fractionated into cytosolic and membrane components. Figures show expression levels relative to normal CD34+ bone marrow cells for which representative data are shown (n = 4); see also note Figure 4. (B) Quantitative analysis of total PDK1 and phospho-PKC expression in AML blasts (normalized to mean expression in CD34+ cells; n = 5). Dashed lines indicate statistical threshold of normal expression (mean ± 2 SD of normal CD34+ cells). (C) Expression of PDK1 related to AML FAB subtype.
Discussion
Ras is classically viewed as a pro-proliferative oncogene and in the hematopoietic context evidence indicates it can induce a myeloproliferative condition in mouse models.36–38 In normal human myeloid progenitors we found no overt indication of a pro-proliferative role for this oncogene. What was more evident was a disturbance of the pattern of lineage specification, as well as in the development of the monocytic lineage. Our initial observations showed an increase in the frequency of monocyte colonies, which occurred largely at the expense of granulocyte colony formation. These data suggested that Ras was able to influence lineage selection in favor of monocyte production and this was borne out by direct measurement of monocyte progenitor frequency (based on the expression of M-CSFR). During subsequent culture, the relative frequency of these cells remained unchanged, which again is in concordance with an initial effect on lineage-uncommitted progenitors. Ras appeared to have no significant effect on the proliferation of these cells with subsequent development, but consistent with previous reports,21–23,39 it did promote the differentiation of these cells in terms of both accelerated developmental marker expression and morphology. In another sense, however, these data are paradoxical because promoting the rate of differentiation would be expected to reduce proliferative capacity. It has been argued that Ras signaling also regulates cell cycle exit during monocytic development (though induction of the Ets suppressor METS),40 so one possible explanation for these data are that aberrant signaling from oncogenic Ras may inhibit cell cycle exit of monocytic cells, despite their relatively differentiated phenotype. Another factor that has been suggested23 is that the degree of Ras hyperactivation may determine its effect on proliferation and self-renewal, with low levels promoting these processes (and high levels having the opposite effect). Given that retroviral promoters tend to become less efficient as cells reach maturity, this may have had the indirect consequence of inhibiting cell cycle exit as the level of Ras expression falls. Further support for the role of Ras in monocytic differentiation comes from the observation that Gab2, which acts as an upstream activator of Ras in hematopoietic cells,41 also has the capacity to promote differentiation of this lineage.42
We also examined the effect of Ras on lineage selection in multipotent and bipotent subsets. The clearest evidence of monocyte lineage selection was observed in bipotential (GMP) progenitors, suggesting that the level of Ras activation may act as a lineage discriminator in this population. In CMP and MEP uncommitted progenitors the main effect of Ras was to reduce colony-forming efficiency, possibly due to the inhibition of erythroid colony formation (which we have previously described).12,16 The role of Ras in regulating cell fate also shows precedent in other developmental model systems.43–45 Together, these data support a role for Ras in lineage specification in the hematopoietic compartment and is consistent with the monocytosis seen in myeloproliferative disorders associated with dysregulated Ras. Hyperactivation of Ras is particularly common in both CMML1 and juvenile myelomonocytic leukemia (JMML),46,47 and these conditions are characterized by increased numbers of monocytic cells but without inhibition of differentiation.48 Furthermore, in AML, Ras mutations are also preferentially associated with monocytic subtypes.3
This study also identified PKC isoenzymes as key mediators of Ras in promoting monocyte commitment. Ras promoted the activity of both classical and novel isoforms of PKC; furthermore, inhibition of PKC activity using inhibitors was able to restore the normal distribution of colony formation. The role of PKC in promoting monocytic differentiation is well known from the ability of PKC agonists to promote differentiation of cell lines with monocytic potential (such as U937 and HL6049 ). It has already been shown that PKC activity can act as a lineage discriminator for myeloid cells: overexpression of constitutively active PKCα in murine bipotential granulocyte/macrophage cells results in a shift to macrophage colony formation, even under conditions favoring granulocyte colony formation.26 Activated PKCα alone was not, however, capable of inducing the degree of monocyte lineage commitment observed in these experiments, suggesting additional PKC isoforms may be important in lineage determination in these cells. Indeed, both PKCα and PKCδ have been reported to be essential for monocytic development in cell lines.27,28 In general, these data highlight the importance of PKC in influencing cell fate decisions in hematopoietic development; a role in keeping with the broader function of these signaling molecules in regulating mammalian development.50,51 There is also evidence that PKC directly influences the activity of transcriptional complexes linked with developmental programming52 as well as transcription factors such as HoxA9 and Pu.1, which are associated with myelopoiesis.53,54
One issue raised by these observations is the mechanism by which Ras promoted the expression and activity of PKC. As we have previously demonstrated in primary erythroid cells, Ras promoted the phosphorylation of PKC.12 Phosphorylation is critical to the stability and activity of c/nPKC isoforms and PDK1 activity is known to be essential for this process.31 Current understanding suggests that PDK1 is responsible for initiating PKC phosphorylation, with subsequent autophosphorylation giving rise to the mature protein, and indeed we found a corresponding increase in the level of PDK1 in cells expressing Ras. Up-regulation of PDK1 by Ras has not been previously reported, though this may only be pertinent in primary cells, because myeloid cell lines already express abundant levels of this kinase (R.L.D., unpublished data, July 2006). These data naturally suggested that the effects on PKC may be mediated through PDK1 overexpression; however, ectopic expression of wtPDK1 was without significant effect, suggesting that if PDK1 is causally significant, it requires coactivating signals emanating from Ras. The regulation of PDK1 is complex and there are reports that its activity is influenced by phosphorylation,55–57 membrane localization,58 coactivating lipids,59,60 and substrate conformation.15 In these experiments, we found that Ras promoted membrane association of PDK1 and the alterations in migration are also suggestive of changes in the phosphorylation of this protein through Ras, which have been previously reported.34 The observation that overexpression of a membrane-anchored form of PDK1 promoted both PKC translocation and monocyte colony formation is suggestive that the increase in membrane-associated PDK1 is significant. On the other hand, that the myristolated construct was less active in this regard than Ras itself, suggests that Ras also drives alternative mechanisms of PDK1 or PKC activation. One possibility that we have examined is that Ras also promotes the formation of diacylglycerol (DAG), which is required for the activity of c/nPKC isoforms.61–63 However, use of DAG kinase inhibitors to promote DAG levels in myrPDK1-expressing cells did not further augment PKC activity or monocyte colony formation in this context (R.L.D., unpublished data, June 2006), so the nature of this alternative mechanism is currently unclear. In summary, although it is apparent that Ras influences the expression, membrane localization and probably the phosphorylation of PDK1, the mechanism by which Ras brings about these changes is currently unclear. The fact that overexpression of membrane-localized PDK1 in part mimics the effect of Ras, indicates that these changes are significant; however, our data also indicate that activation of PDK1 may not be the exclusive mechanism though which Ras influences monocyte lineage selection. This is the first report that PDK1 can influence lineage specification of myeloid cells; however, it has been previously shown to influence T-cell development.64
Our studies of AML patients indicate that dysregulation of PDK1 and phospho-PKC are also significant in the disease context. Overexpression of PDK1 and associated activation of PKCα has previously been shown to drive transformation of epithelial cells.65 This is the first demonstration of PDK1 overexpression in a hematologic malignancy and the association with hyperphosphorylation of PKC suggests that PDK1 is also driving PKC phosphorylation in this context. The fact that PDK1 is overexpressed in subtypes other than those with a predominant monocytic phenotype implies that its role extends beyond that of influencing lineage specification. In the broader context, PDK1 activity appears to be central to the mechanism whereby cancer cells become resistant to apoptotic death and as such is a promising target for antineoplastic drug design.66 We are currently assessing the prognostic implications of PDK1 overexpression in AML.
Authorship
Contribution: L.P. performed all experiments; J.F. performed cell sorting; A.K.B. contributed patient data and resources; and R.L.D. designed the experiments, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard L. Darley, Department of Haematology, School of Medicine, Cardiff University, Cardiff, United Kingdom CF14 4XN; e-mail: darley@cf.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Leukaemia Research Fund of Great Britain and the Medical Research Council.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal